|
|
1.IntroductionmRNA localization is a widespread posttranscriptional mechanism for targeting protein synthesis to specific subcellular locations.1 It is believed that the specific localization of mRNAs plays a key role in the compartmentalization of protein synthesis in the cytoplasm, and facilitates protein-protein interactions.2 Clearly, mRNA localization requires the formation of mRNA ribonucleoprotein particles (mRNPs) that can be targeted to specific regions of a cell3 where polyribosomes (also referred to as polysomes) are assembled. It has also been shown that both mRNP transport and polyribosome formation involve the cytoskeleton.4, 5, 6 However, many important questions remain open concerning mRNA localization. For example, little is known about subcellular localization and organelle association of mRNAs. Although it was suggested that 70 to 80% of mRNPs in a cell are colocalized with the cytoskeleton, such a colocalization has been confirmed for only a very small number of mRNAs.7, 8 We report the direct visualization of subcellular localization of K-ras and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNAs using molecular beacons, and reveal their colocalization with mitochondria. It has been shown that K-ras proteins bind to microtubules,9 which may play an important role for K-ras trafficking. GAPDH proteins, on the other hand, have been shown to be associated with mitochondria10 as well as the cell cytoskeleton. However, there has been no report of organelle association of K-ras and GAPDH mRNAs in living cells. Our unexpected observations of colocalization of K-ras and GAPDH mRNAs with mitochondria, therefore, may provide new insight into the functions of mRNA, as well as motivations for future studies of the localization, transport, and dynamics of mRNAs in living cells. 2.Materials and Methods2.1.Molecular Beacon Design, Synthesis, and DeliveryTwo dual fluorescence resonance energy transfer (FRET) molecular beacon pairs were designed: one to target K-ras mRNA, and the other a negative control pair (“random” sequence molecular beacons), whose specific target sequences do not match with any mammalian gene (Table 1 ). In a previous study we demonstrated that by using the dual probes and detecting the FRET signal, spurious background signal can be significantly reduced.11 Single (unpaired) peptide-linked molecular beacons targeting GAPDH (see Table 1) were designed such that rapid cellular delivery of the probe can be realized. For comparison and as a further control, we designed a molecular beacon to target 28S rRNA. Molecular beacons labeled with Cy5 were synthesized by Biosource International (Camarillo, California) and MWG Biotech (High Point, North Carolina). The Cy3-labeled random beacon and all of the synthetic targets were synthesized by Integrated DNA Technologies, Incorporated (Coralville, Iowa). All molecular beacons, including target K-ras and GAPDH, as well as the “random sequence” control beacons, were first tested in solution to ensure high specificity and signal-to-background ratio.11, 12 The signal-to-background ratios for different molecular beacons used were about 10 to 20. Table 1Design of molecular beacons.
Molecular beacons targeting K-ras mRNA and 28S rRNA, as well as negative control beacons (random beacons), were delivered into living cells using a reversible permeabilization method with Streptolysin O (SLO).11 Specifically, SLO was activated first by adding of Tris (2–carboxyethyl) phosphine hydrochloride (TCEP) to of SLO for at . Cells grown in 24-well plates were incubated for in of serum free medium containing of activated SLO ( of SLO per cells) and of each molecular beacon type for cell permeabilization and beacon delivery. Cells were then resealed by adding of the typical growth medium and incubated for at before performing fluorescence microscopy imaging. Although SLO is a cytotoxic agent, its effect on gene expression is negligible when low concentrations of SLO are used.11, 13 As an alternative delivery approach, molecular beacons targeting GAPDH mRNA were linked to the TAT-1 peptide via a disulfide bridge and delivered into cells.12 TAT-1 peptide has been shown to traverse biological membranes with high efficiency. Although TAT-1 peptide can deliver cargo into the cell nucleus, the subcellular distribution of the cargos is dependent on the affinity of the cargo for its target.14 Specifically, to link the TAT-1 peptide to a molecular beacon, the quencher arm of the stem of a molecular beacon was modified to include a nucleotide dT-amine group with a 6-carbon spacer. The amine-modified molecular beacon was then reacted with TCEP to create a free thiol (-SH) functional group. The TAT-1 peptide was modified with a cysteine at its C terminus. The thiolated beacon was then reacted with cysteine-modified peptide to form a cleavable disulfide bridge.12 We expect the disulfide bridge linking peptide to probe to be cleaved once they enter the reducing environment of the cytoplasm, thereby delivering the probe to the cytoplasm. 2.2.Fluorescence In-Situ HybridizationNormal HDF cells were fixed in 100% methanol at for , allowed to dry, and kept at for . In-situ hybridization assays were performed by first washing the fixed cells for in Phosphate Buffered Saline (PBS) and then incubating them overnight at in PBS containing of fluorescently labeled linear probes targeting wild-type K-ras and GAPDH. The cells were imaged after removing the hybridization solution with washing and adding PBS. 2.3.Organelle Labeling and Fluorescence ImagingMitoFluor Green (Molecular Probes) was used to label mitochondria as directed by Molecular Probes. Fluorescence imaging of live and fixed HDF cells was performed using a Zeiss Axiovert 100 TV epifluorescence microscope coupled to a cooled, Cooke Sensicam SVGA cooled CCD camera or a Zeiss Axiovert LSM-100 confocal microscope. An exposure time of was used for all imaging assays. Specifically, fluorescence imaging of dual FRET molecular beacons were performed using an epifluorescence microscopy with a long-pass filter at for emission detection under Cy3 excitation . Imaging assays with peptide-linked, GAPDH-targeting molecular beacons were performed using a Zeiss confocal microscope with a filter for emission detection under excitation at . The fluorescence signal of MitoFlour Green was detected using a bandpass filter of under the excitation wavelength of . 3.Results and Discussion3.1.mRNA Localization and Colocalization with MitochondriaIn our previous studies we discovered that both K-ras and GAPDH mRNAs have a filamentous localization pattern in live HDF cells.11, 12 To gain insight into subcellular association of mRNA with cytoplasmic structures, we used a cell permeable dye MitoFluor Green to specifically label the mitochondria in stimulated HDF cells, followed by the delivery of dual FRET K-ras-targeting molecular beacons into the same cells using SLO. The HDF cells were first starved for and then stimulated with serum for before MitoFluor Green and molecular beacon delivery. Fluorescence signals due to specific labeling of mitochondria as well as FRET between the donor (Cy3) and acceptor (Cy5) fluorophores of the molecular beacon pair were observed, respectively, after delivery. Unexpectedly, we found that the localization pattern of K-ras mRNA was strikingly similar to that of mitochondria in HDF, as demonstrated clearly by Figs. 1(a) and 1(b) . Overlapping of the fluorescence images in Figs. 1(a) and 1(b) suggests that most of the beacon-accessible K-ras mRNA molecules colocalize with mitochondria [Fig. 1(c)]. To reveal if the observed mRNA-mitochondria colocalization is limited to one gene, we delivered GAPDH-targeting single molecular beacons (with Cy3 as the reporter, see Table 1) into HDF cells using the cell penetrating peptide TAT-1 and imaged the resulting fluorescence signal [Fig. 1(d)]. Imaging of mitochondria in the same cell was also performed [Fig. 1(e)]. Note that here a different targeting (single beacon) and delivery (peptide-based) strategy was used to cross-validate the results obtained using dual-probe and SLO-based delivery. The rapid internalization of peptide-linked molecular beacons into the cells allowed for fluorescence imaging of GAPDH mRNA within of beacon delivery, avoiding possible degradation of molecular beacons. Therefore, single (unpaired) molecular beacons were used. As can be seen from Fig. 1(e), due to the cleavable design of the peptide-linked molecular beacons,12 and the high affinity of GAPDH-targeting molecular beacons to their targets in the cell cytoplasm, only minimal nuclear uptake of the probe occurred. Overlapping of the fluorescence images in Figs. 1(d) and 1(e) again suggests that most of GAPDH mRNA and mitochondria are colocalized, as indicated by Fig. 1(f). Note that the confocal images in Figs. 1(d) and 1(e) represent 2-D cross sections of fluorescence emission, while the epifluorescence images in Figs. 1(a) and 1(b) better reflect the 3-D features of mitochondria and K-ras mRNA due to the larger depth of field. Since MitoFlour Green has essentially no excitation at (Cy3 excitation) and no emission at (Cy5 emission detection), labeling of mitochondria had no effect on the FRET signal. Further, there is very limited excitation of Cy3 at and no fluorescence emission of Cy3 nor Cy5 at ; therefore, detection of fluorescence signal due to molecular beacons had no effect on the image of mitochondria shown in Fig. 1(a). Likewise, signal contamination was minimized in detecting the fluorescence signal of mitochondria labeled with MitoFlour Green and that of GAPDH mRNA using Cy3-labeled single beacons. 3.2.Control Studies for mRNA LocalizationTo confirm that the mRNA localization patterns shown in Figs. 1(b) and 1(e) resulted from specific probe-target hybridization, control studies were performed by delivering negatively charged Cy3 or Cy5 dyes into HDF via SLO and detecting the corresponding fluorescence signal. As shown in Figs. 2(a) and 2(b) , although the signals of Cy3 and Cy5 show some degree of localization due to nonspecific binding to certain cellular structures, they did not give the filamentous localization pattern of mitochondria. This suggested that the Cy3 and Cy5 reporter dyes were not attracted specifically to mitochondria, whose surfaces are negatively charged. Therefore, the fluorescence signal of Cy3- and Cy5-labeled molecular beacons in live HDF cells shown in Figs. 1(b) and 1(e) was due to their specific hybridization to the mRNA targets, not an artifact of the reporter dyes. Fig. 2Control studies of mRNA localization. In (a) and (b), the fluorescence of Cy3 and Cy5 dyes alone inside HDF cells is shown respectively, indicating that free Cy3 and Cy5 dyes inside a living cell did not result in a filamentous mRNA localization pattern shown in Figs. 1(b) and 1(e). In (c), the overlap of a white-light image of an HDF cell and the fluorescence signal of Cy3-labeled, Tat-peptide conjugated 24-mer poly-A oligonucleotides in an HDF cell is shown. Note that most of the labeled oligonucleotides accumulated in the perinuclear region due to the very low affinity of the probe for any target in the cytoplasm. In (d), a confocal fluorescence image of the localization of GAPDH mRNA is shown as the result of SLO-based delivery. Clearly, SLO- and peptide-based delivery gave very similar filamentous patterns of GAPDH mRNA localization. 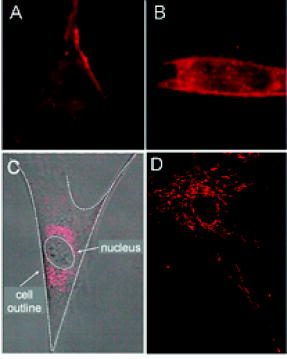 In our previous studies we delivered dual FRET, random-sequence molecular beacons using SLO as well as peptide-blinked single Cy3-labeled random-sequence molecular beacons into HDF cells, and detected the resulting fluorescence signal. This serves as a negative control in that, if the images in Fig. 1 showing colocalization of the K-ras and GAPDH mRNAs with mitochondria were resulted from nonspecific opening of the molecular beacons near mitochondria, the same would occur with random beacons. However, using the same excitation and emission detection optics, the random beacons gave very low background signal as compared with the true signal,11, 12 indicating that the fluorescence signal from the K-ras and GAPDH beacons was indeed a result of specific mRNA detection. Fig. 1Colocalization of K-ras and GAPDH mRNAs with mitochondria in HDF cells. (a) and (b) display respectively the epifluorescence images of mitochondrial staining using MitoFluor Green and K-ras mRNA detected using dual FRET molecular beacons in the same HDF cell. Shown in (c) is the overlapping of the mitochondria and K-ras mRNA images, strongly suggesting that most of K-ras mRNA molecules are colocalized with mitochondria. The confocal images of mitochondrial staining and GAPDH mRNA detection in the same HDF cell are shown respectively in (d) and (e), and their overlap is shown in (f). It appears that GAPDH mRNA and mitochondria are colocalized as well. 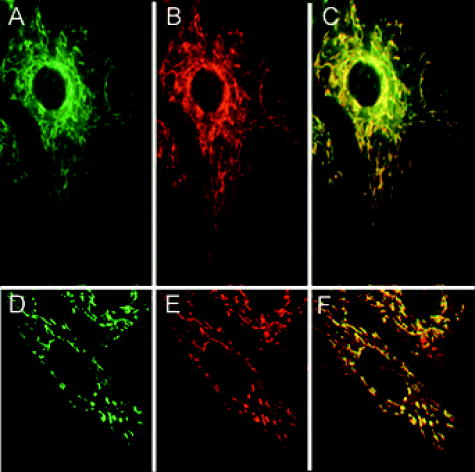 As a further control, we delivered into HDF cells Cy3-labeled linear 24-mer poly-A oligonucleotide conjugated with the Tat-1 peptide through a stable chemical linkage.12 As shown in Fig. 2(c), after delivery, this fluorescent oligonucleotide construct accumulated in the perinuclear region of the cell due to the very low affinity for a target in the cytoplasm. This clearly demonstrates that, with Tat-1 peptide as the delivery vehicle, labeled oligonucleotide may localize differently depending on the target, further confirming that the filament-like localization GAPDH mRNA shown in Fig. 1 was a result of specific targeting of molecular beacons. In addition to peptide-based delivery, we internalized GAPDH-targeting molecular beacons into HDF cells using Streptolysin O and imaged the resulting fluorescence using confocal microscopy. As can be seen from Fig. 2(d), GAPDH mRNAs show a clear filamentous pattern, similar to the results obtained using peptide-linked, GAPDH-targeting molecular beacons. The detection of GAPDH mRNAs using two different delivery methods (both peptide-based and SLO) served as a cross-check of the results shown in Fig. 1. As a positive control, we performed fluorescence in situ hybridization (FISH) assays to see if the filamentous pattern of mRNA localization could be observed in fixed cells. The epifluorescence images of mRNA detection using FISH are displayed in Figs. 3(a) and 3(b) , respectively, for K-ras and GAPDH, indicating that their localization patterns in the cytoplasm are similar to that in living cells as shown in Figs. 1(b) and 1(e). In the FISH assays, methanol was employed for fixation, which is often used to maintain the subcellular structure and retain molecules associated with microtubules.15, 16 Taken together, these positive and negative control studies suggest that the mRNA localization patterns shown in Figs. 1(b) and 1(e) are true. Fig. 3Fluorescence in situ hybridization studies. In (a) and (b), the FISH results of K-ras and GAPDH mRNA detection are shown respectively as positive controls, further confirming the filamentous mRNA localization pattern displayed in Figs. 1(b) and 1(c). (c) A FISH image of the 28S rRNA beacon displaying significant localization with the ER. 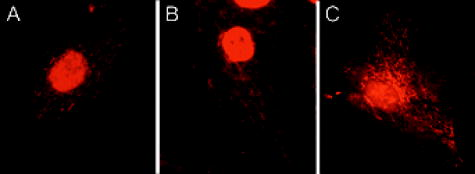 3.3.Localization of 28S rRNA and Its Colocalization with Rough Endoplasmic ReticulumIt has been shown that 28S ribosomal RNA molecules accumulate to rough endoplasmic reticulum (ER), and do not colocalize with mitochondria nor have filament-like localization patterns.17, 18, 19 To demonstrate a different RNA localization pattern using molecular beacons, we designed a molecular beacon to target the human 28S ribosomal RNA (rRNA) with the same hybridization domain as used by Paillasson 17 and Molenaar, 18 where living cell detection of 28S rRNA was performed and the accessibility of target sequence was demonstrated. Detection of 28S rRNA provides an excellent control for subcellular RNA localization, because most of the 28S rRNA molecules in the cytoplasm are known to be either localized to the rough ER or bound to free ribosomes.19 We first performed fluorescence in situ hybridization (FISH) using 28S rRNA-targeting molecular beacons in methanol fixed cells. As can be seen from the FISH image displayed in Fig. 3(c), the fluorescence intensity is quite high in the rough ER (the waffle-like pattern in the cytoplasm), which is different from the localization pattern of the K-ras and GAPDH mRNAs shown in Figs. 3(a) and 3(b). We delivered molecular beacons targeting 28S rRNA into live HDF cells using SLO11 and performed fluorescence imaging of the resulting signal using a confocal microscope. As shown in Fig. 4(a) , most of the fluorescence signal as a result of beacon-rRNA hybridization was near the cell nucleus. Imaging the cell with white light [Fig. 4(b)] and then overlapping the two images demonstrates that the beacon-induced signal in the cytoplasm was localized with the rough ER, as shown in Fig. 4(c). This pattern of 28S rRNA colocalization with the rough ER is significantly different from those displayed in Fig. 1, where the filamentous localization of K-ras and GAPDH mRNAs seems to coincide with mitochondria. We performed a negative control study to delivery single random-sequence molecular beacons into HDF cells using SLO; the resulting fluorescence signal is shown in Fig. 4(d), along with the white-light image of the same cells shown in Fig. 4(e); the overlap of the images in Figs. 4(d) and 4(e) is given in Fig. 4(f). The very low background signal displayed in Fig. 4(f) after of incubation confirms that the image shown in Fig. 4(a) was indeed due to specific detection of 28S rRNA by molecular beacon. Fig. 4A control study for the localization of the molecular beacons in live cells. (a) The fluorescence signal from 28S rRNA-targeting single molecular beacons in HDF cells. (b) A white-light image of the same cell using a confocal microscope. (c) The overlap of the images in (a) and (b), indicating 28S rRNA colocalization with the rough ER. The results of a negative control assay using a Cy3-labeled random beacon are given in (d), (e), and (f), showing respectively the fluorescence signal of random beacons, the white-light image of the same cell, and their overlap. The very low background signal of random beacons confirms the specificity of 28S rRNA detection using molecular beacons. 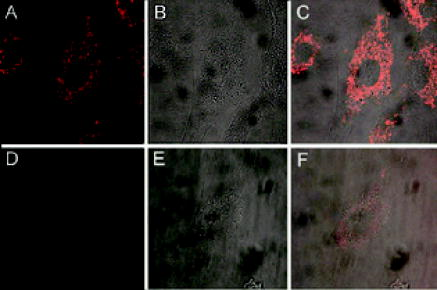 Our results shown in Figs. 4(a), 4(b), 4(c) are consistent with what reported by Molenaar 18 using a FISH assay to image 28S rRNA. Specifically, most of the signal in the cell cytoplasm was concentrated near the cell nucleus, suggesting a colocalization of 28S rRNA with rough ER. However, in the same study, the live-cell imaging of 28S rRNA using microinjection failed to detect 28S rRNA in the cytoplasm, largely due to the delivery method (microinjection) that has been shown to concentrate oligonucleotides rapidly into the cell nucleus.20 Positive detection of 28S rRNA in the cytoplasm of live HeLa cells has also been reported,17 although issues such as the signal-to-background ratio and the concentration of the linear probes render the direct comparison difficult. Taken together, our observations strongly suggest that K-ras and GAPDH mRNA molecules are colocalized with mitochondria, which are in turn associated with the ER and microtubules. GAPDH proteins, which are essential to glycolysis and the production of pyruvate, have been shown to be associated with mitochondria.10, 21 Moreover, it has been shown that K-ras proteins can mediate both antiapoptotic and proapoptotic pathways.22 For example, Bcl-2 and Ras proteins, including K-ras, interact in mitochondria in an IL-2 dependent fashion.23 Therefore, colocalization of K-ras and GAPDH transcripts with mitochondria may facilitate the localization of their corresponding proteins to mitochondria. Although the physical basis of the association of K-ras and GAPDH mRNAs with the ER, mitochondria, and microtubules needs to be further explored, our unexpected finding may stimulate further studies of the transport, localization, and organelle association of mRNA. For example, it will be important to study the localization of mRNAs corresponding to different classes of proteins, and to reveal the biological significance of mRNA-mitochondria association. Molecular-beacon-based RNA detection in living cells may provide a novel approach for studying the dynamics of subcellular localization of specific RNAs and revealing their biological functions. Unlike FISH studies in which cells need to be fixed, living cell RNA detection using molecular beacons enables real-time dynamic measurements of RNA localization, including mRNA trafficking and transport to and from mitochondria, as well as changes in mRNA localization patterns in response to drug molecules and external stimuli such as growth factors and applied shear stress. For example, the structural dynamics of localized mRNA can be studied using molecular beacons in conjunction with fluorescence recovery after photobleaching (FRAP).24 It should also be possible to visualize alterations in mRNA localization as a result of the deletion of specific sequences (e.g., “zipcode”) on a target mRNA, which may help uncover the molecular mechanisms underlying mRNA localization. AcknowledgmentsThis work was supported in part by NSF (BES-0222211) and by NIH/NIGMS (1 P20 GM072069-01). ReferencesM. Kloc,
N. R. Zearfoss, and
L. D. Etkin,
“Mechanisms of subcellular mRNA localization,”
Cell, 108 533
–544
(2002). 0092-8674 Google Scholar
M. Kloc and
S. M. Bilinski,
“RNA localization and its role in the spatially restricted protein synthesis,”
Folia Histochem. Cytobiol., 41 3
–11
(2003). 0239-8508 Google Scholar
R. P. Jansen,
“RNA-cytoskeletal associations,”
FASEB J., 13 455
–466
(1999). 0892-6638 Google Scholar
G. J. Bassell,
R. H. Singer, and
K. S. Kosik,
“Association of poly(A) mRNA with microtubules in cultured neurons,”
Neuron, 12 571
–582
(1994). 0896-6273 Google Scholar
J. Hesketh,
D. Jodar,
A. Johannessen,
K. Partridge,
I. Pryme, and
A. Tauler,
“Enrichment of specific mRNAs in cytoskeletal-bound and membrane-bound polysomes in Chinese hamster ovary cells,”
Biochem. Soc. Trans., 24 187S
(1996). 0300-5127 Google Scholar
P. Mahon,
J. Beattie,
L. A. Glover, and
J. Hesketh,
“Localisation of metallothionein isoform mRNAs in rat hepatoma (H4) cells,”
FEBS Lett., 373 76
–80
(1995). 0014-5793 Google Scholar
J. W. Wiseman,
L. A. Glover, and
J. E. Hesketh,
“Evidence for a localization signal in the untranslated region of myosin heavy chain messenger RNA,”
Cell Biol. Int., 21 243
–248
(1997). 1065-6995 Google Scholar
B. Russell and
D. J. Dix,
“Mechanisms for intracellular distribution of mRNA: in situ hybridization studies in muscle,”
Am. J. Physiol., 262 C1
–C8
(1992). 0002-9513 Google Scholar
Z. Chen,
J. C. Otto,
M. O. Bergo,
S. G. Young, and
P. J. Casey,
“The C-terminal polylysine region and methylation of K-Ras are critical for the interaction between K-Ras and microtubules,”
J. Biol. Chem., 275 41251
–41257
(2000). 0021-9258 Google Scholar
R. Ishitani,
M. Tanaka,
K. Sunaga,
N. Katsube, and
D. M. Chuang,
“Nuclear localization of overexpressed glyceraldehyde-3-phosphate dehydrogenase in cultured cerebellar neurons undergoing apoptosis,”
Mol. Pharmacol., 53 701
–707
(1998). 0026-895X Google Scholar
P. J. Santangelo,
B. Nix,
A. Tsourkas, and
G. Bao,
“Dual FRET molecular beacons for mRNA detection in living cells,”
Nucleic Acids Res., 32 e57
(2004). 0305-1048 Google Scholar
N. Nitin,
P. J. Santangelo,
G. Kim,
S. Nie, and
G. Bao,
“Peptide-linked molecular beacons for efficient delivery and rapid mRNA detection in living cells,”
Nucleic Acids Res., 32 e58
(2004). 0305-1048 Google Scholar
E. L. Barry,
F. A. Gesek, and
P. A. Friedman,
“Introduction of antisense oligonucleotides into cells by permeabilization with streptolysin O,”
BioTechniques, 15 1016
–1020
(1993). 0736-6205 Google Scholar
A. Joliot and
A. Prochiantz,
“Transduction peptides: from technology to physiology,”
Nat. Cell Biol., 6 189
–196
(2004). 1465-7392 Google Scholar
F. Gergely,
C. Karlsson,
I. Still,
J. Cowell,
J. Kilmartin, and
J. W. Raff,
“The TACC domain identifies a family of centrosomal proteins that can interact with microtubules,”
Proc. Natl. Acad. Sci. U.S.A., 97 14352
–14357
(2000). 0027-8424 Google Scholar
L. Berrueta,
S. K. Kraeft,
J. S. Tirnauer,
S. C. Schuyler,
L. B. Chen,
D. E. Hill,
D. Pellman, and
B. E. Bierer,
“The adenomatous polyposis coli-binding protein EB1 is associated with cytoplasmic and spindle microtubules,”
Proc. Natl. Acad. Sci. U.S.A., 95 10596
–105601
(1998). 0027-8424 Google Scholar
S. Paillasson,
M. Van De Corput,
R. W. Dirks,
H. J. Tanke,
M. Robert-Nicoud, and
X. Ronot,
“In situ hybridization in living cells: detection of RNA molecules,”
Exp. Cell Res., 231 226
–233
(1997). 0014-4827 Google Scholar
C. Molenaar,
S. A. Marras,
J. C. Slats,
J. C. Truffert,
M. Lemaitre,
A. K. Raap,
R. W. Dirks, and
H. J. Tanke,
“Linear O-Methyl RNA probes for the visualization of RNA in living cells,”
Nucleic Acids Res., 29 E89
–9
(2001). 0305-1048 Google Scholar
P. B. Moore and
T. A. Steitz,
“The structural basis of large ribosomal subunit function,”
Annu. Rev. Biochem., 72 813
–850
(2003). 0066-4154 Google Scholar
J. P. Leonetti,
N. Mechti,
G. Degols,
C. Gagnor, and
B. Lebleu,
“Intracellular distribution of microinjected antisense oligonucleotides,”
Proc. Natl. Acad. Sci. U.S.A., 88 2702
–2706
(1991). 0027-8424 Google Scholar
M. F. Liaud,
C. Lichtle,
K. Apt,
W. Martin, and
R. Cerff,
“Compartment-specific isoforms of TPI and GAPDH are imported into diatom mitochondria as a fusion protein: evidence in favor of a mitochondrial origin of the eukaryotic glycolytic pathway,”
Mol. Biol. Evol., 17 213
–223
(2000). 0737-4038 Google Scholar
A. Rebollo and
C. Martinez-A,
“Ras proteins: recent advances and new functions,”
Blood, 94 2971
–2980
(1999). 0006-4971 Google Scholar
A. Rebollo,
D. Perez-Sala, and
C. Martinez-A,
“Bcl-2 differentially targets K-, N-, and H-Ras to mitochondria in IL-2 supplemented or deprived cells: implications in prevention of apoptosis,”
Oncogene, 18 4930
–4939
(1999). 0950-9232 Google Scholar
N. Nitin and
G. Bao,
(2004) Google Scholar
|

