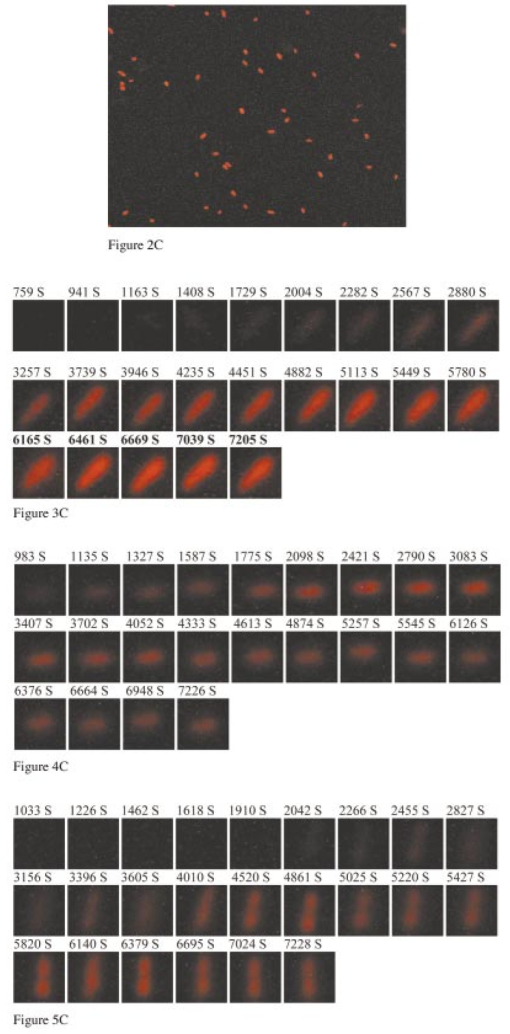
1.IntroductionMany living organisms such as bacteria, yeasts, molds, tumors, and viruses are equipped with xenobiotic efflux pumps that protect the cells from hazardous and noxious compounds.1 2 3 Pseudomonas aeruginosa is renowned for its intrinsic resistance to a wide spectrum of substrates (e.g., detergent, dyes, antibiotics, etc.) and is a ubiquitous Gram-negative bacterium. P. aeruginosa has emerged to be the major opportunistic human pathogen and the leading cause of nosocomial infections in cancer, transplantation, burn, and cystic fibrosis patients. These infections are impossible to eradicate in part because of their intrinsic resistance to the wide spectra of structurally and functionally unrelated antibiotics.3 4 Recent studies have described the interplay between the MexAB-OprM efflux system and the outer membrane barrier in multi-drug resistance and suggested that an effective approach for overcoming the resistance would include the combination of inhibition of the antibiotic efflux pump and enhancement of permeability of the outer membrane.5 6 7 8 9 10 11 Several efflux systems including MexAB-OprM, MexCD-OprJ, MexEF-OprN, and MexXY-OprM, have been reported in P. aeruginosa. 4 5 The MexAB-OprM pump is the major efflux pump in wild-type (WT) cells. This pump consists of two inner membrane proteins (MexA and MexB) and one outer membrane protein (OprM).4 5 The MexB protein consists of 1046 amino acid residues and is assumed to extrude the xenobiotics utilizing the proton motive force as the energy source.6 7 Studies show that it seems most likely that the inner membrane subunits specify the substrates to be transported.8 9 Several models have been proposed in an attempt to describe the possible mechanism of the pump and how membrane pump proteins (e.g., MexAB-OprM) recognize the specific substrates and transport these substrates out of cells.8 9 10 11 12 13 The primary methods for the study of bacteria resistance mechanisms are by measuring the accumulation of quinolone antibiotics in bacteria using radioactively labeled (14 C and 3 H) quinolones or the natural fluorescence of quinolones for detection. A review article of these methods has been published.14 Given that a common characteristic of multi-antibiotic resistance of mutants in Pseudomonas aeruginosa is their broad resistance to quinolones, a popular method for assessing multi-antibiotic resistance is based on determination of the time course of quinolone accumulation in the cell using fluorescence spectroscopy.14 15 16 Fluorescent quinolones are particularly suitable to be used as probes for these experiments because these probes emit weak fluorescence in aqueous solution (outside cells) and become strongly fluorescence in nonpolar and hydrophobic environments (inside cells). Time-course fluorescence spectroscopy has been used as one popular tool for real-time monitoring of the substrates accumulated by bulk bacterial cells. The bulk measurement represents the average behavior of a massive number of cells. It is impossible to use such an average measurement to follow the real-time mechanism of the influx and efflux kinetics because each cell and each extrusion system has its independent kinetics. Furthermore, these techniques are unable to offer the spatial information needed for real-time monitoring of the location of substrates. Recently, the staining assays have been used to study the multi-drug resistance mechanism in eukaryotic cells17 18 using fluorescence probes and microscopy. Unfortunately, these techniques are quite often applied with cell fixation and using high concentration of fluorescence dyes, that lead to cell death. As a result, the fluorescent staining assays for dead-cell imaging are unable to offer the real-time temporal information. Because the current techniques cannot provide the real-time spatial and temporal information of intrinsic resistance of the bacteria at single cell resolution, despite extensive research over decades, the mechanism and structure of the intrinsic resistance of bacterial cells remain incompletely understood and many questions remain unanswered.3 8 9 10 11 12 13 For example, how do the cells sense such a wide range of diverse substrates? How do these membrane protein molecules assemble and recognize these diverse substrates and extrude them selectively out of cells? Do unidentified extrusion pump proteins exist? How rapidly do the substrates diffuse into cells? How quickly does the pump recognize the substrates and extrude them out of the cells? Do substrates stay temporarily or store permanently in the periplasmic space where the membrane pumps are located? Are the influx and efflux kinetics regulated? Does a degradative drug resistance mechanism exist?19 These are the key questions to be addressed in order to fully understand the mechanism of intrinsic multi-drug resistance and design more efficacious drugs to treat infection. To address some of these problems, one must develop a new tool that can provide both temporal and spatial information for real-time monitoring of resistance mechanism of single live bacterial cells. The challenges of such a study include: (i) how to overcome the rapid motion of bacterial cells (2×0.5×0.5 μm) in solution so that the cells can be confined in solution and continuously monitored for hours while still alive; (ii) how to simultaneously monitor a group of cells at single cell resolution that can offer statistical information of bulk cells at single cell resolution; (iii) how to develop a sensitive detection means that allows the low concentration of fluorescence probes to be used for live cell imaging. We report here that we have overcome these challenges and have developed and applied a microchannel system that was able to confine live bacterial cells in solution with no need to immobilize cells. We were able to simultaneously image bulk live cells at single cell resolution using both dark-field optical and fluorescence microscopy equipped with a charge coupled device (CCD) camera. The CCD detector consists of an array of photodiodes (pixels) and this feature makes it possible for real-time monitoring of many single live cells simultaneously. Furthermore, the CCD detector is able to quantitatively measure the fluorescence intensity of fluorescence probes at low concentration, far below the minimum inhibitory concentration (MIC), and hence it is able to provide real-time temporal and spatial information of fluorescent substrates accumulated by live bacterial cells. We used ethidium bromide (EtBr) as a fluorescent probe to study the resistance mechanism of P. aeruginosa using our real-time live cell fluorescence microscopy. EtBr is one of the common fluorescence probes used in previous studies14 15 16 and it is especially suitable for this experiment because EtBr emits weaker fluorescence emission in aqueous environments and becomes strongly fluorescent in nonpolar or hydrophobic environments. The quantum yield of EtBr intercalated with DNA is about ten times higher than that of EtBr in buffer solution.20 EtBr enters the live bacterial cells through passive diffusion and is extruded out of the cell via active efflux pump.15 In addition, the MIC of EtBr for mutants of P. aeruginosa is higher than 1 mM.15 21 We used the MexAB-OprM efflux pump as a working model and investigated the roles of these membrane proteins on extrusion systems by real-time monitoring of EtBr accumulation kinetics of mutants of over-expression or deletion of these subunits. This work demonstrates the possibility of real-time monitoring of resistance mechanisms of single live bacterial cells with temporal and spatial information and may advance the understanding of efflux pump mechanism. 2.Materials and Methods2.1.Chemicals and SuppliesThe EtBr solution was prepared in ultrapure water (Nanopore, 18 MΩ, sterilized) and triple filtered using 0.2 μm sterilized membrane filters (Costar) before EtBr solution was incubated with cells. Quartz slides (25×75 mm), coverslips (22×30×0.08 mm), Nuclepore membrane (25 mm in diameter with 6 μm thickness) were purchased from VWR, Edmund Scientific and Corning Corporation, respectively. The 50 mM phosphate buffer saline (PBS) buffer solution containing 150 mM NaCl pH=7.0 was prepared using ultrapure water (Nanopore, 18 MΩ, sterilized). All chemicals, EtBr, Na 3 PO 4, Na 2 HPO 4, NaH 2 PO 4, and NaCl, were purchased from Sigma and used without further purification or treatment. 2.2.Cell Culture and PreparationP. aeruginosa cell lines,9 16 PAO4290 (a wild-type expression level of MaxAB-OprM), TNP076 (ΔABM, MexAB-OprM deficient mutant), and TNP030#1 (nalB-1, MexAB-OprM over expression mutant), were used. Cells were grown in L-broth medium containing 1 tryptone, 0.5 yeast extract, and 0.5 NaCl at pH 7.2 while rotating at 230 rpm and 37 °C. Cells were first pre-cultured to ensure a full growth and then cultured for an additional 8 h. Cells were harvested by centrifugation at 7500 rpm and 23 °C for 10 min, washed three times with 50 mM phosphate buffer pH=7.0 and then suspended again in the same buffer to have the cell concentration at A600 nm =0.1. The cell solution containing 10 μ M EtBr was prepared by mixing 2703 μL of the cell solution at A600 nm =0.1 with 187 μL ultrapure water and 110 μL of 274 μM EtBr while a timer was started simultaneously to record the incubation time. 2.3.Fluorescence Spectroscopic Measurement of Fluorescence Intensity of Bulk Cells in EtBr SolutionThe time course of fluorescence intensity of EtBr at 590±10 nm from 3.0 mL cell solution containing 10 μ M EtBr was directly recorded by a fluorescence spectrometer (Perkin-Elmer LS50B) with a time-drive mode at a 3 s data acquisition interval and 488 nm excitation (Figure 1). Fluorescence intensity of 10 μM EtBr in each type of cell solution and buffer was recorded in the same day. The experiment was repeated at least three times using fresh cell solutions in other days. The 3.0 mL cell solution containing 10 μ M EtBr was prepared as described above. Figure 1Determination of EtBr accumulation kinetics in intact cells of P. aeruginosa: a time-course of fluorescence intensity of EtBr at 590±10 nm (a) in the PBS buffer and in the presence of intact cells of P. aeruginosa, (b) nalB-1, (c) WT and (d) ΔABM, acquired directly from a 3.0 mL cell solution containing 10 μ M EtBr by a fluorescence spectrometer using a time-drive mode with a 3 s data acquisition interval and 488 nm excitation. The 3.0 mL cell solution containing 10 μ M EtBr was prepared as described in the text. Fluorescence spectra of EtBr showed that the emission peak at 590±10 nm remained almost unchanged during the entire incubation time. 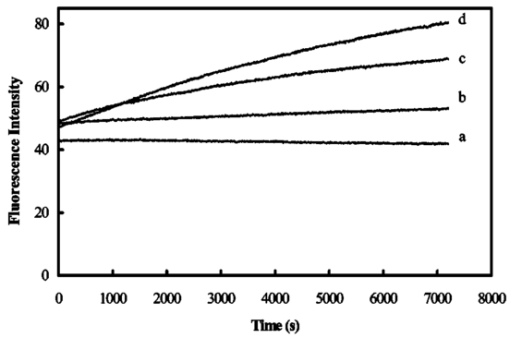 2.4.Dark-field optical and epi-fluorescence microscopy for single live cell imagingThe dark-field optical and epi-fluorescence microscope was equipped with an oil dark-field condenser (oil 1.43–1.20, Nikon), a 100x objective (Nikon Plan fluor 100x oil, iris, SL. NA 0.5–1.3, W.D. 0.20 mm), a microscope illuminator (Halogen lamp, 100 W) for dark-field optical imaging, a super high-pressure mercury lamp (Nikon) for fluorescence imaging, a CCD camera (Micromax, 5 MHz Interline, PID 1030×1300, Roper Scientific) for high-resolution cell imaging, and a color digital camera (Coolpix 990, Nikon) for real-color imaging. Both cameras were added to the microscope through a quad adapter (Nikon) for real-time imaging. Each pixel of the CCD represents a square with 0.067 μ m edges, as calibrated by the microscope stage micrometer (0.1 mm per 50 division movements) via the 100× objective. The epi-fluorescence configuration was applied for real-time fluorescence imaging with a filter cube including a bandpass excitation filter (488 ± 10 nm, Coherent), bandpass emission filter (600 ± 30 nm, Coherent), and a dichroic mirror (565 nm, Chroma Tech). The 20 μ L of the cell solution containing 10 μ M EtBr was added into a 6 μm thickness microchannel and directly imaged using a CCD camera and color digital camera through dark-field optical and epi-fluorescence microscopy. The 6 μm thickness microchannel was constructed by sandwiching the cell solution between a quartz microscope slide (VWR, 1 mm thickness) and a cover slip (0.08 mm thickness) with Nuclepore membrane (Corning) as a spacer (∼6 μm) and then sealing the slide and the cover slip with glue (Deco Cement). The microscope slides and cover slips were cleaned by methanol and dried with nitrogen. 3.Imaging Data Analysis and StatisticsThe full-frame CCD image (Figure 2) was able to simultaneously monitor ∼60 cells from the cell solution (A600 nm =0.1) in the microchannel. We monitored optical and fluorescence intensity of these cells at single cell resolution by acquiring time-course sequence images for 2 h (Figures 3 4 5) using dark-field optical and epi-fluorescence microscopy. Each experiment was repeated at least three times and totally ∼200 cells of each strain of P. aeruginosa were studied and analyzed using both Winview (Roper Scientific) and MetaMorph software (Universal Imagine) that allowed quantitative analysis of analog and digital counts (ADC) of each pixel. The ADC can be converted to photoelectron count (10× for our CCD camera) that represents the digitized electrons accumulated in each pixel and hence reflects the photons collected by the pixel during the CCD camera exposure. Hence ADC is an arbitrary unit for the relative quantitative measurement of intensity of specimen imaged by the CCD camera. Each sequence of images in Figures 3 4 5 was presented using the same scale of ADC and hence the intensity variation in each sequence of images was directly visualized. We selected one representative cell from each strain and presented that sequence of images to illustrate the spatial and temporal information of accumulation kinetics (Figures 3 4 5). Figure 2Representative full frame dark-field optical and fluorescence images showing real-time monitoring of many single cells simultaneously: (A) Representative real-time dark-field optical images of live WT cells, Pseudomonas aeruginosa, incubated with 10 μ M EtBr in the microchannel and directly recorded by a CCD camera through the dark-field optical microscope. The intensity scale of the image is 428–1200 analog and digital counts (ADC). The ADC can be converted to photoelectron count (10× for our CCD camera) and represents the photons collected by each pixel of CCD. Hence ADC can be used as a unit for the relative quantitative measurement of intensity of specimen imaged by CCD camera. Representative real-time fluorescence images of live WT cells, P. aeruginosa, directly recorded by (B) the CCD camera and (see color on page 583) (C) color digital camera using the epi-fluorescence microscopy with a fluorescence filter cube including a band-pass excitation filter (488±10 nm, Coherent), a bandpass emission filter (600±30 nm, Coherent), and a dichroic mirror (565 nm, Chroma Tech). The intensity scale in (B) is 118–170 ADC. Representative contour plots of (D) dark-field optical and (E) fluorescence intensity of a single cell selected from the image (solid-line square) in (A–B). The x axis and y axis represent the CCD pixel array where the cell was located. Each pixel represents 0.067 μm. The z axis represents intensity with the ADC unit. In the dark-field optical image, the periplasmic space appeared brighter than the cytoplasmic space. In the fluorescence image, the periplasmic space appeared as dim as background (EtBr in buffer solution) and the cytoplasmic space was much brighter than the periplasmic space showing that EtBr was accumulated in the cytoplasmic space. Images in (A)–(C) were acquired from the same cell solution almost simultaneously. The cell solution containing 10 μ M EtBr was prepared by mixing 2703 μL of the cell solution (A600 nm =0.1) with 187 μL ultrapure water and 110 μL of 274 μM EtBr. A timer was started simultaneously to record the incubation time. The 20 μL of this cell solution containing 10 μ M EtBr was added into the microchannel and directly imaged using dark-field optical and epi-fluorescence microscopy. 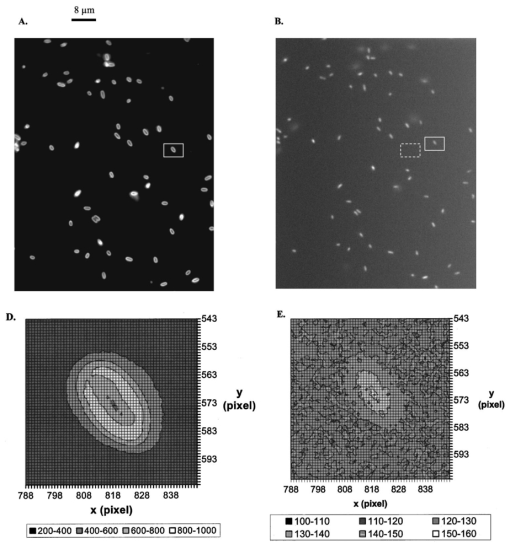 Figure 3Representative time-sequence dark-field optical and fluorescence images and intensity of single WT cells, selected from 60 cells in the same images similar to the one in Figure 2 showing both temporal and spatial information of EtBr accumulation kinetics in live WT cells: Representative snapshots of one of the single cells (WT): real-time (A) dark-field optical images and (B) fluorescence images recorded by CCD camera, and (see color on page 583) (C) real-color fluorescence images recorded almost simultaneously by a digital color camera, for ∼2 h through the dark-field optical and epi-fluorescence microscope. The 60 cells in each sequence image, similar to the one in Figure 2(A), were analyzed. The single cell was selected from these 60 cells as a representative example. The intensity scale is 428–1200 ADC for all sequence images in (A) and 118–170 ADC for all sequence images in (B). (D) Plots of normalized subtracted integrated fluorescence intensity of single WT cells vs time. The cells were similar to the one shown in (A) and selected from the images including 60 cells, similar to one in Figure 2(B). The subtracted integrated fluorescence intensity was obtained by subtracting the integrated fluorescence intensity of the pixel area [dashed-line square area in Figure 2(B)] where no cell was present (EtBr in solution) from the same size of pixel area, in the same image, where the cell was present (EtBr in the cell). This subtraction was used to overcome possible the dark noise of the CCD camera or photodecomposition of EtBr and to ensure that the fluorescence intensity reflects the amount of EtBr accumulated by the cell. Fluorescence was not observed from the cells in the absence of EtBr. The subtracted integrated fluorescence intensity of the cell was normalized with the maximum intensity of that cell in 2 h incubation time. The cell solution containing 10 μ M EtBr was prepared as described in Figure 2. 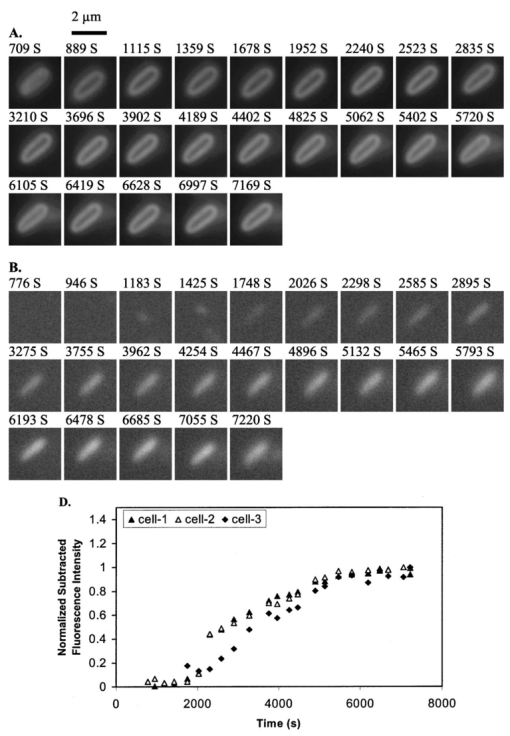 Figure 4Representative time-sequence dark-field optical and fluorescence images and intensity of single nalB-1 cells, selected from 60 cells in the same images similar to the one in Figure 2 showing both temporal and spatial information of EtBr accumulation kinetics in live nalB-1 cells: Representative snapshots of one of the single cells (nalB-1): real-time (A) dark-field optical images and (B) fluorescence images recorded by CCD camera, and (see color on page 583) (C) real-color fluorescence images recorded almost simultaneously by a digital color camera, for ∼2 h through the dark-field optical and epi-fluorescence microscope. The 60 cells in each sequence image, similar to the one in Figure 2(A), were analyzed. The single cell was selected from these 60 cells as a representative example. The intensity scale is 300–980 ADC for all sequence images in (A) and 118–170 ADC for all sequence images in (B). (D) Plots of normalized subtracted integrated fluorescence intensity of single nalB-1 cells versus time demonstrating individual cell showed a differentiated intensity profile that is distinguished from WT and bulk analysis of nalB-1 in Figure 1. The normalized subtracted integrated fluorescence intensity was calculated as described in Figure 3(D). The cells were similar to the one shown in (A) and selected from the sequence images, each including 60 cells. The cell solution containing 10 μ M EtBr was prepared as described in Figure 2. 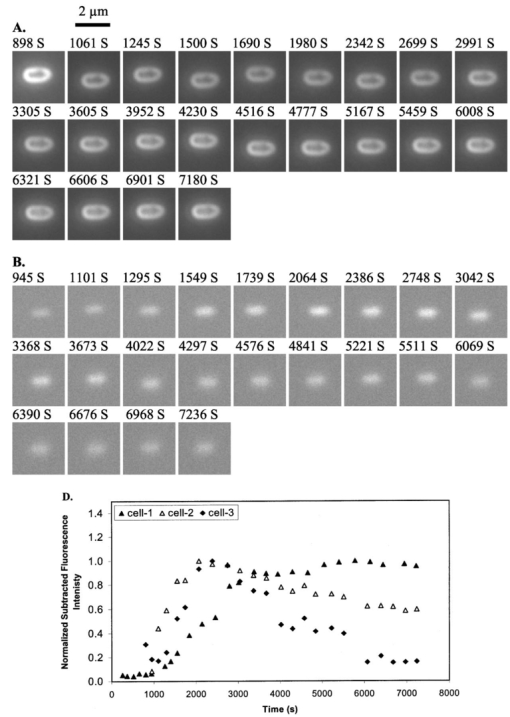 Figure 5Representative time-sequence dark-field optical and fluorescence images and intensity of single ΔABM cells, selected from 60 cells in the same images similar to the one in Figure 2 showing both temporal and spatial information of EtBr accumulation kinetics in live ΔABM cells: Representative snapshots of one of the single cells (ΔABM): real-time (A) dark-field optical images and (B) fluorescence images recorded by CCD camera, and (see color on page 583) (C) real-color fluorescence images recorded almost simultaneously by a digital color camera, for ∼2 h through the dark-field optical and epi-fluorescence microscope. The 60 cells in each sequence image, similar to the one in Figure 2(A), were analyzed. The single cell was selected from these 60 cells as a representative example. The intensity scale is 835–3083 ADC for all sequence images in (A) and 118–170 ADC for all sequence images in (B). (D) Plots of normalized subtracted integrated fluorescence intensity of single ΔABM cells versus time demonstrating the individual cell showed a differentiated intensity profile that is distinguished from WT and nalB-1 and bulk analysis of ΔABM (Figure 1). The normalized subtracted integrated fluorescence intensity was calculated as described in Figure 3(D). Fluorescence from the cell solution in the absence of EtBr was not observed. Hence, the emission of cells containing no EtBr showed less emission intensity than EtBr in the buffer solution. The subtracted fluorescence intensity of EtBr in the solution from that of the cells was negative. The cells were similar to the one shown in (A) and selected from the sequence images, each including 60 cells. The cell solution containing 10 μ M EtBr was prepared as described in Figure 2. 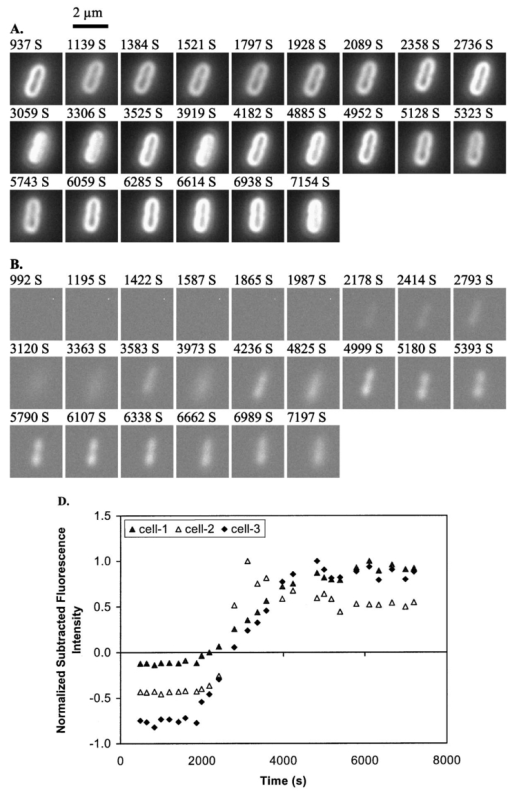 Plots of fluorescence intensity of single cells versus time were analyzed using MetaMorph software (Universal Imagine). We selected an area (60×60 pixel) that contained an individual cell and an equal sized pixel area that contained no cells (solution) in the same frame as shown in Figure 2(B). We then analyzed the integrated fluorescence intensity of each area and subtracted the integrated intensity of area where no cell was present (EtBr in solution) from that of cell. This subtracted integrated fluorescence intensity of an individual cell was then normalized with the maximum fluorescence intensity of that cell in the entire incubation time. Plots of this normalized subtracted fluorescence intensity of representative single cells versus time were used to monitor the accumulation kinetics of single live cells in real time. We selected three representative plots of the cells showing representative accumulation kinetics to illustrate distinct accumulation kinetic profiles of individual cells [Figures 3(D), 4(D), and 5(D)]. 4.Results and DiscussionThe time courses of fluorescence intensity of 10 μM EtBr in the cell solutions of WT, nalB-1 and ΔABM in Figure 1 were measured using a fluorescence spectrometer. The result clearly showed that fluorescence intensity of ΔABM (mutant with deficient pump) increased slowly prior to ∼1000 s and then increased most rapidly showing the highest intensity after 1000 s, whereas that of nalB-1 (mutant with over-expression pump) increased least rapidly and showed the lowest intensity in 2 h incubation. Fluorescence spectra of EtBr showed that the peak emission at 590±10 nm remained almost unchanged during the entire incubation time. This demonstrated the fluorescence intensity at 590±10 nm was suitable to be used to follow EtBr accumulation kinetics in the live cells. Fluorescence intensity of EtBr in the buffer solution remained almost constant for 2 h demonstrating the photostability of EtBr in 2 h incubation. EtBr emits a weak fluorescence in aqueous solution (outside of the cells) whereas it emits a strong fluorescence in hydrophobic environments (e.g., intercalation with DNA). Such changes in fluorescence intensity by EtBr indicated accumulation of EtBr in intact cells. Thus, the result in Figure 1 demonstrates that, among three mutants, ΔABM accumulated EtBr most rapidly, whereas nalB-1 accumulated EtBr most slowly. Two mutants were constructed from the same strain of P. aeruginosa, PAO4290 (a wild-type expression level of MaxAB-OprM). The only distinct feature between them is the expression level of MexAB-OprM.22 Since mutants are differentiated only by the expression level of MexAB-OprM pump proteins, the distinct accumulation kinetic profiles of mutants are most likely attributed to the extrusion kinetics. Therefore, this approach of monitoring of the accumulation of substrates in intact cells using time-course fluorescence intensity of substrates has been widely used as a successful tool to monitor the efflux kinetics of intact cells in real time.14 15 16 This approach offers the average accumulated kinetic information from a huge population of cells and hence it is most likely that the true accumulation kinetic information of single cells was buried in such an ensemble average measurement because the individual cell acts independently and it is very unlikely that each cell had the synchronized accumulation kinetics. Furthermore, this approach is unable to provide spatial information about where substrates are accumulated in intact cells and hence it is unable to address several main questions about the location and mechanism of extrusion kinetics. To visualize the location of EtBr inside live cells and address whether EtBr stays on the cellular membrane before being extruded out of cells, we directly imaged the accumulation of EtBr by live cells in the microchannel using dark-field optical and fluorescence microscopy equipped with the CCD camera and color digital camera. The 6 μm thickness microchannel was able to confine the live bacterial cells to the same location in solution and allowed the same cells to be directly imaged for hours. The representative dark-field optical and fluorescence images of live cells in 10 μM EtBr solution in Figure 2 show that many single cells were monitored simultaneously in real time at the single cell level. The well-dispersed single live cells in the microchannel shown in Figure 2 demonstrated that the cell concentration was suitable for single cell imaging. Fluorescence images of individual cell in Figure 2(B) demonstrate we were able to detect the fluorescence of EtBr in single cells and image bulk cell solution in the same environment at the single cell level simultaneously. The color fluorescence images recorded by the color digital camera in Figure 2(C) further verified the fluorescence emission of EtBr. In dark-field optical microscopy, only scattered light from the specimen captured by the objective creates images and thus the images emerge bright against a dark background.23 The species scattering light most efficiently generates the brightest images. This feature has proven dark-field microscopy well suited for the imaging of species with small sizes, such as live bacterial cells with size at 2×0.5×0.5 μm. Optical images of single cells acquired by dark-field microscopy [Figures 2(A) and 2(D)] clearly illustrated the intensity contours of the periplasmic space (bright) and cytoplasmic space (dim). The cytoplasmic space is surrounded by the periplasmic space and hence the illumination needs to penetrate through the periplasmic space in order to reach the cytoplasmic space. As a result, the intensity of illumination was weaker in the cytoplasmic space than that in the periplasmic space because the periplasmic space absorbed and scattered light. The fluorescence images [Figures 2(B), 2(C), and 2(E)] displayed the bright cytoplasmic space and the dim periplasmic space showing that the fluorescence intensity of EtBr was primarily observed in the cytoplasmic space where biological molecules, such as DNA and RNA, appeared. By comparing dark-field optical images with fluorescence images, it appears that the fluorescence intensity of EtBr was observed in the cytoplasmic space of cells [Figures 2(B) and 2(C)], whereas no obvious fluorescence intensity in the periplasmic space was observed. If EtBr were accumulated in the periplasmic space, we would observe the fluorescence intensity of EtBr in the periplasmic space change with time. This result suggests that EtBr seemed not to be accumulated in the periplasmic space, which is in an excellent agreement with previous studies and models.8 9 10 11 12 13 Fluorescence spectra of EtBr in the cell showed that the emission peak at 590±10 nm remained almost unchanged during the entire incubation time and hence the fluorescence intensity at 590 ± 10 nm was used to follow EtBr accumulation kinetics in WT. To clearly illustrate both spatial and temporal information of EtBr accumulation kinetics in WT, as an example, we selected a representative single WT cell from the image including about 60 cells and presented its sequence dark-field optical and fluorescence images in Figure 3. The time-course of fluorescence intensity of three representative cells, also selected from 60 cells in the same frame, is shown in Figure 3(D). The time-course images and fluorescence intensity of the cells unambiguously demonstrate that the fluorescence intensity of WT cells increased as the incubation time increased and the fluorescence intensity increased more rapidly at the earlier incubation time prior to 4000 s when accumulated EtBr concentration in cytoplasmic space was zero or low. By comparing both dark-field optical and fluorescence images, it is obvious that EtBr molecules accumulated in the cytoplasmic space, but appeared not to be accumulated in periplasmic space. As EtBr molecules accumulated in the cytoplasmic space, the accumulation rate of EtBr in WT began to decrease. This is either due to a decreasing influx rate of EtBr, an increasing efflux rate of EtBr or possible saturation of the cytoplasmic space with EtBr. By comparing the results with that of 40, 100, 400 μM EtBr, the possible saturation of the cytoplasmic space (chromosome and RNA) was ruled out because we observed much higher fluorescence intensity of single WT cells in 40, 100, 400 μM EtBr.21 Plots of the fluorescence intensity of single WT cells (∼60 cells in one image) in 10 μM EtBr versus time were analyzed. The representative plots of cell 1-3 from that of 60 cells monitored in the same image similar to the one in Figure 2(B) are presented in Figure 3(D). These plots showed that similar accumulation kinetics in individual WT cells were observed while some cells (e.g., cell 1-2) accumulated EtBr slightly more rapidly than others (e.g., cell 3). The overall result of real-time measurement of accumulation kinetics in single WT cells using fluorescence microscopy agreed well with that of the bulk measurement using a fluorescence spectrometer (Figure 1). To investigate the role of MexAB-OprM pump proteins in EtBr accumulation kinetics, similar approaches for the study of the WT cells were used to study the mutants of WT, nalB-1 (MexAB-OprM over-expression mutant) and ΔABM (MexAB-OprM deficient mutant). Like WT, representative time-course images of nalB-1 in 10 μM EtBr illustrate EtBr was accumulated in cytoplasmic space and no obvious fluorescence emission was observed in the periplasmic space (Figure 4). Time courses of fluorescence intensity of three representative cells, selected from 60 cells in the same image, indicate that individual nalB-1 cells (cell 1-3 in Figure 4) showed distinguished accumulation kinetic profiles. For some nalB-1 cells (e.g. cell-3), fluorescence intensity increased slowly as the incubation time increased and then reached a constant. This kinetic profile is in an excellent agreement with bulk measurement (Figure 1). For other nalB-1 cells (e.g., cell 1-2), fluorescence intensity increased rapidly prior to 3000 s and then decreased as the time continued. These nalB-1 cells were prepared in the same solution and imaged simultaneously. Therefore, the differing accumulation kinetic profiles from individual cells are real. This result demonstrates individual cells and extrusion systems acted independently and cells and extrusion systems were not synchronized. This result also suggests that the accumulation of EtBr in the cytoplasmic space may somehow trigger MexAB-OprM efflux pump of nalB-1 to extrude the influx of EtBr. Because nalB-1 is a mutant with over-expression MexAB-OprM, the efflux rate is much more pronounced in nalB-1 than that in WT. Hence, the dominating efflux mechanism in nalB-1 led to the decreasing of fluorescence intensity. In comparison with WT, the accumulation kinetic profiles of single cells (nalB-1) were sharply distinguished from that of bulk measurement. It is clear that the accumulation kinetic information of single cells was lost in bulk measurement of the massive population of cells. Like WT and nalB-1 cells, representative time-course images of ΔABM in 10 μM EtBr illustrate EtBr was accumulated in cytoplasmic space and no significant fluorescence emission was observed from the periplasmic space (Figure 5). Plots of fluorescence intensity of three representative ΔABM cells, selected from 60 cells in the same image, indicate that subtracted fluorescence intensity of EtBr in the solution from that of the cells was less than zero at the earlier incubation time (prior to 2000 s), then increased with time more rapidly than either WT or nalB-1 (Figures 3 and 4), and reached a maximum intensity after ∼4000 s. Fluorescence was not observed from the cells in the absence of EtBr so that the emission of cells containing no EtBr showed less emission intensity than EtBr in the buffer solution. Hence, the subtracted fluorescence intensity of EtBr in the solution from that of the cells was negative. This suggested that ΔABM had the lower permeability of EtBr at the earlier incubation time (prior to 2000 s) than WT and nalB-1. However, once EtBr molecules entered ΔABM cells, the accumulation rate of EtBr by ΔABM was accelerated showing a higher accumulation rate than that of WT or nalB-1 (Figures 3 and 4). This suggests that ΔABM, the mutant deficient in the MexAB-OprM efflux pump, was unable to extrude EtBr out of cells and hence accumulated EtBr in the cytoplasmic space more rapidly than WT and nalB-1. As EtBr accumulated, fluorescence intensity reached a maximum at ∼4000 s and remained relatively constant and hence the accumulation rate approached zero. We observed much higher fluorescence intensity of ΔABM in 40 and 100 μM EtBr solution21 demonstrating that the cytoplasmic space of ΔABM was not saturated in 10 μM EtBr. Therefore, this plateau of fluorescence emission intensity may be attributed to the maximum influx capacity of ΔABM or the reduction of cellular membrane permeability. It is also possible that the accumulated EtBr in ΔABM triggered the assembly of unidentified efflux pumps that were able to extrude EtBr out of cells in order to equilibrate the influx of EtBr. This assumption of the efflux mechanism in ΔABM agrees with our recent real-time measurement of sizes and dynamics of single efflux pump of ΔABM using nano-optics24 and the study of the dependence of efflux kinetics on substrate concentrations.21 The recent genome sequencing result also showed that P. aeruginosa encodes more than a dozen unidentified efflux pump proteins.19 The observation of a low accumulation rate of ΔABM at the earlier incubation time in Figure 5 agreed with that from bulk analysis in Figure 1. However, the plateau of fluorescence emission intensity in Figure 5(D) reflecting the influx and efflux equilibrium was not observed by bulk analysis in Figure 1. This may be explained by the average measurement of bulk analysis that masked the differing accumulation kinetic profiles of individual cells. Taken together, the results suggest that the different accumulation kinetics in WT, nalB-1 and ΔABM may be attributed to the expression level of MexAB-OprM and real-time transformation of membrane permeability and efflux pump machinery. It appeared that the accumulated EtBr in the cytoplasmic space of WT and nalB-1 somehow triggered the efflux kinetics of WT and nalB-1 cells, whereas the accumulated EtBr in the cytoplasmic space of the ΔABM cells promoted the influx of EtBr into the ΔABM cell at the earlier incubation time of 2000–4000 s and inhibited the further accumulation of EtBr at the late incubation time (after ∼4000 s) and then the accumulated EtBr remained unchanged. Unlike the bulk measurement (Figure 1),15 21 this result demonstrated that the influx and efflux kinetics of P. aeruginosa were also associated with individual cells even for the same type of mutant. This result indicated the importance of studying the resistance mechanism at single cell resolution because the bulk measurement only represents an average behavior of a massive number of cells. It is impossible to use such an average measurement to follow the real-time resistance mechanism because each cell and each efflux pump apparently acted independently. Furthermore, it is very likely that influx and efflux kinetics is highly regulated. To further depict the resistance mechanism at the molecular level, efforts are in progress to measure the influx and efflux kinetics of substrates by single pumps in single live cells using single-molecule detection. 5.ConclusionWe have developed and applied single live cell imaging for the real-time monitoring of accumulation kinetics of three mutants of P. aeruginosa, WT, nalB-1 and ΔABM. Time-course dark-field optical and fluorescence images of these three mutants showed that EtBr was accumulated in the cytoplasmic space whereas no EtBr accumulation appeared to be observed in the periplasmic space. This suggested that EtBr molecules passed through the cellular membrane rapidly. The differing accumulation kinetics of EtBr in three mutants were directly observed at the single cell level showing that, unlike the bulk measurement, single cell imaging overcame the ensemble measurement of bulk analysis and demonstrated the individual cell had its distinguished accumulation kinetic profile. By comparing the accumulation kinetic profiles of three mutants, the influx and efflux kinetics of three mutants were studied at single cell resolution. The result showed that the resistance mechanism was associated with the expression level of MexAB-OprM and that influx and efflux kinetics were highly regulated. This opens up new opportunities for real-time monitoring of highly regulated multi-drug resistance mechanisms using single live cell imaging. AcknowledgmentsWe thank Taiji Nakae (Tokai University School of Medicine, Japan) for three strains of Pseudomonas aeruginosa. The support of this work in part by NIH (RR15057-01), Old Dominion University, in the form of start-up, and Amideast Scholarship and Starr Foundation Scholarship (S.V.K.) and Old Dominion Honor College Undergraduate Research Fellowship (M.E.N.), is gratefully acknowledged. REFERENCES
T. Nakae
,
“Multiantibiotic resistance caused by active drug extrusion in Pseudomonas aeruginosa and other gram-negative bacteria,”
Microbiologia , 13
(3), 273
–284
(1997). Google Scholar
S. P. Cole
,
G. Bhardwaj
,
J. H. Gerlach
,
J. E. Mackie
,
C. E. Grant
,
K. C. Almquist
,
A. J. Stewart
,
E. U. Kurz
,
A. M. Duncan
, and
R. G. Deeley
,
“Over expression of a transporter gene in a mutidrug-resistant human lung cancer cell line,”
Science , 258 1650
–1654
(1992). Google Scholar
B. M. Byan
,
T. J. Dougherty
,
D. Beaulieu
,
J. Chuang
,
B. A. Dougherty
, and
J. F. Barrett
,
“Efflux in bacteria: what do we really know about it?,”
Expert Opin. , 10
(7), 1409
–1422
(2001). Google Scholar
D. Ma
,
D. N. Cook
,
J. E. Hearst
, and
H. Nikaido
,
“Efflux pumps and drug resistance in gram-negative bacteria,”
Trends Microbiol. , 2
(12), 489
–493
(1994). Google Scholar
H. Maseda
,
H. Yoneyama
, and
T. Nakae
,
“Assignment of the substrate-selective subunits of the MexEF-OprN multidrug efflux pump of Pseudomonas aeruginosa,”
Antimicrob. Agents Chemother. , 44
(3), 658
–664
(2000). Google Scholar
S. R. Morshed
,
Y. Lei
,
H. Yoneyama
, and
T. Nakae
,
“Expression of genes associated with antibiotic extrusion in Pseudomonas aeruginosa,”
Biochem. Biophys. Res. Commun. , 210
(2), 356
–362
(1995). Google Scholar
Y. Lei
,
K. Sato
, and
T. Nakae
,
“Ofloxacin-resistant Pseudomonas aeruginosa mutants with elevated drug extrusion across the inner membrane,”
Biochem. Biophys. Res. Commun. , 178
(3), 1043
–1048
(1991). Google Scholar
T. Nakae
,
E. Yoshihara
, and
H. Yoneyama
,
“Multiantibiotic resistance caused by active drug extrusion in hospital pathogens,”
J. Infect. Chemother , 3 173
–183
(1997). Google Scholar
T. Nakae
,
“Role of membrane permeability in determining antibiotic resistance in Pseudomonas aeruginosa,”
Microbiol. Immunol. , 39
(4), 221
–229
(1995). Google Scholar
A. Lee
,
W. Mao
,
M. S. Warren
,
A. Mistry
,
K. Hoshino
,
R. Okumura
,
H. Ishida
, and
O. Lomovskaya
,
“Interplay between efflux pumps may provide either additive or multiplicative effects on drug resistance,”
J. Bacteriol. , 182
(11), 3142
–3150
(2000). Google Scholar
M. Germ
,
E. Yoshihara
,
H. Yoneyama
, and
T. Nakae
,
“Interplay between the efflux pump and the outer membrane permeability barrier in fluorescent dye accumulation in Pseudomonas aeruginosa,”
Biochem. Biophys. Res. Commun. , 261
(2), 452
–455
(1999). Google Scholar
T. Kitahara
,
H. Yoneyama
, and
T. Nakae
,
“Antibiotic diffusion pathways in the outer membrane of Pseudomonas aeruginosa,”
Biochem. Biophys. Res. Commun. , 238
(2), 457
–461
(1997). Google Scholar
N. Masuda
,
E. Sakagawa
,
S. Ohya
,
N. Gotoh
,
H. Tsujimoto
, and
T. Nishino
,
“Substrate specificities of MexAB-OprM, MexCD-OprJ, and MexXY-oprM efflux pumps in Pseudomonas aeruginosa,”
Agents Chemother. , 44
(12), 3322
–3327
(2000). Google Scholar
P. G. Mortimer
and
L. J. Piddock
,
“A comparison of methods used for measuring the accumulation of quinolones by Enterobacteriaceae, Pseudomonas aeruginosa and Staphylococcus aureus,”
J. Antimicrob. Chemother. , 28
(5), 639
–653
(1991). Google Scholar
A. Ocaktan
,
H. Yoneyama
, and
T. Nakae
,
“Use of fluorescence probes to monitor function of the subunit proteins of the MexA-MexB-oprM drug extrusion machinery in Pseudomonas aeruginosa,”
J. Biol. Chem. , 272
(35), 21964
–21969
(1997). Google Scholar
H. Yoneyama
,
H. Maseda
,
H. Kamiguchi
, and
T. Nakae
,
“Function of the membrane fusion protein, MexA, of the MexA, B-OprM efflux pump in Pseudomonas aeruginosa without an Anchoring Membrane,”
J. Biol. Chem. , 275
(7), 4628
–4634
(2000). Google Scholar
H. Morjani
,
N. Aouali
,
R. Belhoussine
,
R. J. Veldman
,
T. Levade
, and
M. Manfait
,
“Elevation of glucosylceramide in multidrug-resistant cancer cells and accumulation in cytoplasmic droplets,”
Int. J. Cancer , 94
(2), 157
–65
(2001). Google Scholar
E. Crivellato
,
L. Candussio
,
A. M. Rosati
,
F. Bartoli-Klugmann
,
F. Mallardi
, and
G. Decorti
,
“The fluorescent probe Bodipy-FL-verapamil is a substrate for both P-glycoprotein and multidrug resistance-related protein (MRP)-1,”
J. Histochem. Cytochem. , 50
(5), 731
–734
(2002). Google Scholar
C. K. Stover
,
X. Q. Pham
,
A. L. Erwin
,
S. D. Mizoguchi
,
P. Warrener
,
M. J. Hickey
,
F. S. Brinkman
,
W. O. Hufnagle
,
D. J. Kowalik
,
M. Lagrou
,
R. L. Garber
,
L. Goltry
,
E. Tolentino
,
S. Westbrock-Wadman
,
Y. Yuan
,
L. L. Brody
,
S. N. Coulter
,
K. R. Folger
,
A. Kas
,
K. Larbig
,
R. Lim
,
K. Smith
,
D. Spencer
,
G. K. Wong
,
Z. Wu
, and
I. T. Paulsen
, Nature (London) , 406 959
–964
(2000). Google Scholar
I. T. Paulsen
,
M. K. Sliwinski
, and
M. H. Saier Jr.
, J. Mol. Biol. , 277 573
–592
(1998). Google Scholar
A. R. Morgan
,
J. S. Lee
,
D. E. Pulleyblank
,
N. L. Murray
, and
D. H. Evans
,
“Review: ethidium fluorescence assays. Part 1. Physicochemical studies,”
Nucleic Acids Res. , 7
(3), 547
–569
(1979). Google Scholar
K. Saito
,
H. Yoneyama
, and
T. Nakae
,
“nalB-type mutations causing the overexpression of the MexAB-OprM efflux pump are located in the mexR gene of the Pseudomonas aeruginosa chromosome,”
FEMS Microbiol. Lett. , 179
(1), 67
–72
(1999). Google Scholar
X.-H. N. Xu
,
J. Chen
,
R. Jeffers
, and
S. Kyriacou
,
“Direct measurement of sizes and dynamics of single living membrane transporters using nano-optics,”
Nano Lett. , 2 175
–182
(2002). Google Scholar
|
CITATIONS
Cited by 25 scholarly publications.
Luminescence
Resistance
Live cell imaging
CCD cameras
Microscopy
Proteins
Fluorescence spectroscopy