|
|
1.IntroductionElucidation and modulation of the intricate mechanisms underlying basic cellular functions often require delivery of compounds and macromolecules into living cells, including various nucleic acids [e.g., DNA plasmids, aptamers, siRNA (small interfering RNA), etc.], biosensors, proteins, ions,1 and semiconductor nanocrystals.2 However, these substances generally do not pass unassisted through intact cell membranes. Consequently, efficient methods for delivering macromolecules into living cells are indispensable in many areas of biological research and medicine. The importance of macromolecule delivery, and lack of an ideal robust procedure that works well with a range of macromolecules and cell types, has resulted in the development of numerous methods for delivering macromolecules into cells. The most commonly used methods include cationic reagents,3 liposomes,4, 5 electroporation,6 viruses,3, 7 biolistics,8 and microinjection.9 Each of these methods has found utility within certain application areas. However, with the exception of microinjection (which is very slow and laborious), none of these methods are broadly applicable to many different classes of macromolecules and cell types. Unfortunately, most of these delivery methods also exhibit varying degrees of toxicity, limiting their use to robust cell types (e.g., HeLa, 293, etc.). Thus, a significant and unmet need exists to develop benign and versatile methods for delivering substances into traditionally refractory cells of direct physiological relevance (e.g., primary cells, neuronal, B cell, and T cell lines, etc.). The use of lasers to deliver plasmid DNA into cells was first reported by Tsukakoshi 10 in 1984 and was later confirmed by others.11, 12, 13 In these reports, tightly focused laser irradiation was found to transiently permeabilize the cell membrane, enabling plasmid delivery to occur. Despite these initial promising reports, very little is known about optoinjection and it has not been widely adopted, most likely due to the slow and laborious process required to manually locate, target and irradiate individual cells with a laser on a microscope. We recently reported the development of an automated system for high-throughput cell imaging and laser-mediated cell purification.14 This novel laser-enabled analysis and processing (LEAP™) system enables cell imaging at rates up to , coupled with laser-irradiation of individual cells at rates exceeding . These unique capabilities of LEAP were used to explore and characterize parameters that impact the efficiency and toxicity of optoinjection, and then to demonstrate successful high-throughput optoinjection of a variety of compounds and macromolecules into a diverse range of cell types demonstrating broad utility. 2.Methods2.1.Cell Culture and ReagentsHeLa, NIH/3T3, 293T, HepG2, NTERA-2, PFSK-1, SU-DHL-4, CEM, and 184A1 cells were obtained through the American Type Culture Collection (ATCC; Manassas, Virginia) and grown according to published guidelines. MO-2058 (gift from Dr. Timothy C. Meeker) were grown in Iscove’s modified Dulbecco’s medium with 10% fetal bovine serum (FBS). Primary rat cardiomyocytes were a gift from Dr. Andrew D. McCulloch. Sytox Green , Sytox Blue , Cell Tracker dyes , Hoechst , RhodZin1 , and Dextran-tetramethylrhodamine were from Invitrogen (Carlsbad, California). The green fluorescent protein (GFP)-encoding plasmid, phrGFP-1, was purchased from Clontech (Palo Alto, California). Fluorescent semiconductor nanocrystals (gift from SC BioSciences; Tokyo, Japan) were used at . The Cdc42 binding domain of WASP conjugated to an I-SO dye was a gift from Dr. Klaus M Hahn.15 2.2.LEAP SystemA schematic representation of the optical setup used for these experiments is shown in Fig. 1 , and more detailed descriptions of LEAP can be found in prior publications and patents.14, 16 A custom Nd:YAG pulsed laser (JDS Uniphase; San Jose, California) emitting light in three harmonics (1064, 532, and ) delivers pulses of duration at up to frequency. The output of the laser (a maximum of ) was measured using a NOVA II power meter with a PD10-PJ-SH sensor (Ophir; Portland, Oregon). The laser spot profile at the sample plane was determined to be Gaussian by measuring transmission through moving Ronchi ruling grids (Edmund Optics; Barrington, New Jersey), and the nominal diameter of the beam [i.e., the diameter which contained 63% of the irradiance] was determined at specific beam expander settings. Fig. 1Schematic representation of the key components of LEAP used to carry out these experiments. An arc lamp provides excitation illumination through an excitation filter wheel, off a dichroic filter (in a wheel; not shown), through an galvanometer mirror assembly, through a large F-theta scanning lens [0.25 numerical aperture (NA)], and into the cell sample. Emitted light returns along the same path, but passes through the dichroic filter (in the wheel; not shown), an emission filter wheel, and into a camera. Laser pulses pass through a filter wheel (to select the wavelength; not shown), a variable neutral density wheel (to control the energy delivered; not shown), a beam expander (to control the laser spot diameter; not shown), and then through the galvanometer mirror assembly, the F-theta scanning lens, and finally into the cell sample. Small, rapid (i.e., ) movements of the galvanometer mirrors are used to image different sections of the plate, and to irradiate different cells, all without having to move the plate. 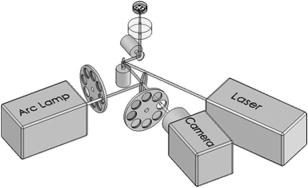 2.3.OptoinjectionCells destined for optoinjection were first stained in T-flasks with Cell Tracker Orange or Green in growth medium for at , followed by two rinses with phosphate-buffered saline (PBS), and trypsinization for removal in the case of adherent cells. Cells were counted and plated at 300 to 500 cells per well in custom 384-well plates that were designed for flatness and high transmission of the laser without damage (Cyntellect; San Diego, California). Cells were allowed to settle and grow overnight, then rinsed twice with optoinjection buffer (Cyntellect), and finally placed in of optoinjection buffer containing the compound to be optoinjected. Plates were loaded onto LEAP for optoinjection, which was programmed to automatically focus on each well within a plate, image the well, identify and target individual cells based on Cell Tracker dye fluorescence, and then fire the laser according to user specified parameters. In the case of grid-based irradiation, the steps of imaging and individual cell targeting were not necessary. After optoinjection, cells were rinsed twice with growth medium. Cells optoinjected with siRNA were incubated and assayed later by polymerase chain reaction (PCR) and Western analysis. Cells optoinjected with fluorescent indicator molecules were incubated with Sytox Blue for at for determination of cell viability. Cells were rinsed four times with PBS and then of PBS was added to each well. Cells were then imaged on LEAP or a fluorescent microscope for analysis. Viability was based on the percentage of cells that resisted subsequent Sytox Blue staining. Optoinjection efficiency was calculated as the percentage of total viable cells remaining that demonstrated obvious nuclear staining with optoinjected Sytox Green. 2.4.Lipid-Mediated TransfectionNonspecific siRNA and siRNA targeted to the cytoplasmic dynein heavy chain, sequence -AAG GCC AAG GAG GCG CTG GAA- , were purchased from Dharmacon (Lafeyette, Colorado) and used at a concentration of for both lipid-mediated transfection and optoinjection. Oligofectamine™ (Invitrogen) transfection was optimized according to manufacturer instructions. Plates were incubated at for and then the transfection mixture was replaced with three washes of growth medium. Plates were incubated for before being processed for reverse transcription-PCR and Western analysis. 2.5.PCR AnalysisFive wells of a 384-well plate were pooled for each data point. Approximately 5000 HeLa cells were processed after siRNA delivery for RNA extraction using an RNeasy Mini Kit (Qiagen; Valencia, California). RNA concentration and quality was determined by taking and readings. Reverse transcription-PCR reactions were performed using the SuperScript III Platinum One-Step qRT-PCR System (Invitrogen). Reaction volumes were containing RNA and of each primer in thin wall MicroAmp tubes (Applied Biosystems; Foster City, California). Primer sequences for cytoplasmic dynein heavy chain were forward -ACC TCC GAT GCA GTG ACC TTC ATC- and reverse -GTG GGA ACT GGA ACC TTT GCT TTT C- . Primer sequences for -actin were forward -AAC GGC TCC GGC ATG TGC AA- and reverse -TCT GAC CCA TGC CCA CCA TCA C- . Thermal cycling was carried out on a Primus (MWG Biotech; High Point, North Carolina) at for , for , for , for a total of 20, 25, 30, 35, and . Reactions were run on a 2% agarose gel, stained with ethidium bromide, and visualized for densitometry analysis of PCR products on an Alpha Innotech imager (San Leandro, California). Reactions were optimized so that produced densitometry results within a linear range. Individual data points from densitometry analysis were normalized against -actin and then all data were normalized to the nonspecific siRNA controls. 2.6.Western DetectionFive wells of a 384-well plate were pooled for each data point. Wells were rinsed twice with PBS, replaced with of NuPAGE LDS Sample Buffer (Invitrogen), and pooled together in tubes. Samples were incubated at for , then was run on a 12-well NuPAGE 4 to 12% Bis-Tris gel (Invitrogen). Protein was then transferred to nitrocellulose membrane. Western analysis was carried out using the Western Breeze detection kit (Invitrogen). Primary antibodies against cytoplasmic dynein heavy chain (Santa Cruz Biotechnology; Santa Cruz, California) and GAPDH (Santa Cruz Biotechnology) were used at 1:500 dilution. Blots were visualized for densitometry analysis on an Alpha Innotech imager. Individual data points from densitometry analysis were normalized against GAPDH, and then all data were normalized to the nonspecific siRNA controls. 3.Results3.1.Characterization of the Optoinjection Process3.1.1.Cells individually targeted with a single laser pulseThe cell-impermeable nuclear-staining dye Sytox Green was used for characterization of optoinjection with HeLa cells using single laser pulses of different energies. Optoinjection efficiency and cell viability were dependent on the radiant exposure delivered to each targeted cell [Fig. 2a ]. Above a critical radiant exposure , a rapid decrease in cell viability was observed, consistent with prior published results.14 Interestingly, the maximum optoinjection efficiency was achieved near the critical radiant exposure. At very high radiant exposures, optoinjection efficiency declined to zero because no viable cells remained. Optoinjection throughput was using this approach. Fig. 2Optoinjection of Sytox Green with a single laser pulse. (a) HeLa cells were optoinjected with Sytox Green on LEAP by individually targeting each cell with the laser using different radiant exposure settings. (b) to (d) Single laser pulses were fired in a 2-D grid pattern covering the entire area of a well without regard to specific cell locations. Three different grid spacings were tested with each radiant exposure setting, including (b) 14, (c) 28, and (d) . Mean values [± SEM (standard error of the mean)] from five independent experiments (each performed in triplicate) are shown. See Fig. 7b for representative image of cells optoinjected with Sytox dyes. 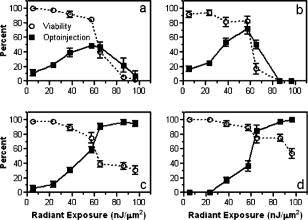 Fig. 7Optoinjection of a variety of compounds into diverse cell types. (a) NIH/3T3 cells were pre-loaded with the -sensitive dye RhodZin-1, buffer was added, and cells were optoinjected using grid-based laser irradiation. The grid was placed over a small defined region within the bottom left of the field of view to demonstrate specificity of optoinjection. (b) HeLa cells were counterstained with Cell Tracker Orange. A subset of cells was optoinjected in a ring pattern in the presence of Sytox Green. Cells were rinsed, and then another subset of cells was optoinjected in a small circle in the presence of Sytox Blue. (c) HeLa cells were optoinjected with dextran conjugated to tetramethylrhodamine. Cells were counterstained with Cell Tracker Green and nonviable cells were detected with Sytox Blue. (d) 293T cells were optoinjected with a GFP-encoding plasmid and assessed the next day for GFP expression. (e) 293T cells were optoinjected with a protein consisting of the Cdc42 binding domain of WASP conjugated to an I-SO dye (red) and were counterstained for viable cells using calcein green. Fluorescence of the I-SO dye was increased on binding to activated Cdc42.15 (f) HepG2 (human hepatocyte) cells were optoinjected with semiconductor nanocrystals that fluoresce at . Cells were counterstained with Cell Tracker Orange and nonviable cells were detected with Sytox Blue. 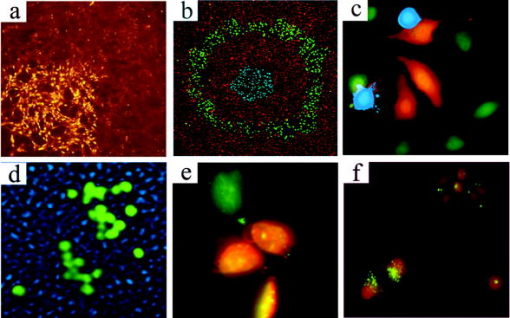 3.1.2.Single pulses fired in a grid patternTo improve throughput by eliminating the cell imaging and image processing steps needed to locate cells, single laser pulses were fired in a 2-D grid pattern covering the entire area of a well without regard to specific cell locations. A grid spacing with a nominal diameter laser beam gave results that were analogous to direct cell targeting [Fig. 2b versus Fig. 2a], which was not surprising since every cell should have been directly hit approximately once given that the space between adjacent spots in the grid was equivalent to the beam diameter. However, the grid-based approach did show improved optoinjection efficiency relative to the degree of cell death incurred when compared with the direct cell-targeting approach. Noting that the distribution of energy within the laser spot was Gaussian, and further, that 37% of the laser’s energy falls outside the nominal -diam spot, it was hypothesized that direct shots at cells with the center of the beam (i.e., the peak irradiance spot) were more lethal than cell irradiance that occurred away from the center of the beam. This hypothesis led to testing larger grid spacings of 28 and , thereby reducing the number of direct cell hits with the center of the beam while still allowing cells to be irradiated with substantial peripheral energy. The grid spacing improved optoinjection efficiency to near 100% with cell viability improved to 40% [Fig. 2c]. The grid spacing using higher radiant exposure per point produced the best results, achieving 85 to 100% optoinjection efficiency with up to 80% cell viability. Using this grid-based approach, more than one cell was affected with each laser pulse, improving throughput of the process to (depending on plated cell density). 3.1.3.Kinetics and toxicity of optoinjectionBecause cell morphology appeared normal during and after optoinjection, laser-induced permeabilization was believed to be transient in nature. Experiments were therefore performed in which the addition of Sytox Green was delayed by following irradiation to assess the duration of the permeabilization event (Fig. 3 ). Cell permeability was rapidly increased following the laser pulse, followed by an exponential return to the original nonpermeable state over a period. Kinetics were identical for individually targeted cells and grid-based irradiation, and were also independent of the compound added (not shown) or the cell type investigated. These results indicate that the cell membrane quickly returns to a normal state after optoinjection, demonstrating limited cell damage at sublethal laser doses. Importantly, a variety of optoinjected cell types (HeLa, NTERA-2, PFSK-1) exhibited growth rates equivalent to nonoptoinjected control cells, whereas the same cells exposed to a commonly used lipid delivery reagent (Oligofectamine™) exhibited significantly retarded growth (Fig. 4 ). This demonstration of reduced cell toxicity (i.e., initial viability and long-term growth) with optoinjection versus cationic lipids has not been previously reported. Fig. 3Kinetics of optoinjection. HeLa cells and SU-DHL-4 (human B-cells) were irradiated using a grid spacing with a single laser pulse per point at a radiant exposure setting of . Sytox Green was then added to the well at various times postirradiation. All data were normalized to the zero time point. The best-fit line was an exponential decay with a time constant of . Mean values (± SEM) from three independent experiments (each performed in triplicate) are shown. 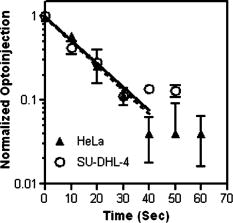 Fig. 4Toxicity of optoinjection. Growth of HeLa (∎), PFSK-1 (●), and NTERA-2 (◆) cells was measured following either mock transfection with Oligofectamine or laser irradiation using a grid spacing at a radiant exposure of . Exponential growth was not affected by optoinjection, whereas Oligofectamine inhibited cell growth long after it was removed from the culture, particularly with the more sensitive PFSK-1 and NTERA-2 cell lines. 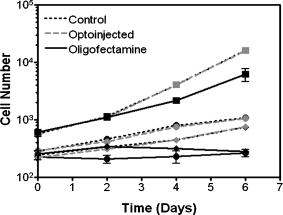 3.1.4.Multiple sequential pulses of lower irradianceThe rapid kinetics of optoinjection, combined with the knowledge that lethal laser effects are not additive at this wavelength,14 led to the hypothesis that increased optoinjection efficiency might be achieved with repeated sublethal laser pulses without affecting cell viability. Cells were therefore individually targeted for optoinjection using multiple pulses of the laser at relatively low radiant exposures. Experiments were performed in which a varied number of laser pulses were delivered to cells in intervals. Groupings of laser pulses were also repeated a various number of times with intervals between pulse groupings. Repeat intervals greater than produced similar results (not shown). Results from a radiant exposure setting of per pulse are shown (Fig. 5 ), which previously gave only modest optoinjection efficiency with high cell viability when delivered as a single pulse [Fig. 2a]. Optoinjection efficiency increased significantly as the number of laser pulses was increased. At 30 pulses, optoinjection efficiency was increased twofold with only a 15% decline in cell viability as compared with one pulse. Beyond 30 pulses in one grouping, increased optoinjection efficiency was offset by an equivalent decrease in cell viability. However, smaller groupings of laser pulses repeated several times achieved greater than 90% optoinjection efficiency while maintaining very high cell viability (Fig. 5). Extremely high levels of cumulative radiant exposure, over , could be delivered to the cells by this approach. These data indicate that lower radiant exposures delivered in multiple small groupings of repeated pulses produced more desirable optoinjection results than higher radiant exposure delivered in a single pulse, or than a single large grouping of low radiant exposures. Fig. 5Optoinjection of Sytox Green using multiple sequential pulses. HeLa cells were individually targeted for optoinjection of Sytox Green using multiple pulses of the laser at a radiant exposure of . Pulses were spaced apart. Repeated groupings of pulses were spaced apart. Mean values (± SEM) from five independent experiments (each performed in triplicate) are shown. 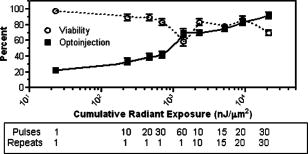 3.2.Versatility of Optoinjection3.2.1.Optoinjection of traditionally refractory cell linesA pressing need in macromolecule delivery is successful application to more physiologically relevant cell types that exhibit significant toxicity with current delivery methods. Efficiency and toxicity of optoinjection was therefore measured with a variety of cell lines generally known to exhibit low transfection efficiencies and/or high toxicity with conventional transfection methods (e.g., see Fig. 4). These cell lines include NTERA-2 (human testicular embryonic cells), MO-2058 (human B-cells), PFSK-1 (human neuronal cells), CEM (human T-cells), and 184-A1 (human mammary epithelial cells). Optoinjection efficiency of Sytox Green was 79 to 91% with these cell types (Fig. 6 ). Importantly, cell viability of 81 to 93% was obtained under these conditions. These data demonstrate the general utility of optoinjection for use with difficult cell types, in addition to more robust cell lines. Fig. 6Optoinjection of Sytox Green in refractory cell types. NTERA-2, MO-2058, PFSK-1, 184-A1, and CEM cells were optoinjected using the methods of Fig. 5. Mean values (± SEM) from three independent experiments (each performed in triplicate) are shown. 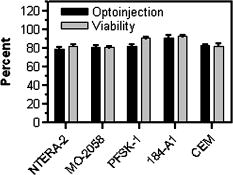 3.2.2.Variety of compounds and macromolecules optoinjected into diverse cell typesNumerous cell types were successfully optoinjected with different classes of substances to demonstrate broad applicability of optoinjection. Different ions ( and ) were optoinjected into NIH/3T3 [ shown in Fig. 7a ], 293T, HeLa, and primary rat cardiomyocytes (not shown). Studying the effects of changes in intracellular ion concentrations on cell physiology has broad applications,1, 17 and optoinjection represents the only reversible (i.e., transient) and targeted method for rapid delivery of ions into specific cells. Also shown is sequential targeted optoinjection of two different molecules (Sytox Green and Sytox Blue) into different subsets of cells within the same well [Fig. 7b], and optoinjection of tetramethylrhodamine-dextran (MW 3000) into HeLa cells [Fig. 7c]. A GFP-encoding plasmid was successfully optoinjected into 293T cells [Fig. 7d]. A biosensor protein of was successfully optoinjected into 293T cells as well [Fig. 7e]. To demonstrate the capability of optoinjection with even the most unusual macromolecules, fluorescent semiconductor nanocrystals18 were optoinjected into HepG2 [human hepatocyte; Fig. 7f] and PC-3 (human prostate epithelial, not shown) cells. These data demonstrate the broad utility of optoinjection for use with diverse classes of substances and types of cells. 3.2.3.Comparison of optoinjection with conventional lipid-mediated transfectionThe vast majority of existing cell delivery methods has been developed for delivery of nucleic acids, therefore suggesting a suitable comparison point. A particular type of nucleic acid macromolecule of significant current interest is siRNA for the targeted silencing of gene expression at the messenger RNA (mRNA) level through RNA interference (RNAi).19, 20 Applications of RNAi range from basic research to applied therapeutics, but its utility is hampered by inefficient delivery into important cell types and toxic effects of the delivery method, which can obscure the target gene knockdown effects. To address this important application, siRNA-mediated silencing of cytoplasmic dynein heavy chain expression was used as a model system to compare optoinjection with lipid-mediated transfection. Results from PCR analysis of mRNA expression [Fig. 8a ] and Western blot analysis of protein expression [Fig. 8b] show specific knockdown of cytoplasmic dynein heavy chain expression in HeLa cells. These data demonstrate that optoinjection of siRNA produced better results than common cationic lipid transfection in this robust cell type. The optoinjection process itself was inherently less toxic to cells than lipid-based transfection, as shown by the diminished NS effects seen with optoinjection versus those effects seen with Oligofectamine. For example, Oligofectamine transfection of NS siRNA caused a reduction in both mRNA and protein versus null untreated cells, whereas optoinjection of NS siRNA did not cause a significant change versus null untreated cells [Figs. 8a and 8b]. Numerous other functional siRNAs have been efficiently optoinjected into NIH/3T3, 293T, HeLa, PFSK-1, SU-DHL-4, and embryonic C166 cells (manuscript in preparation). Fig. 8Comparison of optoinjection with conventional lipid-mediated delivery of siRNA. HeLa cells were either optoinjected or transfected by Oligofectamine with siRNA directed against cytoplasmic dynein heavy chain and then assessed by (a) PCR analysis and (b) Western blot. Cells were either untreated (Null) or subjected to optoinjection/transfection in the absence of siRNA (No siRNA), a nonspecific siRNA (NS siRNA), or siRNA targeted against cytoplasmic dynein heavy chain (Dyn siRNA). Mean values (± SEM) from three independent experiments are shown. Data were normalized to the NS siRNA controls, defined as 1.0. 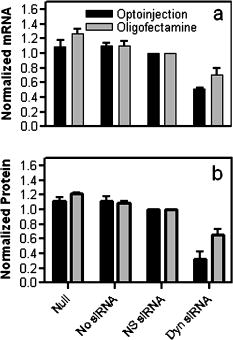 4.DiscussionThis is the first report establishing a broad utility of optoinjection, a novel method for targeted delivery of a substantially diverse array of compounds and macromolecules into a variety of different cell types with high efficiency and low toxicity, thereby overcoming many limitations of existing delivery methods. The optoinjection process was studied in considerable detail using large numbers of cells employing the novel capabilities of LEAP, elucidating key parameters for optimization. The results showed that particular importance must be given to the method of laser energy delivery, namely, that relatively low radiant exposures applied in a series of multiple pulses leads to efficient optoinjection and high cell viability. Grid-based targeting achieved better results by simultaneously irradiating several cells with each shot, many of which were outside the center peak-irradiance portion of the Gaussian beam, which appeared to induce more cell damage. This result will lead directly to greatly increased optoinjection throughput, as the laser beam diameter is adjustable and can be made large enough to encompass many cells with each pulse of the laser. Also demonstrated was the unique ability to spatially localize delivery of different macromolecules into different cells within a well via sequential macromolecule addition and optoinjection steps [Fig. 7b]. The mechanism underlying optoinjection is unknown. It has been hypothesized that optoinjection takes place through a single physical hole formed in the membrane by the laser when it is tightly focused on a portion of the cell.21 The data presented here, and elsewhere,14 do not support this mechanism of action as the laser spot used was often larger than the target cell, and further, did not require a direct hit on the target cell with the center of the beam. However, it is possible that numerous micropores are formed in the membrane, a mechanism that would be distinct from the single-hole hypothesis. Optoporation is a method that is related to optoinjection in which the laser is focused on the culture substrate to induce photoablation, resulting in a shock wave that causes a temporary permeabilization in the membranes of nearby cells. Optoporation can load numerous cells with a single laser pulse. However, significant cell death occurs near ground zero, and cells at varying distances from the shock wave epicenter are loaded to different extents.22, 23 The methods of the current study, including the grid-based approach, did not produce data consistent with the shock wave model since the laser irradiances used were far below the photoablation threshold24 and no gradients of cell loading were observed. This study, in which repeated sublethal energy pulses delivered to entire cells (or simultaneously to multiple cells) induced transient changes in overall cell membrane permeability, appears to represent a mechanism of action that is distinct from the hole-punching or shock wave models. Kinetic measurements demonstrating the same transient response from a variety of cell lines suggest that optoinjection is a physicochemical process. The unique combination of laser wavelength and radiant exposures used in this study are far outside of ranges known to cause biological damage to cells and tissues24 (other than retinal), consistent with the low degree of toxicity observed. Practical implementation of optoinjection requires the ability to perform it in a high-throughput setting. A limitation of previously published optoinjection approaches10, 11, 12, 13, 25, 26 was the relatively low throughput that can be achieved with microscope-based instrumentation. Additionally, we observed that significant cell motion and/or damage are induced by laser pulses using the previously published experimental conditions (not shown), thereby making orderly and efficient cell processing challenging, if not impossible. These limitations have been overcome here through several innovations. The F-theta scanning lens provides a large field of regard that encompasses a large area of a well plate, containing up to cells. Galvonometer mirrors enable high-throughput imaging and rapid laser beam steering anywhere within the field of regard without moving the plate, thereby increasing throughput. This optical configuration was then combined with software to locate and target cells based on multiple criteria, thereby automating the entire process. Finally, the grid-based optoinjection approach, using a laser beam larger than the cell to affect many cells simultaneously with multiple low-radiant exposures, was a significant departure from published approaches. This grid-based approach provided the highest throughput with the lowest amount of cell damage under the studied conditions, although such conditions should be applicable to targeted optoinjection with some modifications. Targeted in situ optoinjection linked with laser-mediated cell purification14 represents a powerful combination of cell manipulation capabilities that has not been available before. The ability to selectively deliver different macromolecules into specific cells in a well and eliminate unwanted cells (e.g., untransfected, or lacking a certain phenotype or desired response), all performed in situ in an iterative fashion, opens up a new realm of experimental possibilities. Furthermore, the success of the grid-based approach establishes the prospect of implementing nontargeted optoinjection on a nonimaging system, which can be of significantly lower cost and complexity, thereby potentially increasing access for a larger base of researchers. Although conventional transfection methods, such as lipid-based reagents and electroporation, can potentially be implemented in high-throughput fashion, these methods have many shortcomings. Conventional methods work with a limited class of molecules (i.e., nucleic acids), tend to be toxic, are refractory to use in several important cell types, and certainly cannot be used for specific targeted delivery. The applicability of optoinjection to a wide range of substances and cell types has broad implications for different fields of study. For example, the increasing interest in siRNA has resulted in new demand for oligonucleotide delivery methods, particularly for cell types refractory to existing methods. In these RNAi-based studies, the goal is to specifically determine the function of a single gene. Any background gene modulation due to the delivery method, as has been shown to occur by others27, 28, 29 and in Fig. 8 of this paper, will interfere with the goal of the study. Further, many important genes of interest are active only in cell types that are difficult to transfect (e.g., immune cells, neurons, etc.), making them inaccessible to RNAi-based approaches. Another example of an emerging need for macromolecule delivery is the use of semiconductor nanocrystals, many applications of which require intracellular delivery.2 Optoinjection has the potential to address needs in these rapidly growing fields of research. In summary, there is broad and growing interest in delivering cell-impermeable compounds and macromolecules into living cells for use as biosensors, to elucidate basic cell mechanisms, and to investigate potential drug-like effects. With the advent of sequencing the human genome, the ensuing cataloging of the human proteome and other related technological developments, coping with the consequences of such definition for our understanding of human cell physiology and treatment of pathological conditions are likely to be profound. The challenge that the life science community now faces is the functional assessment of gene products and candidate therapeutic molecules, not only in isolation, but also in the context of a living cell. The simple, robust, and versatile loading of compounds and macromolecules into living cells via optoinjection provides a unique tool for addressing these complex biological issues. AcknowledgmentsThis material is based in part on work supported by the National Science Foundation (NSF) under Award No. DMI-0321740 and by the National Institutes of Health (NIH) under Award Nos. R44RR15374 and R44HG002985. Any opinions, findings, and conclusions or recommendations expressed in this publication are those of the authors and do not necessarily reflect the views of NSF or NIH. The authors are employees or direct affiliates of Cyntellect, Inc. ReferencesR. Rudolf,
M. Mongillo,
R. Rizzuto, and
T. Pozzan,
“Looking forward to seeing calcium,”
Nat. Rev. Mol. Cell Biol., 4 579
–586
(2004). 1471-0072 Google Scholar
X. Gao,
Y. Cui,
R. M. Levenson,
L. W. K. Chung, and
S. Nie,
“In vivo cancer targeting and imaging with semiconductor quantum dots,”
Nat. Biotechnol., 22 969
–976
(2004). https://doi.org/10.1038/nbt994 1087-0156 Google Scholar
J. Sambrook,
E. F. Fritsch, and
T. Maniatis, Molecular Cloning: A Laboratory Manual, 2nd ed.Cold Spring Harbor Press, Cold Spring Harbor, NY
(1989). Google Scholar
J. R. Eldstrom,
K. La, and
D. A. Mathers,
“Polycationic lipids translocate lipopolysaccharide into HeLa cells,”
BioTechniques, 28 510
–516
(2000). 0736-6205 Google Scholar
A. Hirko,
F. Tang, and
J. A. Hughes,
“Cationic lipid vectors for plasmid DNA delivery,”
Curr. Med. Chem., 10 1185
–1193
(2003). 0929-8673 Google Scholar
S. J. Beebe,
P. M. Fox,
L. J. Rec,
E. L. Willis, and
K. H. Schoenbach,
“Nanosecond, high-intensity pulsed electric fields induce apoptosis in human cells,”
FASEB J., 17 1493
–1495
(2003). 0892-6638 Google Scholar
A. J. Bridge,
S. Pebernard,
A. Ducraux,
A. L. Nicoulaz, and
R. Iggo,
“Induction of an interferon response by RNAi vectors in mammalian cells,”
Nat. Genet., 34 263
–264
(2003). 1061-4036 Google Scholar
J. O’Brien and
S. C. Lummis,
“An improved method of preparing microcarriers for biolistic transfection,”
Brain Res. Brain Res. Protoc, 10 12
–15
(2002). 1385-299X Google Scholar
K. D. Jensen,
A. Nori,
M. Tijerina,
P. Kopeckova, and
J. Kopecek,
“Cytoplasmic delivery and nuclear targeting of synthetic macromolecules,”
J. Controlled Release, 87 89
–105
(2003). 0168-3659 Google Scholar
M. Tsukakoshi,
S. Kurata,
Y. Nominya,
Y. Ikawa, and
T. Kasuya,
“A novel method of DNA transfection by laser microbeam cell surgery,”
Appl. Phys. B: Photophys. Laser Chem., 35 135
–140
(1984). https://doi.org/10.1007/BF00697702 0721-7269 Google Scholar
W. Tao,
J. Wilkinson,
E. J. Stanbridge, and
M. W. Berns,
“Direct gene transfer into human cultured cells facilitated by laser micropuncture of the cell membrane,”
Proc. Natl. Acad. Sci. U.S.A., 84 4180
–4184
(1987). 0027-8424 Google Scholar
Y. Guo,
H. Liang, and
M. W. Berns,
“Laser-mediated gene transfer in rice,”
Physiol. Plant., 93 19
–24
(1995). https://doi.org/10.1034/j.1399-3054.1995.930104.x 0031-9317 Google Scholar
G. Palumbo,
M. Caruso,
E. Crescenzi,
M. F. Tecce,
G. Roberti, and
A. Colasanti,
“Targeted gene transfer in eukaryotic cells by dye-assisted laser optoporation,”
J. Photochem. Photobiol., B, 36 41
–46
(1996). https://doi.org/10.1016/S1011-1344(96)07335-6 1011-1344 Google Scholar
M. R. Koller,
E. G. Hanania,
J. Stevens,
T. M. Eisfeld,
G. C. Sasaki,
A. Fieck, and
B. O. Palsson,
“High-throughput laser-mediated in situ cell purification with high purity and yield,”
Cytometry, 61A 153
–161
(2004). 0196-4763 Google Scholar
P. Nalbant,
L. Hodgson,
V. Kraynov,
A. Toutchkine, and
K. M. Hahn,
“Activation of endogenous Cdc42 visualized in living cells,”
Science, 305 1615
–1619
(2004). 0036-8075 Google Scholar
B. O. Palsson,
M. R. Koller, and
T. M. Eisfeld,
“Method and apparatus for selectively targeting specific cells within a mixed cell population,”
(2003). Google Scholar
X. H. T. Wehrens and
A. R. Marks,
“Novel therapeutic approaches for heart failure by normalizing calcium cycling,”
Nat. Rev. Drug Discovery, 3 565
–573
(2004). 1474-1776 Google Scholar
W. C. Chan,
D. J. Maxwell,
X. Gao,
R. E. Bailey,
M. Han, and
S. Nie,
“Luminescent quantum dots for multiplexed biological detection imaging,”
Curr. Opin. Biotechnol., 13
(1), 40
–46
(2002). https://doi.org/10.1016/S0958-1669(02)00282-3 0958-1669 Google Scholar
S. M. Elbashir,
J. Harborth,
W. Lendeckel,
A. Yalcin,
K. Weber, and
T. Tuschl,
“Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells,”
Nature (London), 411 494
–498
(2001). 0028-0836 Google Scholar
T. Tuschl,
“RNA interference and small interfering RNAs,”
ChemBioChem, 2 239
–245
(2001). 1439-4227 Google Scholar
S. Kurata,
M. Tsukakoshi,
T. Kasuya, and
Y. Ikawa,
“The laser method for efficient introduction of foreign DNA into cultured cells,”
Exp. Cell Res., 162 372
–378
(1986). https://doi.org/10.1016/0014-4827(86)90342-3 0014-4827 Google Scholar
T. B. Krasieva,
C. F. Chapman,
V. J. LaMorte,
V. Venugopalan,
M. W. Berns, and
B. J. Tromberg,
“Mechanisms of cell permeabilization by laser microirradiation,”
Proc. SPIE, 3260 38
–44
(1998). 0277-786X Google Scholar
J. S. Soughayer,
T. Krasieva,
S. C. Jacobson,
J. M. Ramsey,
B. J. Tromberg, and
N. L. Allbritton,
“Characterization of cellular optoporation with distance,”
Anal. Chem., 72 1342
–1347
(2000). https://doi.org/10.1021/ac990982u 0003-2700 Google Scholar
M. H. Niemz, Laser-Tissue Interactions: Fundamentals and Applications, Springer-Verlag, Berlin
(1996). Google Scholar
Y. Shirahata,
N. Ohkohchi,
H. Itagak, and
S. Satomi,
“New technique for gene transfection using laser irradiation,”
J. Investig. Med., 49 184
–190
(2001). 1081-5589 Google Scholar
U. K. Tirlapur and
K. Konig,
“Targeted transfection by femtosecond laser,”
Nature (London), 418 290
–291
(2002). https://doi.org/10.1038/418290a 0028-0836 Google Scholar
L. J. Scherer and
J. J. Rossi,
“Approaches for the sequence-specific knockdown of mRNA,”
Nat. Biotechnol., 21 1457
–1465
(2003). 1087-0156 Google Scholar
P. C. Scacheri,
O. Rozenblatt-Rosen,
N. J. Caplen,
T. G. Wolfsberg,
L. Umayam,
J. C. Lee,
C. M. Hughes,
K. S. Shanmugam,
A. Bhattacharjee,
M. Meyerson, and
F. S. Collins,
“Short interfering RNAs can induce unexpected and divergent changes in levels of untargeted proteins in mammalian cells,”
Proc. Natl. Acad. Sci. U.S.A., 101 1892
–1897
(2004). https://doi.org/10.1073/pnas.0308698100 0027-8424 Google Scholar
M. Scherr,
K. Battmer,
A. Ganser, and
M. Eder,
“Modulation of gene expression by lentiviral-mediated delivery of small interfering RNA,”
Cell Cycle, 2 251
–257
(2003) Google Scholar
|

