|
|
1.IntroductionA central challenge facing biologists is to understand how the genetic blueprint contained within an organism’s cells is deployed in a three-dimensional (3-D) network over a tissue or organ.1 Meeting this challenge, particularly in the context of disease, involves integrating genetic and morphological information over multiple length scales from the subcellular level to entire organs and organism.2 Currently, the gold standard in analyzing tissues is histology. Despite its strengths, histology is inherently a two-dimensional (2-D) technique, which does not provide full 3-D structural information about a tissue, and is typically limited to examining the morphology and organization of a relatively small population of cells, limiting the statistical power of any measurement. Furthermore, it is often prohibitively labor intensive, requiring the imaging of hundreds to thousands of 2-D sections and successively aligning them into a single 3-D image. We describe a new technique, two-photon tissue cytometry, which images organs and tissues with sufficient detail to reveal 3-D tissue and cell morphology as well as underlying gene and protein expression patterns, thus helping bridge the divide between the genotype and phenotype of an organ. The choice of an appropriate imaging technology is critical for the development of 3-D tissue image cytometry. Important criteria include spatial resolution, sensitivity, depth penetration, molecular specificity, and data throughput. Many methods to image 3-D thick tissues have been developed, but most of these techniques do not have subcellular resolution or the necessary molecular sensitivity. While high-resolution magnetic resonance imaging (MRI) is capable of imaging whole animals, its resolution is limited3 to 10 to and it is not compatible with common fluorescent markers. Optical imaging methods, in general, provide the highest resolution. Optical coherence tomography has a few-micrometer level resolution and a penetration depth of a few millimeters; however, optical coherence tomography does not provide reliable subcellular level imaging today.4 Optical projection tomography is more compatible with molecular imaging and can study fluorescent and nonfluorescently stained tissues up to approximately .5, 6 However it also does not possess sufficient resolution to resolve details of individual cells and has difficulty in imaging opaque tissues containing opaque materials such as bone or cartilage. Selective plane illumination microscopy has demonstrated the ability to provide detailed images of over a millimeter of relatively transparent samples such as embryos5 but has limitations with opaque samples. Common 3-D imaging techniques that possess subcellular resolution include surface imaging microscopy (SIM), confocal microscopy, and two-photon microscopy. SIM is a block face technique whose major strength is the isotropic resolution, which can be obtained by careful tuning of the embedding medium.7 The fixation and embedding procedure is typically harsher on the protein environment of the sample than standard histology and may lead to greater distortions in the tissue morphology. A promising additional block face technique recently introduced employs scanning electron microscopy (SEM) imaging and microtome sectioning to construct 3-D images, and has demonstrated voxel sizes . However, voxel resident times are of the order of , which would make it impractical for imaging large tissue volumes. Fluorescence confocal microscopy8 is capable of providing high-resolution 3-D images, but typically it is not practical to image beyond into a tissue sample due to scattering, and it can cause significant photodamage to out-of-focus planes. Among 3-D tissue optical imaging techniques, two-photon microscopy (TPM) is particularly promising. TPM is a fluorescent optical microscopy technique developed by Denk 9 It features submicrometer resolution, low phototoxicity, excellent penetration depth, and inherent 3-D sectioning capability. The excellent depth penetration of TPM in tissues is due to lower scattering and absorption of the infrared excitation wavelength employed10 and the lack of the need for a detection pinhole, enabling greater signal collection efficiency than in confocal microscopy. In addition, like all fluorescence-based techniques, it provides high molecular specificity in mapping gene and protein expression profiles, and has clearly demonstrated its utility for visualizing gene activity in vivo with green fluorescent protein (GFP) over the past decade.11 Despite these strengths, traditional TPM cannot be directly applied to organ level imaging due to three factors. First, the depth penetration of TPM is still limited to a few hundred micrometers for most tissue types, presenting a barrier to imaging tissue or organs that have a substantive axial extent. Second, the typical field of view of standard TPM has a linear dimension of only a few hundred micrometers, insufficient for whole organ imaging. Third, standard TPM systems have an image acquisition speed that is prohibitively slow for imaging macroscopic 3-D tissues. A typical TPM system would require approximately 60 days at a minimum to image a 5-mm cube of tissue at a resolution of and pixel residence time. 2.Materials and Methods2.1.Tissue PreparationThe heart tissue samples were prepared with an intravital labeling protocol followed by fixation and paraffin embedding. The mice were first anesthetized with anesthesia (7.5% Nembutal, 18.5% ethyl alcohol, 74% saline solution) and then injected with of Hoechst 33342 ( , Molecular Probes, Inc., H-3570) followed later with of Alexa594 lectin ( , Molecular Probes, Inc., I-21411) and allowed to circulate for followed by sacrifice of the animal with a lethal dose of anesthesia. To label the boundaries of the myocytes, the mouse was anesthetized as already described and injected via a tail vein injection with of Texas Red maleimide (Molecular Probes Inc., T6008) solution ( of saline mixed with Texas Red maleimide in DMEM). The animal was sacrificed with a lethal dose of anesthesia after 0 to . The hearts were then surgically removed and fixed in 4% paraformaldehyde overnight and then dehydrated with successive 70, 90, and 100% ethanol solutions before paraffin embedding. The sample was then mounted on the microscope and imaged. 2.2.InstrumentThe experiments were performed on a video-rate two-photon microscope using a scanning polygonal mirror. The instrument is described in detail elsewhere.12 Briefly, the instrument is based around a Zeiss (Axioscope, Zeiss Thornwood, New York) microscope. The 780-nm laser light from a Ti-sapphire laser (Mira 900, Coherent, Palo Alto, California) is directed into the microscope by a rotating polygonal scanner (Lincoln Laser, Phoenix, Arizona) and a galvanometer scanner (Cambridge Technology, Watertown, Massachusetts). A Zeiss PlanNeoFluar 0.8 numerical aperture (NA) objective focused the laser light onto the sample and directed the fluorescent signal to the emission path of the microscope. The pixel residence time was approximately . Significantly higher pixel residence times can be achieved with multifoci systems, while still maintaining high frame rates. The emission optics consisted of a 530-nm dichroic (Chroma), which split the fluorescence into two channels; a 525 lowpass filter was used in the blue channel and a 600/50 bandpass filter for the red channel. The detectors were two photomultiplier tubes running in analog mode (R3896, Hamamatsu, Bridgewater, New Jersey). The 3-D scanning was enabled by moving the objective with respect to the sample using an objective piezocontroller (P-721.00, Physik Instrument, Waldbronn, Germany). To image areas larger than the field of view of the objective and translate the sample after mechanical sectioning, a robotic stage (H101, Prior Scientific, Rockland, Massachusetts) was used. The same stage was used to translate the sample from the objective to a home-built milling machine that had been integrated into the microscope system to section the tissue sample. The milling machine consisted of a 3/8-in. mill bit driven by a dc motor rotating at . The translation speed was approximately across the sample, which was empirically arrived at to produce smooth [ root mean square (rms)] cuts at the surface of the sample determined by TPM imaging. For the current application, the sectioning depth was and the imaging depth was , enabling a axial registration overlap. 2.3.Image ProcessingThe images were visualized using the Amira 3.1 visualization package (Mercury Computer Systems, San Diego, California). The entire heart image was constructed by first applying a normalization image to the individual 3-D images to account for uneven sample illumination and axial intensity falloff. Incorporating techniques to homogenize the sample illumination, such as the use of refractive beam shapers, will offer significant advantages and reduce the necessity to postprocess the data.13, 14, 15 A median filter was applied to the data to reduce shot noise, while still preserving the edge features. The large heart image was constructed by downsampling the individual cubes by a factor of 16 in the radial directions and by a factor of 8 in the axial direction. Neighboring cubes were overlapped by a constant offset given by the step size of the robotic stage and an intensity averaging was performed in the overlapped regions. The higher resolution images were constructed similarly but without the downsampling of the data. The nuclei staining was a factor 10 to 15 brighter than the weaker blue/green autofluorescence from the heart tissue, enabling a segmenting of the nuclei from the tissue by an intensity thresholding. 3.Results3.1.Construction of a Two-Photon Tissue CytometerA two-photon 3-D tissue image cytometer using multiphoton excitation is presented in Fig. 1 . To overcome the depth penetration limitation of TPM in studying thicker specimens, we integrated an automated microtome into a high-speed TPM system. By alternating and overlapping optical sectioning with mechanical sectioning, it is possible to rapidly image samples with arbitrary thickness. Once the upper portion of the sample is imaged, the uppermost portion of the tissue sample can be removed by the microtome. The depth of mechanical sectioning is chosen to be less than the imaged depth, ensuring that the region of the sample that is subjected to mechanical sectioning has been already imaged. An advantage of our approach over block face techniques is that any distortions that are introduced by the sectioning procedure do not introduce artifacts in the reconstructed 3-D volume since the cut plane is always preimaged. Further, imaging overlapping regions between successive sections ensures accurate digital registration between sections. To overcome the limitations in data acquisition speed, we deployed two versions of high-speed TPM. One version uses a high-speed polygonal scanner,12 which is capable of imaging at 12 frames/s; a second version uses multifoci excitation providing over 30 frames/s imaging speed with superior image SNR by incorporating parallel data acquisition.16 Both systems are capable of obtaining submicrometer resolution in tissues. To image a 5-mm cubic sample at sampling, the first system requires approximately , whereas the second system requires . To overcome the field of view limitations of the objective, the robotic stage raster scans the sample, enabling a larger image to be constructed from a series of overlapping 3-D volumes. After the section is removed with the microtoming procedure, the entire sample is translated toward the objective. Unlike with serial section reconstruction, where alignment of successive sections is often ambiguous, the integrated precision robotic stage ensures high reliability in registering neighboring volumes both in the axial and radial direction. Finally, note that the microscope system is entirely automated and requires no user intervention once the sample has been mounted. Fig. 1Schematic of a multifoci multiphoton microscope. (a) A lenslet array is used to produce an array of foci that are scanned across the sample in a raster pattern. A dichroic mirror splits the fluorescence into two separate channels (not shown) and a multianode photomultiplier tube (PMT) is used in each channel to detect the fluorescence from the foci. The inset image shows a fluorescent image of a fluorescein sample excited by the multifoci array. (b) An enlarged view of the optical path of the excitation and emission light. Ti:Sap, titanium sapphire laser; T, telescope; MLA, microlens array; DCM 1, dichroic mirror 1; DCM 2, dichroic mirror 2; GSM, galvanometric scanning mirror; MA PMT, multianode PMT; ZP, -piezo; OL, objective lens; TS, tissue sample; M, motor; DH, microtome; TPB, two-photon barrier filter. 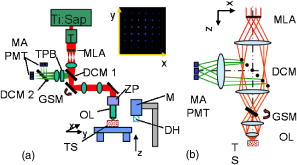 An initial application of this technique focuses on cardiac biology. The mammalian heart has four complex chambers on the macroscopic scale and spiralling asymmetric morphology with interlaced myocytes, fibroblasts, and microvasculature on the microscopic scale. To accurately understand cardiac function requires a detailed understanding of the 3-D spatial arrangement of myocardial tissue at multiple length scales simultaneously from submicrometer to centimeters. Subcellular features such as nuclei and sarcomere architecture have features on the submicrometer level and single cardiomyocyte cells are over long, thus requiring 3-D optical techniques. A simple application of our technique is the study of cardiac hypertrophy. Cardiac hypertrophy is a clinically important early step in the progression to heart failure, and hypertrophy is usually studied by 2-D cross sections of the myocardium. Typical myocytes have dimensions of the order of 10 to in diameter and in length. Therefore, traditional histopathology cannot accurately quantify changes in the 3-D morphology of the cardiomyocyte due to the difficulty of estimating 3-D morphology from thin 2-D slices taken at random orientations with respect to the myocyte axis. In contrast, 3-D morphology can be readily measured using our 3-D technique. Further, myocytes are packed together into myocardial fibers that are arranged into spiralling sheets, which are four to six myocytes thick.17 These sheets have dimensions of the order of hundreds of micrometers and are loosely bound to neighboring sheets by collagen fibers. In many forms of myocardial pathology, increased collagen deposition is an important feature in the development in the progression to heart failure. At an even higher hierarchal level, the coronary vascular system extends across the heart and is heterogeneous at all length scales. Realistic modeling of the heart vasculature requires morphometric information about the microvasculature including capillary diameter, 3-D branching angles, and connectivity. These parameters vary transmurally across the ventricular wall and are important for modeling and understanding coronary blood flow. As a demonstration of whole organ imaging we imaged an entire heart from a mouse with subcellular resolution. To image subcellular structures and mesoscopic tissue structures, intravital perfusion was performed: the cell nuclei of the heart were labeled with Hoechst 33342 and the heart vasculature was labeled with a lectin-Alexa 594 conjugate. In addition to the specific labels, the intrinsic autofluorescence of the tissue allows the identification of cellular morphological features of the heart. Figure 2 shows a digitally reconstructed image of an entire mouse heart imaged from the individual 2-D sections. All the major anatomical regions of the heart were easily identifiable. The image was sampled at in the radial direction and in the axial direction. These sampling values were chosen as a compromise between the resolution necessary to identify cellular nuclei and vasculature and the imaging speed and data set size. The entire organ was approximately in linear extent, thus the data set spanned almost five orders of magnitude in length scale. In contrast, previous high-resolution 3-D studies of hearts were limited to selected regions.18, 19 Fig. 2Isosurface and volume rendering showing a mouse heart at different axial positions reconstructed digitally from a 3-D image. The right and left ventricular chambers can be seen in the figure as well as the descending aortic arch and left atrium. The heart was fixed in 4% paraformaldehyde and embedded in paraffin and mounted on the two-photon tissue image cytometer. The scale bar in the figure is . 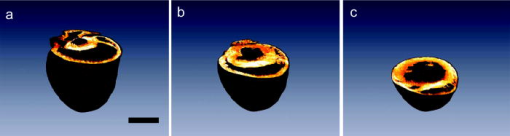 Our technique enables orthogonal slices from the heart to be visualized with progressively higher magnification and increasing levels of detail (Fig. 3 ). Morphology of individual nuclei at micrometer scale can be quantified. The vasculature throughout the 3-D volume of the heart can be readily traced. Figure 4 displays a 3-D reconstruction of cubic portion of the heart with dimensions of . Based on emitted fluorescence spectra, we can isolate three different structures of the heart: vasculature, nuclei, and extracellular matrix. Tissue autofluorescence and Hoechst-33342-labeled nuclei were collected by the blue channel. The intensity of the labeled nuclei was much brighter than the weaker tissue autofluorescence, and we were able to separate the two components based on an intensity thresholding. The Alexa-594-labeled vasculature was collected in the red channel. The 3-D spatial architecture of the heart is clearly identifiable on both the microscopic and macroscopic scales. Fig. 3(a) Three-dimensional axial, coronal, and sagittal orthoslices through the entire heart. The horizontal plane is shown in (b) and successive magnifications of the inner ventricular wall are shown in (c) and (d). We can see that the microvasculature and cardiomyocyte orientation follow the known spiralling architecture of the ventricular wall. The scale bars in the figure are , , and (b), (c), and (d), respectively. 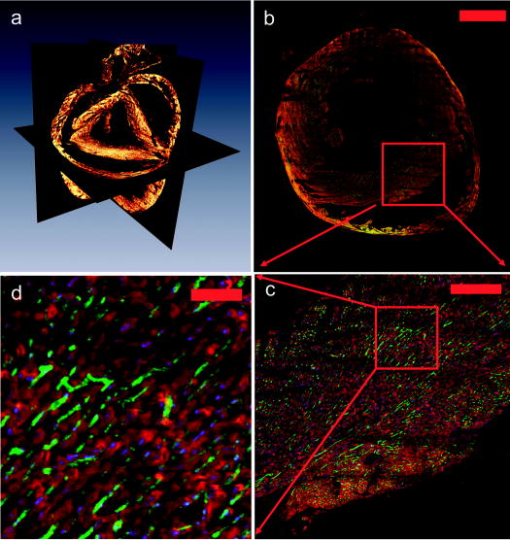 Fig. 4(a) Three-dimensional section of the tissue is enlarged from the apical end of the left ventricle wall; (b) autofluorescence from the heart tissue, the labeled nuclei and vasculature were visualized based on spectroscopic and intensity segmentation, where the scale bar is ; (c) autofluorescence of the myocardium reveals laminar sheets and the cleavage planes were visible as have been noted by other researchers20; (d) the morphology of the 3-D microvasculature; (e) nuclei from the myocytes, fibroblasts, and endothelial cells lining the vasculature; and (f) a section removed from the region outlined in yellow in (b) to better show the tissue constituents; the arrow indicates the space between successive cleavage planes. 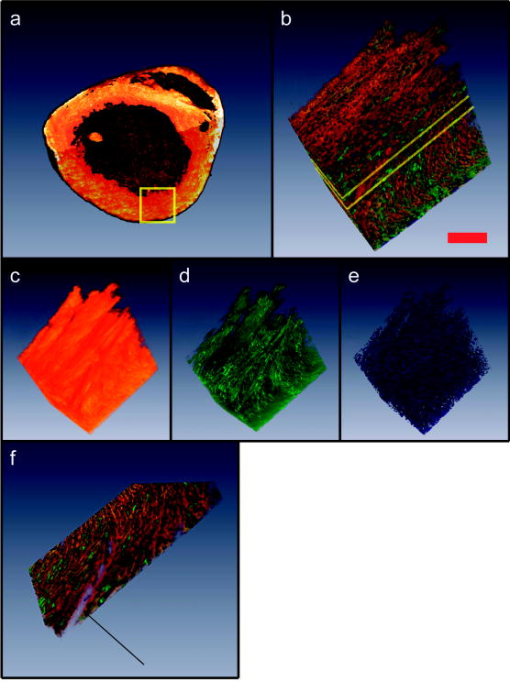 This technology can be applied to study cardiac hypertrophy by accurately quantifying the morphology of individual myocytes. We developed an intravital labeling protocol that labels the extracellular matrix and clearly defines boundaries of individual myocytes. The intravital labeling protocol uses a maleimide conjugate, which labels extracellular sulfhydryls; the conjugate is excluded from the living cells. Figure 5 is cross section of heart tissue with a single myocyte reconstructed in three dimensions. This procedure makes it possible to directly measure myocyte 3D morphology. 4.DiscussionWe demonstrated the 3-D imaging of an entire heart with sufficient detail to capture intracellular features. The approach we implemented is very general in its applicability and can be used to extend almost any type of tissue studies previously investigated with other 3-D fluorescence techniques. Many questions in organ and tissue physiology can be investigated in a more comprehensive and quantitative manner than was practical previously using standard histopathology. It is known that the tissue environment plays an important role mediating many disease processes and these diseases can be fully understood only if studied at the tissue level. For instance, other techniques that focus on the spatial organization of tissues, such as such as laser capture microdissection,21 have proven valuable in uncovering the expression profiles of selected neoplastic cells and spatially distinct cells near the tumor periphery, which mediate the growth of a tumor.22 The biologically important changes in the gene expression profiles would remain hidden if the bulk properties of the heterogeneous tissue were sampled or if the cells had been studied under in vitro conditions. Similarly, two-photon tissue cytometry will be useful for understanding how an organism’s genetic makeup is spatially manifested at the highly relevant 3-D tissue level. A key strength of two-photon tissue cytometry is subcellular resolution. Cancer, cell cycle dynamics, apoptosis, and intracellular protein or drug localization are only a few examples of processes that require information about cell size, shape, and intracellular features, and thus require micrometer-level resolution. Assays involving DNA ploidy analysis, tumor vascularization, and hepatotoxicity will benefit immensely from the subcellular resolution and morphology information that two-photon tissue cytometry (TPTC) provides. Furthermore, processes involving the extracellular matrix, including fibrosis and angiogenesis, require sufficient resolution to quantify extracellular components such as collagen, elastin, and the vasculature. Our present approach couples an integrated micromilling machine to physically remove material from the sample. This can be useful for investigations into very hard materials such as bone, where previous results with confocal microscopy have been promising, 23, 24, 25, 26, 27 and that potentially may be improved with multiphoton microscopy. Also, it is not necessary to embed the fixed tissue into paraffin for mechanical stability using a milling approach. Other sectioning methods such as standard fixed blade microtomy or vibrating blade for unfixed or soft materials can also be used. All optical histology is an elegant technique based on laser ablation that has been developed for deep 3-D tissue imaging.28 This method is inherently slower for sectioning and may not lend itself as well to high-speed data acquisition of large 3-D tissue data sets as mechanical sectioning. However, laser ablation introduces less mechanical deformations into the sample, which can affect the image quality in techniques such as surface image microscopy. We sidestep these distortion issues by preimaging the cut surface. Finally, however, nondestructive methods are preferable to destructive methods and our technique is compatible with preservation of tissue sections after imaging using retrieval of selected slices with standard microtome or vibratome sectioning methods or automated approaches. More detailed analysis by various physical and biochemical techniques can then be performed on the saved sections, or alternatively, the slices can be archived for additional studies at later dates. The choice of an appropriate image resolution is determined by the biological application, with the lower boundary set by the optical diffraction limit of approximately in the radial direction and in the axial direction. There exists a trade-off between the level of detail required and the imaging time and data set size. Increasing the sampling rate by a factor of two in each dimension increases the total data set size and imaging time by approximately a factor of eight. Note that it is not necessary to uniformly image the sample at the same resolution throughout, and higher resolutions can be used for regions of interest requiring more detail. This can be done either interactively as the data is required or resampled postcapture. To realize the full potential of tissue cytometry, significant computational resources will be required to aid the archival, data mining, quantification, and modeling of TPTC data sets. The data presented in this paper were acquired with two channel 16-bit imaging and was in size without compression. Lossless compression reduced this by a factor of 7, and lossy compression systems such as the JPEG 2000 standard currently employed in some medical image applications have realized a factor of 20 in reduction of data set size while maintaining acceptable quality for many medical applications. Furthermore, raw data storage costs have been declining 50 to 60% per year since 1997 and will continue for several more years at least with a terabyte of raw storage cost projected to drop below $50/Tbyte in 2009 based on current trends.29 The steep decline in storage costs and lossless or high-quality lossy compression facilitates the storage of multiple copies of the same data set at different scales to speed data access. We do not anticipate storage requirements to be a limiting factor. On the computational side, many of the analyses appropriate for organ-level data sets lend themselves well to parallelization across multiple processors and subsequent computational increases, and fundamental algorithms can be speeded by hardware implementation. Furthermore, database systems such as SQL and Oracle routinely manage data sets hundreds of gigabytes to terabytes and can be leveraged for the analysis of tissue data sets. In addition, hierarchal data analysis approaches, where regions of interest are selected from larger, lower resolution images and more detailed analyses applied as appropriate, can also introduce substantial decreases in analysis time. We foresee the major challenge being the development of appropriate 3-D segmentation, analysis, cytometric, and data mining algorithms to analyze the data sets to generate biologically relevant results. The ready availability of high-resolution 3-D data sets of macroscopic tissues will facilitate work in this area. A major attraction of flow and image cytometry is the ability to categorize rare subpopulations. Assuming an average cell diameter of , a cubic centimeter of tissue will contain approximately cells. Identifying single cells from similarly sized populations have been shown in traditional cytometry. Achieving the same performance with tissue cytometry depends on three factors: accurate 3-D segmentation algorithms, labeling protocols with high specificity and even distribution, and adequate computational resources to process the 3-D data sets. Currently, 2-D segmentation algorithms are more mature than 3-D algorithms, but this is rapidly changing. Confocal and multiphoton microscopy, MRI, positron emission tomography (PET), and computed tomography (CT) are undergoing major development, and significant resources are being devoted to 3-D segmentation. There are no fundamental barriers facing 3-D segmentation and many algorithms are being generalized from the 2-D case. Tissue labeling is also experiencing rapid progress. Transgenic animals transfected with fluorescent proteins can provide very specific markers of biological function, and new tissue labeling protocols are being actively developed to help study disease in animal models. We have found intravital labeling superior to labeling the tissue in solution as the vasculature aids even distribution. In the previous section, we discussed computational issues, and noted that hierarchal segmentation approaches can help identify rare subpopulations by examining low-resolution images for unique spectral signatures, followed by more detailed high-resolution algorithms. In general, tissue cytometry should achieve results comparable to those seen in flow and image cytometry in identifying rare populations, possibly even a single cell throughout an organ. 5.ConclusionsWe developed a fast, automated method that can generate submicrometer 3-D images of macroscopic tissues and organs and identify unique cells in larger populations as well as reveal information about the biochemical state of the organ. Our initial application is in cardiac biology, but we foresee a broad range of applications in other fields, including tissue engineering, stem cell biology, angiogenesis, and cancer biology. The development of macroscopic engineered tissues requires the formation of a vasculature at both the mesoscopic and microscopic scales to ensure viability. Detailed evaluation of the formation of 3-D microvasculature over the extent of the tissue is possible with this technique. Another promising application area is cancer biology. Cancer metastatic studies are hindered by the difficulty of locating rare metastatic cells within a bulk organ. Multiphoton 3-D imaging has already proven useful in visualizing metastatic cell arrangement with respect to the surrounding vasculature and tissue stroma.30, 31 Our technique can be used to extend these types of studies on an organwide basis and provide additional information about rare metastatic events hidden within the bulk tissue. Additional applications include investigating tissue homeostasis and repair by mapping stem cell distribution throughout entire organs,32 the production of apoptotic maps to better understand disease processes and apoptotic therapies, physiome initiatives,33 high-resolution 3-D mapping of gene expression in whole mount embryos,34 and any area where highly detailed 3-D histological and molecular measurements are required over macroscopic samples. Industrial applications include drug development, where the use of highly physiologically relevant tissue studies is currently hampered by the lack of high-throughput tissue imaging techniques. TPTC will be a significant advance in correlating tissue physiology with the underlying biochemical state and morphology of an organ and will open many new research avenues. AcknowlegmentsTwo of the authors (T.R. and K.B.) were supported in part by NIH, R43 CA097670–01 and R44 CA097670–02. H.H. was supported in part by NIH, R21 EB004646. ReferencesD. Davidson and
R. Baldock,
“Bioinformatics beyond sequence: mapping gene function in the embryo,”
Nat. Rev. Genet., 2
(6), 409
–417
(2001). 1471-0056 Google Scholar
H. Kitano,
“Systems biology: a brief overview,”
Science, 295
(5560), 1662
–1664
(2002). https://doi.org/10.1126/science.1069492 0036-8075 Google Scholar
M. Dhenain,
S. W. Ruffins, and
R. E. Jacobs,
“Three-dimensional digital mouse atlas using high-resolution MRI,”
Dev. Biol., 232
(2), 458
–470
(2001). 0012-1606 Google Scholar
D. Huang,
“Optical coherence tomography,”
Science, 254
(5035), 1178
–1181
(1991). https://doi.org/10.1126/science.1957169 0036-8075 Google Scholar
J. Huisken,
J. Swoger,
F. Del Bene,
J. Wittbrodt, and
E. H. K. Stelzer,
“Optical sectioning deep inside live embryos by selective plane illumination microscopy,”
Science, 305
(5686), 1007
–1009
(2004). https://doi.org/10.1126/science.1100035 0036-8075 Google Scholar
J. Sharpe,
U. Algren,
P. Perry,
B. Hill,
A. Ross,
J. hecksher-Sorensen,
R. Baldock, and
D. Davidson,
“Optical projection tomography as a tool for 3D microscopy and gene expression studies,”
Science, 296
(5567), 541
–545
(2002). https://doi.org/10.1126/science.1068206 0036-8075 Google Scholar
A. J. Ewald,
H. McBride,
M. Reddington,
S. E. Fraser, and
R. Kerschmann,
“Surface imaging microscopy, an automated method for visualizing whole embryo samples in three dimensions at high resolution,”
Dev. Dyn., 225
(3), 369
–375
(2002). https://doi.org/10.1002/dvdy.10169 1058-8388 Google Scholar
T. Wilson, Confocal Microscopy, Academic Press, London (1990). Google Scholar
W. Denk,
J. H. Strickler, and
W. W. Webb,
“Two-photon laser scanning fluorescence microscopy,”
Science, 248
(4951), 73
–76
(1990). https://doi.org/10.1126/science.2321027 0036-8075 Google Scholar
P. T. C. So,
C. Y. Dong,
B. R. Masters, and
K. M. Berland,
“Two-photon excitation fluorescence microscopy,”
Annu. Rev. Biomed. Eng., 2 399
–429
(2000). https://doi.org/10.1146/annurev.bioeng.2.1.399 1523-9829 Google Scholar
M. Chalfie,
Y. Tu,
G. Euskirchen,
W. W. Ward, and
D. C. Prasher,
“Green fluorescent protein as a marker for gene expression,”
Science, 263 802
–805
(1994). https://doi.org/10.1126/science.8303295 0036-8075 Google Scholar
K. H. Kim,
C. Buehler, and
P. T. C. So,
“High-speed, two-photon scanning microscope,”
Appl. Opt., 38
(28), 6004
–6009
(1999). 0003-6935 Google Scholar
D. N. Fittinghoff,
P. W. Wiseman, and
J. A. Squier,
“Widefield multiphoton and temporally decorrelated multifocal multiphoton microscopy,”
Opt. Express, 7
(8), 273
–279
(2000). 1094-4087 Google Scholar
T. Nielsen,
M. Frick,
D. Hellweg, and
P. Andresen,
“High efficiency beam splitter for multifocal multiphoton microscopy,”
J. Microsc., 201 368
–376
(2001). https://doi.org/10.1046/j.1365-2818.2001.00852.x 0022-2720 Google Scholar
L. Sacconi,
E. Froner,
R. Antolini,
M. R. Taghizadeh,
A. Choudhury, and
F. S. Pavone,
“Multiphoton multifocal microscopy exploiting a diffractive optical element,”
Opt. Lett., 28
(20), 1918
–1920
(2003). 0146-9592 Google Scholar
J. Bewersdorf,
R. Pick, and
S. W. Hell,
“Mulitfocal multiphoton microscopy,”
Opt. Lett., 23 655
–657
(1998). 0146-9592 Google Scholar
I. J. LeGrice,
B. H. Smaill,
L. Z. Chai,
S. G. Edgar,
J. B. Gavin, and
P. J. Hunter,
“Laminar structure of the heart: ventricular myocyte arrangement and connective tissue architecture in the dog,”
Am. J. Physiol. Heart Circ. Physiol., 269
(2), H571
–H582
(1995). 0363-6135 Google Scholar
A. A. Young,
I. J. LeGrice,
M. A. Young, and
B. HA. Smaill,
“Extended confocal microscopy of myocardial laminae and collagen network,”
J. Microsc., 192 139
–150
(1998). 0022-2720 Google Scholar
M. Hara,
N. Noiseux, and
V. P. Bindokas,
“High resolution optical imaging of infarction in intact organs,”
BioTechniques, 39
(3), 373
–376
(2005). 0736-6205 Google Scholar
D. A. Hooks,
K. A. Tomlinson,
S. G. Marsden,
I. J. LeGrice,
B. H. Smaill,
A. J. Pullan, and
P. J. Hunter,
“Cardiac microstructure—implications for electrical, propagation and defibrillation in the heart,”
Circ. Res., 91
(4), 331
–338
(2002). https://doi.org/10.1161/01.RES.0000031957.70034.89 0009-7330 Google Scholar
M. R. EmmertBuck,
R. F. Bonner,
P. D. Smith,
R. F. Chuaqui,
Z. P. Zhuang,
S. R. Goldstein,
R. A. Wei, and
L. A. Liotta,
“Laser capture microdissection,”
Science, 274
(5289), 998
–1001
(1996). https://doi.org/10.1126/science.274.5289.998 0036-8075 Google Scholar
L. A. Liotta and
E. C. Kohn,
“The microenvironment of the tumour-host interface,”
Nature (London), 411
(6835), 375
–379
(2001). https://doi.org/10.1038/35077241 0028-0836 Google Scholar
N. D. Sahar,
S. I. Hong, and
D. H. Kohn,
“Micro- and nano-structural analyses of damage in bone,”
Micron, 36
(7-8), 617
–629
(2005). 0968-4328 Google Scholar
I. Marriott,
D. L. Gray,
D. M. Rati,
V. G. Fowler,
M. E. Stryjewski,
L. S. Levin,
M. C. Hudson, and
K. L. Bost,
“Osteoblasts produce monocyte chemoattractant protein-1 in a murine model of Staphylococcus aureus osteomyelitis and infected human bone tissue,”
Bone (N.Y.), 37
(4), 504
–512
(2005). 8756-3282 Google Scholar
S. A. Colopy,
“Response of the osteocyte syncytium adjacent to and distant from linear microcracks during adaptation to cyclic fatigue loading,”
Bone (N.Y.), 35
(4), 881
–891
(2004). 8756-3282 Google Scholar
N. L. Fazzalari,
M. R. Forwood,
B. A. Manthey,
K. Smith, and
P. Kolesik,
“Three-dimensional confocal images of microdamage in cancellous bone,”
Bone (N.Y.), 23
(4), 373
–378
(1998). https://doi.org/10.1016/S8756-3282(98)00111-2 8756-3282 Google Scholar
V. Bentolila,
T M. Boyce,
D. P. Fyhrie,
R. Drumb,
T. M. Skerry, and
M. B. Schaffler,
“Intracortical remodeling in adult rat long bones after fatigue loading,”
Bone (N.Y.), 23
(3), 275
–281
(1998). https://doi.org/10.1016/S8756-3282(98)00104-5 8756-3282 Google Scholar
P. S. Tsai,
“All-optical histology using ultrashort laser pulses,”
Neuron, 39
(1), 27
–41
(2003). https://doi.org/10.1016/S0896-6273(03)00370-2 0896-6273 Google Scholar
R. J. T. Morris and
B. J. Truskowski,
“The evolution of storage systems,”
IBM Syst. J., 42
(2), 205
–217
(2003). 0018-8670 Google Scholar
E. B. Voura,
J. K. Jaiswal,
H. Mattoussi, and
S. M. Simon,
“Tracking metastatic tumor cell extravasation with quantum dot nanocrystals and fluorescence emission-scanning microscopy,”
Nat. Med., 10
(9), 993
–998
(2004). 1078-8956 Google Scholar
X. H. Gao,
Y. Y. Cui,
R. M. Levenson,
L. W. K. Chung, and
S. M. Nie,
“In vivo cancer targeting and imaging with semiconductor quantum dots,”
Nat. Biotechnol., 22
(8), 969
–976
(2004). https://doi.org/10.1038/nbt994 1087-0156 Google Scholar
T. Tumbar,
G. Guasch,
V. Greco,
C. Blanpain,
W. E. Lowry,
M. Rendl, and
E. Fuchs,
“Defining the epithelial stem cell niche in skin,”
Science, 303
(5656), 359
–363
(2004). 0036-8075 Google Scholar
P. J. Hunter,
A. J. Pullan, and
B. H. Smaill,
“Modeling total heart function,”
Annu. Rev. Biomed. Eng., 5
(1), 147
–177
(2003). https://doi.org/10.1146/annurev.bioeng.5.040202.121537 1523-9829 Google Scholar
D. Kosman,
C. M. Mizutani,
D. Lemons,
W. G. Cox,
W. McGinnis, and
E. Bier,
“Multiplex detection of RNA expression in Drosophila embryos,”
Science, 305
(5685), 846
(2004). 0036-8075 Google Scholar
|


