|
|
1.IntroductionThe cell represents a complex biological system capable of generating and responding to forces in its environment. Still, there are relatively few scientific tools that are capable of acquiring information about displacements and forces at the cellular scale. Our laboratory has developed spectral domain phase microscopy (SDPM), a noninvasive, noncontact, optical technique capable of resolving nanometer-scale motions at sample surfaces in real time.1 A potentially important cellular application for SDPM is that of cytoskeletal rheology, which refers to the study of how the cytoskeleton deforms in response to applied forces. The cytoskeleton is crucial for cell survival, playing significant roles not only in cell structure, but also in transport, locomotion, signal transduction, and gene expression.2, 3 The complex structure of the cytoskeleton is not completely characterized, and of the many models put forth to describe its mechanical properties, no single model is universally accepted.4, 5 We show SDPM to be a useful tool for monitoring the cellular response to applied forces, and fit the resulting data to several models for cytoskeletal rheology. We recognize that a generalized rheological model cannot completely account for the regionally varying and highly dynamic properties of the cytoskeleton,5 but such a model allows us to take a step toward a rigorous characterization of the cytoskeleton. There are a variety of techniques for applying a controlled force to a cell: micropipette aspiration,6, 7, 8 compression between microplates,9, 10 cell poking,11 atomic force microscopy,12, 13, 14 optical trapping,15, 16, 17 magnetic trapping,18, 19, 20, 21, 22, 23 and magnetic twisting cytometry.24, 25, 26, 27 These techniques can be divided into several categories. The first two techniques, as well as some optical trapping methods,16 probe global properties of the cell, while the remainder are used to obtain local information. Of the localized techniques, a second major distinction lies in specificity. When attempting to study the properties of the cytoskeleton, it is desirable to be able to isolate the response of the cytoskeleton from that of the rest of the cell. Both optical and magnetic techniques typically apply forces through microspheres attached to the cell surface. By coating these microspheres with an appropriate protein, extracellular forces can be directed to cytoskeletal filaments through integrin receptors.28 We have selected a magnetic tweezer for this study because magnetic techniques allow for a broad range of forces and are capable of applying up to several nN of force to a cell. The most common method for cell visualization used in previous studies of cytoskeletal rheology has been video microscopy. Typically, an attached bead is pulled or twisted, and image processing algorithms are used to track the displacement of the bead centroid in a lateral direction. Using SDPM, we will show the cellular response as the cell is pulled in the vertical direction, i.e., perpendicular to the image plane of the microscope.15, 29 Combined with light microscopy, this technique can provide access to cellular motion in three dimensions. The rheological properties of the cytoskeleton have been modeled using simple combinations of springs and dashpots9, 18, 29 to characterize its viscoelastic response. This approach is intuitive, as both the structure and dynamics of the cytoskeleton are often compared to that of cross-linked polymer networks,30, 31 which are commonly represented by such models.32 The most basic viscoelastic model of this type is the standard linear solid,33 or SLS, in which the elastic component of the cytoskeleton is represented by two linear springs and the viscous component is represented by a dashpot. Figure 1a shows a commonly used model for the cytoskeleton, an SLS body in series with a dashpot.18 Its time-dependent extension, , under an applied force, , is given in Eq. 1 (Ref. 33), where and represent the n'th spring constant and viscous constant, respectively, and is the unit step function: The SLS body reacts in an exponential manner that exhibits both a fast elastic and a slower viscous response to an applied force. Physically, this two-phase response can be accounted for as in any viscoelastic material: the fast response arises from changes in the lengths and angles of the chemical bonds between constituent atoms, while the slower response arises from larger-scale rearrangement of the material.34 One approach to improving the accuracy of this model has been to increase the number of SLS elements,33 thereby increasing the number of exponentials in the curve [Fig. 1b]. In this manner, a double exponential response is described by:Critics of this method maintain that no finite number of time constants is capable of capturing the true response of the cell.10 A significant weakness of both of these models lies in the fact that [in Eqs. 1, 2]. This instantaneous elastic response is a feature of ideal, linear springs but does not make intuitive sense when thinking about cellular data.10, 18Fig. 1(a) A commonly used mechanical model for the cytoskeleton consists of a standard linear solid (SLS) body in series with a dashpot. This model represents changes within the cell as a force, , is applied to an attached magnetic bead. It produces a fast elastic and slower viscous behavior, as seen in Eq. 1. (b) These types of models can be extended to include multiple time constants by adding additional SLS bodies [Eq. 2]. 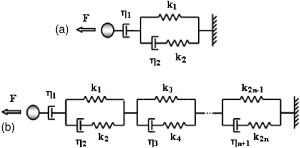 Recently, the description of the cytoskeletal response to an applied force has shifted away from simple mechanical models toward power-law models.10, 26, 35 In these models, it is assumed that the cytoskeleton behaves like a soft glassy material. The governing equation for a power-law response is given by the function: This approach is also biologically justified, providing a possible explanation for the variable mechanical properties of the cell, theorized as a sol-gel transition,35 as well as mimicking responses shown in higher-order tissues.36 It has been suggested that the cell has the ability to alter its mechanical properties; in some instances, it behaves as a solid gel, able to resist external forces and maintain its shape, while in others, it acts as a liquid, able to crawl, divide, or spread onto a substrate.3 This sol-gel transition can potentially be explained by the prediction that the cell is a glassy material close to a phase transition, allowing the system to move between order and disorder.35 In contrast to the preceding linear mechanical models, the power-law model predicts a continuous distribution of time constants.10One ubiquitous characteristic of previous cell rheology studies is that the resulting data are highly variable.5 Cell displacement distributions often span several orders of magnitude,20 and the extracted parameters, most notably from mechanical models, contain significant amounts of error. A great deal of this variability may stem from the fact that individual physical differences between examined cells are often overlooked. We have attempted to reduce the variation in our measurements by accounting for the physical and geometrical differences between each cell undergoing measurement. We have chosen to fit our data to three different models, namely, a single exponential, a double exponential, and a power law. 2.Spectral Domain Phase Microscopy TheoryOptical coherence tomography (OCT) is a noncontact, noninvasive biomedical imaging technique capable of providing high-resolution images of biological samples to a depth of a few millimeters.37 OCT utilizes low-coherence interferometry to achieve axial sectioning of the sample. For a light source with a Gaussian spectral line shape, the axial resolution is inversely proportional to the spectral bandwidth. The most elementary OCT system takes the form of a Michelson interferometer, where source light is split into two optical paths by a beamsplitter. The sample arm path is focused onto the sample, while the reference arm path is repetitively scanned in path length using a translating mirror. Back-reflected reference and sample arm light is recombined at the beamsplitter and incident on a detector. For spectral domain implementations of OCT (known as spectral domain optical coherence tomography), the reference mirror is fixed and the recombined light is spectrally resolved using a spectrometer with a multichannel detector such as a CCD array.38 The interferometric signal is manifested as sinusoidal oscillations in the measured spectrum, where the frequency of the oscillations is directly proportional to the path length difference between the reference and sample reflectors. Thus, Fourier transformation of the spectral interferometric signal results in a depth-resolved profile or A-scan. A two-dimensional (2-D) image can be built up by laterally scanning the sample arm beam. SDPM is a functional extension of spectral domain OCT (SDOCT)39, 40, 41 that allows for the measurement of nanometer-scale motions within each pixel of a SDOCT image in real time.1 The major modification in an SDPM setup from a standard SDOCT setup is the substitution of a common path interferometer for the Michelson interferometer.42 A common path interferometer is essentially a folded Michelson interferometer in which the reference and sample arm occupy the same optical path. In the SDPM common path interferometer, the reflection from the bottom surface of a coverslip serves as the reference reflection, and the cells above the coverslip comprise the sample. The spectrum recorded at the multichannel detector includes an interferometric term that contains both amplitude and phase information about the sample optical field relative to the reference field. We can imagine the sample as being composed of many small reflectors of various reflectivity, each corresponding to an interface within the sample. Each of these reflectors contributes to the spectral interferometric signal as a sinusoid with position-dependent frequency. The spectral interferogram recorded at the detector for a single sample reflector is described by: where denotes the position of the sample reflector defined by the path length difference between it and the reference reflection, is the average group index of refraction, is the detector responsivity, is the source power density function, is the spectrometer spectral resolution, is the spectrometer integration time, and and are the reference and sample reflectivities, respectively. indicates the reflector position to within the axial resolution of the SDOCT system, whereas represents subresolution departures of the reflector position from . These are small displacements that do not lead to substantial frequency shifts in the interferogram; however, they are evident in the phase of the Fourier transform of the signal at a depth corresponding to .Fourier transformation of gives , a one-dimensional (1-D), depth-resolved, complex-valued reflectivity profile, or A-scan (Fig. 2 ). The magnitude of this function has peak values at , corresponding to the reflector position. In order to monitor small motions, SDPM tracks phase changes as a function of time for the interference between the reference and sample reflectors. The interferometric phase is a function of the entire optical path that the sample and reference light traverses. In the SDPM common path interferometer, the reference and sample arm optical paths differ only by the distance between the bottom surface of the coverslip (reference reflection) and the cell. Thus, any changes in the interferometric phase are due to changes in the sample optical path, which is attributed to motion of the surface of the cell. Since phase and displacement are linearly related, relative changes in the subresolution position of the sample reflector can be derived from sequential phase measurements as a function of time. As phase measurements are inherently relative, each subsequent phase measurement is referenced to the initial phase at time . Conversion between phase and displacement is accomplished using Eq. 5, which provides time-dependent displacement profiles, shown in Fig. 2, at selected depth locations within the sample. These depths are specified to within the coherence length of the source, . Thus, we are essentially monitoring average motion within each pixel of a standard SDOCT depth profile. Here, ∠ is the phase operator, and is the source center wave number. It should be noted that the preceding analysis refers to a single reflector within the sample. In reality, a biological sample is composed of many such reflectors. Thus, this technique allows us to track subresolution movements of any reflector within the cell sample. For the application of tracking mechanical deformations in cells, we may be interested in tracking the position of the cell membrane–media interface or, in the current work, of the cell–magnetic bead interface (as shown in Fig. 2).Fig. 2The SDPM system acquires depth-resolved information from a single transverse location on the sample. (a) Illustration depicting the experimental setup. (b) A-scan, or depth profile, plotted as log(magnitude) versus depth. (c) Complex valued M-scan. Both log(magnitude) and phase are plotted versus depth and time. The magnitude of the M-scan gives a series of A-scans, while the phase of the M-scan carries information about small changes in reflector position over time. (d) Phase (displacement) versus time at several depths in the sample. Stationary surfaces, such as the top and bottom of the coverslip, do not show any change in phase over time. 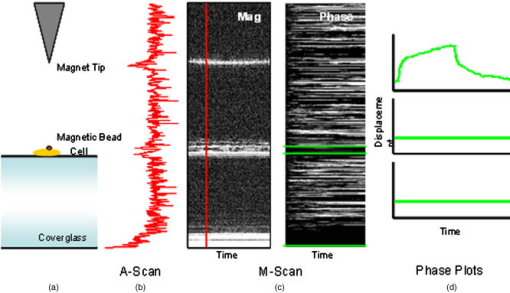 Figure 2 diagrams the information that we acquire with SDPM for the particular cellular arrangement employed in this study. We obtain a complex valued M-scan (depth profile over time) after acquiring and Fourier transforming raw SDPM data from a single transverse location over time. The magnitude of this M-scan gives us a standard SDOCT image built up of many A-scans, where reflections corresponding to the coverslip surface, the cell–magnetic bead interface, and the electromagnet tip can be clearly visualized. We also have access to the phase of this complex M-scan (relative to time ), and by selecting the depth corresponding to a reflection of interest, we extract a time-dependent displacement profile for that specific depth. Figure 2 also illustrates the fact that we do not see any phase changes from depths that correspond to stationary surfaces (i.e., the top and bottom of the coverslip). The resolution of SDPM displacement measurements is limited by the phase stability of . A common path interferometer is used to maximize the phase stability, since much of the phase noise is common-mode due to the shared sample and reference optical paths. With optimal displacement resolution that is less than a nanometer, SDPM is an ideal tool for making localized cellular measurements. The time resolution and sampling time of SDPM, defined by the integration time and readout time, respectively, of the spectrometer ( and in this study), allows for the visualization of the true dynamics of the cell. Last, SDPM is noncontact and fully compatible with light microscopy, potentially allowing for detection of motion in lateral as well as axial planes. There are several advantages to detecting motion in an axial direction. Cells growing on a coverslip surface live in a 2-D environment, implying that cytoskeletal filments should be arranged anisotropically. It is interesting and useful to compare rheological data acquired in orthogonal directions. Additionally, since SDPM is both noncontact and noninvasive, it can potentially be used for monitoring cellular motions in three-dimensional (3-D) growth scaffolds. We note that several other methods have been developed for monitoring cellular motions in the axial direction. Particle tracking techniques allow for the measurement of the mean square displacement (MSD) of fluorescent particles in three dimensions using tools such as cylindrical detection optics43 or two-photon microscopy.44, 45 The MSD of particles in solutions such as cytoplasm can be used to make measurements of viscosity, as well as diffusion constants.43, 44 The advantage of SDPM in comparison to fluorescence-based tracking methods is that we do not need to add exogenous particles, such as fluorescent beads or other markers, to track motions of the cell. (The beads used in this study serve purely for force application.) We have demonstrated SDPM to be capable of monitoring movements of the interface defined by the top surface of the cell.1 With respect to other magnetic microrheometers, our force application setup is quite similar to studies such as Bausch, 18 Alenghat, 20 and Feneberg 22 We are capable of applying forces in the range of 100's of pN to 10's of nN in a single direction using an electromagnet milled to a sharp point. More sophisticated magnetic systems have been developed for force application in multiple directions allowing for oscillatory forces,19, 21 created by placing electromagnet coils on alternate sides of the cell. The advantage of our system lies in displacement data retrieval. We are capable of high-speed measurements limited by the readout time of the CCD ( in this study), which can be as fast as sub ms (Ref. 46). Our current system speed is comparable to many of the reported magnetic microrheometers,18, 19 although there are both faster26 and slower23, 29 systems as well. Additionally, our phase processing is relatively simple. We do not require particle tracking algorithms to follow the motion of the magnetic beads, as is required for many lateral tracking microrheometers. It is illustrative to make a comparison between SDPM and other commonly used interferometric techniques. Here we will focus on reflection interference contrast microscopy (RICM), an interferometric method used to extract nanometer-scale topographical information in the one to few micron range.47, 48, 49, 50 RICM images are typically formed from interfering reflections of monochromatic light from the top surface of a substrate with that from the bottom surface of the target sample, allowing for a surface profile of the sample to be reconstructed. This method has been useful in applications involving the measurement of ultrathin layers,47, 48 as well as in investigating membrane-substrate interactions49 and wetting phenomena.47 RICM is capable of determining the distance between adjacent optical interfaces, as long as they correspond to the dominant reflections in the sample. By using broadband light and taking advantage of the coherence gating properties of OCT, SDPM is capable of determining relative nanometer-scale motions of multiple sample reflectors, as long as they are separated by the coherence length of the light source, on the order of several microns. This feature is particularly useful for characterizing multiple reflections, such as the cell surface and electromagnet tip reflections utilized in this study. Additionally, the sensitive spectral detection employed by SDPM allows for imaging up to several millimeters in depth. 3.Materials and Methods3.1.Cell PreparationMCF-7 human breast cancer cells from the Karmonos Cancer Center, provided by the Duke Cell Culture Facility, were grown in Dulbecco’s Modified Eagle’s Medium (Sigma, St. Louis) with 10% Fetal Bovine Serum (Sigma), Sodium Pyruvate (Gibco, Grand Island, New York), and Non-Essential Amino Acids (Gibco), at and 5% . When approximately 80% confluent, the cells were trypsinized (0.25% Trypsin-EDTA, Gibco) and plated in coverglass dishes coated with an antireflective layer designed to reduce Fresnel reflections from the glass-water interface. Since the cells were unable to adhere to the antireflective coating, the coverglasses were also coated with type I collagen (Sigma). In order to specifically target cytoskeletal filaments, superparamagnetic beads were coated with fibronectin and added to the cell media at a concentration of . These superparamagnetic beads were polystyrene with embedded iron oxides and had a diameter of . Fibronectin (Sigma), a protein capable of binding to integrin receptors on the cell surface, was attached to reactive tosyl groups on the bead surface according to a protocol provided by the manufacturer (Dynal Biotech, Oslo, Norway). Before cell experiments commenced, cells were incubated for approximately to allow for the magnetic beads to attach. 3.2.Electromagnet Construction and CalibrationIn order to apply a controlled force to the cell, an electromagnet was designed and constructed consisting of a Mu Metal core (Ed Fagan, Inc., Franklin Lakes, New Jersey), measuring in length and in diameter. The final centimeter was machined to a sharp point (Fig. 3 ). The core was wrapped with 2500 turns of copper wire. The magnet was powered by a DC power supply and mounted on a micromanipulator for translation in three dimensions. The time constant of the electromagnet was determined to be . Force calibration was performed by using the electromagnet to drag the magnetic beads through a fluid of known viscosity,19 in this case, glycerol. The results can be seen in Fig. 4 for a coil current of . A CCD camera was used to record the motion of the beads as they traveled toward the magnet, and the velocity of a single bead was determined from its position as a function of time. Velocity is related to force through Stoke’s law for low Reynold’s number (Ref. 19), , where is viscosity , is bead diameter, and is velocity. During cell experiments, this curve was used to position the electromagnet to achieve the desired force on the magnetic bead. We chose a force of , on the relatively flat portion of the curve, so that any error in magnet placement would cause only slight changes in the applied force. Fig. 3(a) Experimental setup showing the electromagnet mounted above the cell sample. The tip of the magnet was within the media during actual experiments. (b) Electromagnet schematic. (c) Photomicrograph of an MCF-7 cell with fibronectin-coated magnetic bead attached. 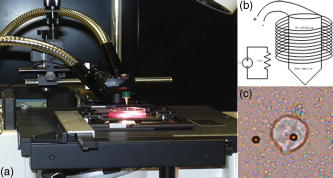 Fig. 4Force calibration results for a coil current of 0.05 A. This analysis allows for the electromagnet to be appropriately positioned to deliver the desired force to the cell. We specifically chose this current to place our desired force, , on the flat portion of the curve to prevent slight variations in magnet position from introducing a significant amount of error in the applied force. This force was not achievable in previous studies using laser tweezers.15, 16, 17 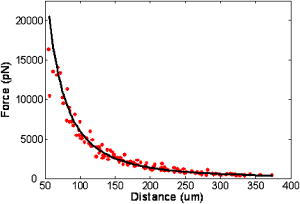 3.3.Optical SetupThe fiber-based SDPM interferometer in Fig. 5 was built using source light from a broadband femtosecond Ti:Sapphire laser (Femtolasers, Vienna, Austria), with a center wavelength of and a full-width half-maximum (FWHM) bandwidth. After passing through a fiber coupler (AC Photonics, Santa Clara, California), SDPM light was relayed into an inverted microscope (Carl Zeiss Axiovert 200, Oberkochen, Germany) through a documentation port using lenses and and focused by a , 0.5 NA objective. The reflection from the coverslip surface closest to the interferometer served as the reference reflection. The resulting interferometric signal was detected using a commercial spectrometer (Ocean Optics USB2000, Dunedin, Florida) with a readout rate using a integration time. Taking advantage of the beamsplitter in the microscope, it was possible to simultaneously acquire both SDPM data and visible light microscopy video, which was obtained using a CCD camera (A642, PixeLink, Ottawa, Canada) mounted on a second documentation port. Fig. 5SDPM common-path implementation adapted to an inverted microscope. Source light from a Ti:Sapphire laser passes through a fiber coupler (FC). The beam is relayed into the microscope via a documentation port (DP). System components include the spectrometer (Spect.), collimating lens , SDPM objective lens , tube lens (TL), microscope objective , coverslip (CS), camera objective , and camera (CCD). 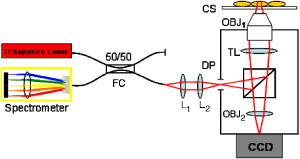 3.4.Cellular ExperimentsThe system was aligned to focus the SDPM beam at the same depth as the microscope visual focus. The focused SDPM beam, visible on the CCD camera as a localized bright spot in diameter, was used to visualize the position on the cell from which data were collected. The electromagnet was then positioned to place the tip, visible in the CCD image, directly above the cell layer at the same lateral position as the SDPM beam. A target cell was located, and the cell sample was translated to place an attached magnetic bead at this location as well. Care was taken to choose cells that were not attached to any other cell, in order to probe only the properties of a single cell of interest. Using the magnet tip reflection on the SDPM A-scan as a guide (Fig. 2), the magnet was translated vertically to apply the desired force. In order to record the cellular response, concurrent SDPM data and light microscopy video were recorded as the magnet applied of force for approximately , and as the cell recovered for an equivalent amount of time. Additionally, experiments were conducted to visualize the response of cells after of treatment with Cytochalasin D, a drug that indirectly causes the depolymerization of actin networks in the cell.3 A stock solution of Cytochalasin D (A.G. Scientific, Inc., San Diego, California) dissolved in dimethyl sulfoxide (DMSO) was combined with cell media to create a solution and heated to . After taking control data as previously described, the cell media was replaced with media containing Cytochalasin D. Completely replacing the media allowed the drug to reach the cells without any dependence on diffusion. Subsequent control experiments were performed to rule out any additional mechanical effects caused by the DMSO. Raw SDPM data were collected and processed in LabVIEW (National Instruments, Austin, Texas). The phase of the Fourier transform of the spectral interferometric signal was extracted at a depth corresponding to the cell surface–magnetic bead interface. Thus, in recording displacements of the magnetic bead, we gain information about the deformation of the cell as it is stretched normal to the substrate. Time-dependent phase profiles were converted to displacements using Eq. 5; the cell’s index of refraction was assumed to be constant. The resulting displacement curves were fit to the three models using a user-defined nonlinear regression in SigmaPlot. Model parameters are reported as deviation, and the coefficient of variation (CV) is listed as a measure of sample-to-sample fluctuation. Images of each cell were analyzed in ImageJ in an attempt to reduce variation in the cellular response by correlating displacement to the physical characteristics of the cell. A student’s -test was performed on extracted parameters to test for differences between treated and untreated cells. Statistical significance was reported at a confidence level of 95% . 4.Results and DiscussionThe resolution of our SDPM system is determined by the interferometric phase stability at the interface of interest. The phase stability is a function of the signal-to-noise ratio (SNR) at a given point and, assuming shot noise limited detection, is determined by the source power, sample reflectivity, and integration time of the detector.1 In the past, we have demonstrated that a similar system could operate at a stability of using a coverslip as a test surface.1 However, the SNR at the cell–magnetic bead interface is significantly less than that at the proximate coverslip surface due to the decreased reflectivity of this interface. We measured the average SNR of the cell–magnetic bead interface from our OCT A-scans [as shown in Fig. 2b] to be . For this SNR, we calculated an expected phase stability of approximately using the sensitivity expression given by Choma1: To determine the actual resolution of our cell displacement measurements, we recorded the standard deviation of the cell surface motion before force application. It is possible that cellular motions larger than our displacement sensitivity could further reduce our ability to discriminate small displacements. Living cells have been shown to exhibit oscillatory motion on a nanometer scale likely linked to activity of molecular motors and other metabolic processes.51, 52 However, we measured a standard deviation of , which is very close to our expected system sensitivity. This value gives the noise floor for future measurements and indicates that any larger displacements were caused by the applied forces.Representative cellular responses for both untreated and treated cells exposed to a force are illustrated in Fig. 6 , and the average maximum cellular displacement is listed. The reported untreated cell data were averaged over 25 individual cells from a total of nine petri dishes. Treated cell data were averaged over 11 individual cells from three dishes. In each dish, the proceeding experimental procedure was performed on three to four cells, chosen at locations isolated from one another in order to reduce any residual effects from previous magnetization. Following data collection, CCD camera images of each cell were examined in order to discard those that were actually a cluster of several cells as opposed to a single cell. After of applied force, the untreated cell surfaces displaced ( deviation). Upon treatment with Cytochalasin D, the maximum surface displacement increased dramatically to , demonstrating a decreased resistance to deformation and thus indicating that the cytoskeletal structure had been compromised. Control experiments showed that the addition of the solvent, DMSO, alone had no significant effect, as all displacements were found to be within the range defined by ± one standard deviation of the untreated responses (Fig. 6). Fig. 6Plot showing representative cellular responses to the application and removal of a force before and after of treatment with Cytochalasin D. Average cell displacement and recovery data for 25 untreated cells and 11 treated cells is shown. A statistically significant increase in cell displacement was seen after of treatment. 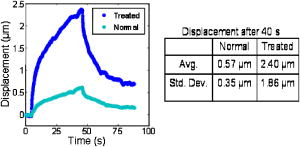 The individual cell response data were fit to each of the three models: the single- and double-exponential linear mechanical models [Eqs. 1, 2] as well as the power-law model [Eq. 3]. A representative cellular response fit to each of the three models can be seen in Fig. 7 . We found that the parameters associated with the double-exponential model were highly dependent on one another. Thus, the use of this model cannot be justified given the experimental data. For the remainder of this discussion, we restrict our attention to the single-exponential and power-law models. Several statistical fit parameters calculated from the nonlinear regression, including the coefficient of determination , the F statistic, the Akaike information criteria (AIC), and the predicted residual error sum of squares (PRESS) statistic, can be seen in Table 1 . These results indicated that from a statistical perspective, the single-exponential model performed better than the power-law model. The averaged extracted parameters from the two models are listed in Table 2 along with their coefficient of variation (CV), given by standard deviation/mean. Fig. 7A representative normal cell response fit to the (a) single-exponential [Eq. 1], (b) double-exponential [Eq. 2], and (c) power-law [Eq. 3] models. The differences in the three fits are most apparent in the first few seconds of the response. The power law model begins at zero, while both exponential models include an “instantaneous” response. All models included significant variation in the extracted parameters. Statistical goodness-of-fit metrics can be found in Table 1. 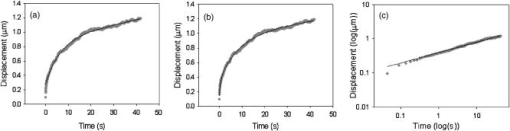 Table 1The two models were compared based on several statistics resulting from the nonlinear regression. The coefficient of determination, or R2 , is a goodness-of-fit parameter in which values closest to 1 indicate the best fit. The X2 statistic is a measure of how close measured data points match with expected values. For both calculated X2 values, there is less than a 0.001 chance that the same results could have been obtained randomly. The F statistic is used to test the null hypothesis that all of the regression coefficients are equal to zero. Both reported F-statistics correspond to a probability of <0.0001 , allowing us to reject the null hypothesis for each model. The Akaike information criteria (AIC) is used to compare various models by selecting the best fitting model while penalizing for increasing complexity.53, 54 The absolute AIC values are unimportant; it is the difference between associated AIC values that is used to rank the models, where the smallest AIC value indicates the best model. Last, the PRESS statistic is indicative of the predictive power of the model, where the minimization of the statistic is optimal.55 Each of the statistical analysis measurements agree in ranking the single exponential better than the power law.
In order to validate our measurements with SDPM, it is necessary to compare our results to previous studies. We have presented our extracted parameters in terms of the cell surface displacement profile, . Thus, our mechanical model data are in the form of spring constants, , and viscous constants, . For the purposes of comparison with previous cytoskeletal rheology studies, it is helpful to convert our measurements to elastic moduli, , and viscous moduli, . These moduli are the constants extracted from a fit to the creep function, which is the ratio of strain to stress: , where is the time-varying strain, is constant stress, is area, is force, and is the initial thickness of the cell. Thus, we are able to back-calculate elastic and viscous moduli from our parameters using the area of the magnetic bead, , and the cell thickness. Based on separate measurements from SDPM A-scans, we estimate the average cell thickness to be approximately . Our averaged single exponential parameters thus become , , , and . Lo, 29 using a similar magnetic tweezers geometry, found elastic moduli ( , ) between 0.2 and and a viscosity of , which fall in the same range as our results. However, these results may not be directly comparable to our own due to the fact that the time resolution of their measurement system was on the order of seconds. Wu ’s14 AFM measurements show an elastic modulus of and a viscosity of , which again are similar to our findings. Both of these studies investigated the local properties of adherent cell lines to forces applied in the normal direction. The bulk of the literature in this field presents widely varying properties and parameters to characterize the cell, often overlooking physical and geometrical differences between examined cells. It is evident that some of this variation is due to a wide array of measurement techniques and experimental setups. However, it is very likely that the individual physical characteristics of each cell contribute to its unique response. We attempted to reduce the variation in our results by accounting for differences in the cellular cross-sectional area as well as the position of the magnetic bead upon the cell. CCD images of each of the 25 normal cells were analyzed using ImageJ to measure the cross-sectional area of the cell as visualized through the microscope. As seen in Fig. 8 , decreasing cell displacement was correlated with decreasing area. Assuming a uniform distribution of integrin receptors on the cell surface, a larger area of attachment suggests that more connections have been formed between the magnetic bead and cytoskeletal filaments that are anchored to the coverslip. Thus, there are more connected pathways to resist the force of the electromagnet. We found that the area trend is best fit by a power law with an exponent of . It is interesting to note that if we consider nodes in a generalized network, graph theory predicts that the number of connections between nodes scales as for large . We found that the strength of the cytoskeletal network scales in a similar manner. It is this increasing number of connections to anchored surfaces that imparts strength to the cell. Using this trend along with knowledge of cell thickness, and assuming a generalized 3-D shape, such as a coin-like cylinder topped by a dome,29 it would be interesting to determine whether a similar trend exists in cell volume. It is likely that a larger volume of cell material would be capable of deforming farther in the vertical direction. Fig. 8The cross-sectional area of the adherent cell is one geometrical factor that determines how far the cell is able to stretch in the vertical direction. The area trend is best fit by a power law with exponent . Rounding to , we were able to correct our cell displacement data by dividing by the area squared. Assuming an even distribution of integrin receptors on the cell surface, the corrected model parameters describe the response of the cell on a “per-connection” basis. 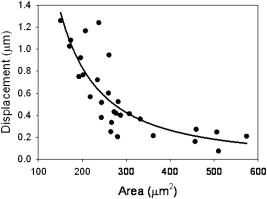 An additional parameter of potential importance is the position of the magnetic bead on the cell surface. When the microspheres were added in solution, they were observed to adhere randomly on the surfaces of the cells. It is appropriate to question whether this position affects the vertical displacement of the bead. We would expect to see more deformation near the center of the cells than near the edges, where the top and bottom surfaces of the cell are directly coupled by the actin cortex. Treating the cell images as essentially circular, a normalized radial bead position was calculated for each cell by dividing the distance from the bead to the center of the cell by the radius of the cell. Thus, a position of 1 corresponded to any edge of the cell, while 0 was the center of the cell. The average cell surface displacement of the six cells from positions between 0 and 0.3 was , and that of the eight cells from positions between 0.7 and 1.0 was . These measurements did not exhibit the anticipated trend. This discrepancy might be attributed to low sample size; we anticipate future studies measuring a larger number of cells. The cell displacement data were corrected based on the area trend found in Fig. 8. Rounding the power-law exponent from to , we divided each displacement profile by . By correcting for the area of the cells, we were indirectly correcting for the number of connections to the coverslip. We were, in essence, extracting new parameter values on a “per-connection” basis. This correction noticeably reduced the spread of our data, decreasing the CV in maximum cell surface displacement by 35.5%. This suggests that a portion of the large distribution of the mechanical properties of cells, observed both by ourselves and by other authors, may be due to distributions in their size and shape. Table 2 also shows the effect of the correction on the individual parameters of each model. In each model, there are parameters that determine the final magnitude of the cellular response, , , , , and , and others that determine how quickly it reaches this final value, and , defining the shape of the curve. We expected to see significant improvement in the parameters that govern the magnitude of displacement or stretch, but little effect upon parameters that control the time dependence of the response. The most significant improvement, a reduction in CV, is seen in , the leading dashpot in the single-exponential model. With the exception of , we saw the CV in all of the displacement-governing parameters decrease by 12.2% to 27.8%. Accordingly, there was very little change in the CV for , 0.75%. However, we still saw a relatively large improvement in the CV for . The area correction was also performed on the treated cell data in order to make a comparison between the two groups. Extracted parameters, plotted in Fig. 9 , clearly show the effects of treatment with Cytochalasin D. Again, we saw significant changes in the parameters that control the magnitude of displacement and little change in those that govern the time dependence of the curve. Fig. 9Comparison of averaged model parameters extracted from fits to Eqs. 1, 3 for 25 untreated and 11 drug treated cellular responses. Treated cells were exposed to a solution of Cytochalasin D for prior to data collection. Error bars indicate ± one standard deviation. Stars (∗) represent statistically significant differences with 95% confidence . 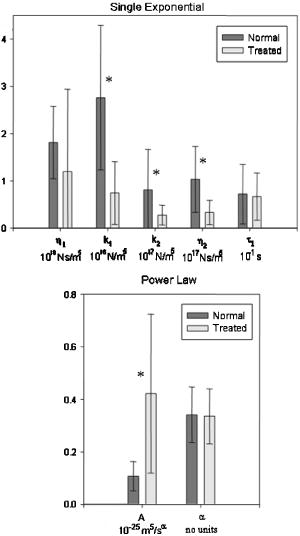 Table 2Raw model parameters extracted from nonlinear regressions to Eqs. 1, 3 were averaged over 25 untreated cellular responses and are labeled “Precorrection.” The maximum cell displacement was found to increase with decreasing cross-sectional area, prompting correction of the cell displacement data using this trend. Assuming a uniform distribution of integrin receptors on the cell surface, postcorrection parameters describe the cell behavior on a per-connection basis and reduce variation in the averaged extracted parameters. All results are reported as mean±standard deviation. The coefficient of variation (CV) is listed as a measure of cell-to-cell variation. This metric, determined as standard deviation/mean, is appropriate due to the face that precorrection and postcorrection parameters differ in units. Last, the percent reduction in the CV is listed for each parameter.
Our comparison of cytoskeletal rheology parameters before and after treatment with Cytochalasin D allows us to test predictions made by each of the postulated models. One hypothesis of previous studies that used linear mechanical models was that there should be a correlation between linear model elements and physical components of the cytoskeleton. If this were true, the drug-induced weakening of actin filaments should preferentially affect the model parameters that correspond to actin filaments and not others. However, our results indicate that the relationship is not as straightforward as a separable correlation between model parameters and physical cell components. Each spring and viscous constant in the single-exponential model appeared to be affected by drug treatment. In fact, with the exception of the leading dashpot, the decrease in each of these constants was statistically significant with . These results indicate that the actin cytoskeleton contributes both an elastic and a viscous response. This finding is in agreement with Feneberg, 22 who found that the actin cortex is responsible for the viscoelastic response of the composite cell envelope. Previous studies have used treatment with Cytochalasin D as a means of inducing a sol-gel transition.15 By weakening and dissolving actin networks, the cell should shift toward a liquid state. According to Fabry, 35 the theory of soft glassy materials predicts that a phase transition occurs as the power-law exponent describing the frequency dependence of the elastic and viscous properties of the cell moves between 1 and 0. Here, an exponent of 1 describes entirely fluid-like behavior, while an exponent of 0 describes entirely solid-like behavior.34 Desprat 10 found that the power-law exponents for both the frequency dependence and the time dependence of these properties are one and the same.10 If Cytochalasin D indeed moves the cell towards a sol-gel transition, we expect to see a corresponding increase in the exponent . The data in Fig. 9 show a change only in the parameter , not . This leads us to one of two conclusions. Either Cytochalasin D does not affect the cell in the same manner as a sol-gel transition, or soft glassy materials are not an appropriate model for the cell. 5.ConclusionsIn conclusion, we have used SDPM to monitor local nanometer-scale motions of single adherent cells under a controlled force applied through magnetic tweezers and observed the drug-induced weakening of cytoskeletal structure upon treatment with Cytochalasin D. Cell response data were fit to several cytoskeletal rheological models in order to quantitatively analyze the difference in model parameters between normal and compromised cells. We also successfully correlated the physical and geometrical properties of individual cells to displacement in order to reduce variation in our measurements. Due to its sensitivity and time resolution, SDPM appears to be a useful tool for making local measurements when studying single cell dynamics. AcknowledgmentsThis research was funded by the National Institutes of Health, Grant No. R24-EB000243. The authors thank Marinko Sarunic for assistance with the Ti:Sapphire laser, Greg Applegate for statistical advice, and Changhuei Yang for helpful discussion. ReferencesM. A. Choma,
A. K. Ellerbee,
C. H. Yang,
T. L. Creazzo, and
J. A. Izatt,
“Spectral-domain phase microscopy,”
Opt. Lett., 30
(10), 1162
–1164
(2005). https://doi.org/10.1364/OL.30.001162 0146-9592 Google Scholar
B. Alberts,
D. Bray,
J. Lewis,
M. Raff,
K. Roberts, and
J. D. Watson, Molecular Biology of the Cell, Garland Publishing, New York
(2005). Google Scholar
H. Lodish,
A. Berk,
L. S. Zipursky,
P. Matsudaira,
D. Baltimore, and
J. Darnell, Molecular Cell Biology, W. H. Freeman, New York
(2000). Google Scholar
D. Stamenovic and
D. E. Ingber,
“Models of cytoskeletal mechanics of adherent cells,”
Biomechanics and Modeling in Mechanobiology, 1
(1), 95
–108
(2002). Google Scholar
S. R. Heidemann and
D. Wirtz,
“Towards a regional approach to cell mechanics,”
Trends Cell Biol., 14
(4), 160
–166
(2004). https://doi.org/10.1016/j.tcb.2004.02.003 0962-8924 Google Scholar
E. Evans and
A. Yeung,
“Apparent viscosity and cortical tension of blood granulocytes determined by micropipet aspiration,”
Biophys. J., 56
(1), 151
–160
(1989). 0006-3495 Google Scholar
M. Sato,
D. P. Theret,
L. T. Wheeler,
N. Ohshima, and
R. M. Nerem,
“Application of the micropipette technique to the measurement of cultured porcine aortic endothelial-cell viscoelastic properties,”
ASME J. Biomech. Eng., 112
(3), 263
–268
(1990). 0148-0731 Google Scholar
F. Guilak,
G. R. Erickson, and
H. P. Ting-Beall,
“The effects of osmotic stress on the viscoelastic and physical properties of articular chondrocytes,”
Biophys. J., 82
(2), 720
–727
(2002). 0006-3495 Google Scholar
O. Thoumine and
A. Ott,
“Time scale dependent viscoelastic and contractile regimes in fibroblasts probed by microplate manipulation,”
J. Cell. Sci., 110 2109
–2116
(1997). 0021-9533 Google Scholar
N. Desprat,
A. Richert,
J. Simeon, and
A. Asnacios,
“Creep function of a single living cell,”
Biophys. J., 88
(3), 2224
–2233
(2005). https://doi.org/10.1529/biophysj.104.050278 0006-3495 Google Scholar
C. Pasternak,
S. Wong, and
E. L. Elson,
“Mechanical function of dystrophin in muscle cells,”
J. Cell Biol., 128
(3), 355
–361
(1995). https://doi.org/10.1083/jcb.128.3.355 0021-9525 Google Scholar
R. E. Mahaffy,
S. Park,
E. Gerde,
J. Kas, and
C. K. Shih,
“Quantitative analysis of the viscoelastic properties of thin regions of fibroblasts using atomic force microscopy,”
Biophys. J., 86
(3), 1777
–1793
(2004). 0006-3495 Google Scholar
J. Alcaraz,
L. Buscemi,
M. Grabulosa,
X. Trepat,
B. Fabry,
R. Farre, and
D. Navajas,
“Microrheology of human lung epithelial cells measured by atomic force microscopy,”
Biophys. J., 84
(3), 2071
–2079
(2003). 0006-3495 Google Scholar
H. W. Wu,
T. Kuhn, and
V. T. Moy,
“Mechanical properties of L929 cells measured by atomic force microscopy: effects of anticytoskeletal drugs and membrane crosslinking,”
Scanning, 20
(5), 389
–397
(1998). 0161-0457 Google Scholar
C. H. Lee,
C. L. Guo, and
J. Wang,
“Optical measurement of the viscoelastic and biochemical responses of living cells to mechanical perturbation,”
Opt. Lett., 23
(4), 307
–309
(1998). 0146-9592 Google Scholar
J. Guck,
R. Ananthakrishnan,
H. Mahmood,
T. J. Moon,
C. C. Cunningham, and
J. Kas,
“The optical stretcher: a novel laser tool to micromanipulate cells,”
Biophys. J., 81
(2), 767
–784
(2001). 0006-3495 Google Scholar
F. Wottawah,
S. Schinkinger,
B. Lincoln,
R. Ananthakrishnan,
M. Romeyke,
J. Guck, and
J. Kas,
“Optical rheology of biological cells,”
Phys. Rev. Lett., 94
(9), 098103
(2005). https://doi.org/10.1103/PhysRevLett.94.098103 0031-9007 Google Scholar
A. R. Bausch,
F. Ziemann,
A. A. Boulbitch,
K. Jacobson, and
E. Sackmann,
“Local measurements of viscoelastic parameters of adherent cell surfaces by magnetic bead microrheometry,”
Biophys. J., 75
(4), 2038
–2049
(1998). 0006-3495 Google Scholar
F. Ziemann,
J. Radler, and
E. Sackmann,
“Local measurements of viscoelastic moduli of entangled actin networks using an oscillating magnetic bead micro-rheometer,”
Biophys. J., 66
(6), 2210
–2216
(1994). 0006-3495 Google Scholar
F. J. Alenghat,
B. Fabry,
K. Y. Tsai,
W. H. Goldmann, and
D. E. Ingber,
“Analysis of cell mechanics in single vinculin-deficient cells using a magnetic tweezer,”
Biochem. Biophys. Res. Commun., 277
(1), 93
–99
(2000). https://doi.org/10.1006/bbrc.2000.3636 0006-291X Google Scholar
M. Keller,
J. Schilling, and
E. Sackmann,
“Oscillatory magnetic bead rheometer for complex fluid microrheometry,”
Rev. Sci. Instrum., 72
(9), 3626
–3634
(2001). https://doi.org/10.1063/1.1394185 0034-6748 Google Scholar
W. Feneberg,
M. Aepfelbacher, and
E. Sackmann,
“Microviscoelasticity of the apical cell surface of human umbilical vein endothelial cells (HUVEC) within confluent monolayers,”
Biophys. J., 87
(2), 1338
–1350
(2004). https://doi.org/10.1529/biophysj.103.037044 0006-3495 Google Scholar
B. D. Matthews,
D. R. Overby,
R. Mannix, and
D. E. Ingber,
“Cellular adaptation to mechanical stress: role of integrins, Rho, cytoskeletal tension, and mechanosensitive ion channels,”
J. Cell. Sci., 119
(3), 508
–518
(2006). https://doi.org/10.1242/jcs.02760 0021-9533 Google Scholar
M. Puig-de-Morales,
M. Grabulosa,
J. Alcaraz,
J. Mullol,
G. N. Maksym,
J. J. Fredberg, and
D. Navajas,
“Measurement of cell microrheology by magnetic twisting cytometry with frequency domain demodulation,”
J. Appl. Physiol., 91
(3), 1152
–1159
(2001). 8750-7587 Google Scholar
N. Wang and
D. E. Ingber,
“Probing transmembrane mechanical coupling and cytomechanics using magnetic twisting cytometry,”
Biochem. Cell Biol., 73
(7–8), 327
–335
(1995). 0829-8211 Google Scholar
G. Lenormand,
E. Millet,
B. Fabry,
J. P. Butler, and
J. J. Fredberg,
“Linearity and time-scale invariance of the creep function in living cells,”
J. R. Soc., Interface, 1
(1), 91
–97
(2004). https://doi.org/10.1098/rsif.2004.0010 1742-5689 Google Scholar
B. Fabry,
G. N. Maksym,
S. A. Shore,
P. E. Moore,
R. A. Panettieri,
J. P. Butler, and
J. J. Fredberg,
“Signal transduction in smooth muscle—selected contribution: time course and heterogeneity of contractile responses in cultured human airway smooth muscle cells,”
J. Appl. Physiol., 91
(2), 986
–994
(2001). 8750-7587 Google Scholar
D. E. Ingber,
“Tensegrity II. How structural networks influence cellular information processing networks,”
J. Cell. Sci., 116
(8), 1397
–1408
(2003). https://doi.org/10.1242/jcs.00360 0021-9533 Google Scholar
C. M. Lo and
J. Ferrier,
“Electrically measuring viscoelastic parameters of adherent cell layers under controlled magnetic forces,”
Eur. Biophys. J., 28
(2), 112
–118
(1999). https://doi.org/10.1007/s002490050190 0175-7571 Google Scholar
D. A. Head,
A. J. Levine, and
E. C. MacKintosh,
“Deformation of cross-linked semiflexible polymer networks,”
Phys. Rev. Lett., 91
(10), 108102
(2003). https://doi.org/10.1103/PhysRevLett.91.108102 0031-9007 Google Scholar
D. Boal, Mechanics of the Cell, Cambridge University Press, New York
(2002). Google Scholar
L. H. Sperling, Introduction to Physical Polymer Science, John Wiley and Sons, New York
(1992). Google Scholar
Y. C. Fung, Biomechanics: Mechanical Properties of Living Tissues, Springer-Verlag, New York
(1993). Google Scholar
D. Roylance, Engineering Viscoelasticity,
(2001) http://web.mit.edu/course/3/3.11/www/modules/visco.pdf Google Scholar
B. Fabry,
G. N. Maksym,
J. P. Butler,
M. Glogauer,
D. Navajas, and
J. J. Fredberg,
“Scaling the microrheology of living cells,”
Phys. Rev. Lett., 8714
(14), 148102
(2001). https://doi.org/10.1103/PhysRevLett.87.148102 0031-9007 Google Scholar
B. Suki,
A. L. Barabasi, and
K. R. Lutchen,
“Lung tissue viscoelasticity—a mathematical framework and its molecular basis,”
J. Appl. Physiol., 76
(6), 2749
–2759
(1994). 8750-7587 Google Scholar
D. Huang,
E. A. Swanson,
C. P. Lin,
J. S. Schuman,
W. G. Stinson,
W. Chang,
M. R. Hee,
T. Flotte,
K. Gregory,
C. A. Puliafito, and
J. G. Fujimoto,
“Optical coherence tomography,”
Science, 254
(5035), 1178
–1181
(1991). https://doi.org/10.1126/science.1957169 0036-8075 Google Scholar
A. F. Fercher,
C. K. Hitzenberger,
G. Kamp, and
S. Y. Elzaiat,
“Measurement of intraocular distances by backscattering spectral interferometry,”
Opt. Commun., 117
(1–2), 43
–48
(1995). https://doi.org/10.1016/0030-4018(95)00119-S 0030-4018 Google Scholar
R. Leitgeb,
C. K. Hitzenberger, and
A. F. Fercher,
“Performance of Fourier domain vs. time domain optical coherence tomography,”
Opt. Express, 11
(8), 889
–894
(2003). 1094-4087 Google Scholar
M. A. Choma,
M. Sarunic,
C. Yang, and
J. A. Izatt,
“Sensitivity advantage of swept source and Fourier domain optical coherence tomography,”
Opt. Express, 11
(18), 2183
–2189
(2003). 1094-4087 Google Scholar
J. F. de Boer,
B. Cense,
B. H. Park,
M. C. Pierce,
G. J. Tearney, and
B. E. Bouma,
“Improved signal-to-noise ratio in spectral-domain compared with time-domain optical coherence tomography,”
Opt. Lett., 28
(21), 2067
–2069
(2003). 0146-9592 Google Scholar
A. B. Vakhtin,
D. J. Kane,
W. R. Wood, and
K. A. Peterson,
“Common-path interferometer for frequency-domain optical coherence tomography,”
Appl. Opt., 42
(34), 6953
–6958
(2003). 0003-6935 Google Scholar
H. P. Kao and
A. S. Verkman,
“Tracking of single fluorescent particles in 3 dimensions—use of cylindrical optics to encode particle position,”
Biophys. J., 67
(3), 1291
–1300
(1994). 0006-3495 Google Scholar
V. Levi,
Q. Q. Ruan, and
E. Gratton,
“3-D particle tracking in a two-photon microscope: application to the study of molecular dynamics in cells,”
Biophys. J., 88
(4), 2919
–2928
(2005). https://doi.org/10.1529/biophysj.104.044230 0006-3495 Google Scholar
T. Ragan,
H. D. Huang,
P. So, and
E. Gratton,
“3D particle tracking on a two-photon microscope,”
J. Fluoresc., 16
(3), 325
–336
(2006). https://doi.org/10.1007/s10895-005-0040-1 1053-0509 Google Scholar
C. Joo,
T. Akkin,
B. Cense,
B. H. Park, and
J. E. de Boer,
“Spectral-domain optical coherence phase microscopy for quantitative phase-contrast imaging,”
Opt. Lett., 30
(16), 2131
–2133
(2005). https://doi.org/10.1364/OL.30.002131 0146-9592 Google Scholar
G. Wiegand,
K. R. Neumaier, and
E. Sackmann,
“Microinterferometry: three-dimensional reconstruction of surface microtopography for thin-film and wetting studies by reflection interference contrast microscopy (RICM),”
Appl. Opt., 37
(29), 6892
–6905
(1998). 0003-6935 Google Scholar
C. Picart,
K. Sengupta,
J. Schilling,
G. Maurstad,
G. Ladam,
A. R. Bausch, and
E. Sackmann,
“Microinterferometric study of the structure, interfacial potential, and viscoelastic properties of polyelectrolyte multilayer films on a planar substrate,”
J. Phys. Chem. B, 108
(22), 7196
–7205
(2004). https://doi.org/10.1021/jp037971w 1089-5647 Google Scholar
N. Fang,
V. Chan,
K. T. Wan,
H. Q. Mao, and
K. W. Leong,
“Colloidal adhesion of phospholipid vesicles: high-resolution reflection interference contrast microscopy and theory,”
Colloids Surf., B, 25
(4), 347
–362
(2002). https://doi.org/10.1016/S0927-7765(01)00336-8 0927-7765 Google Scholar
R. Parthasarathy and
J. T. Groves,
“Optical techniques for imaging membrane topography,”
Cell Biochem. Biophys., 41
(3), 391
–414
(2004). https://doi.org/10.1385/CBB:41:3:391 1085-9195 Google Scholar
A. E. Pelling,
S. Sehati,
E. B. Gralla,
J. S. Valentine, and
J. K. Gimzewski,
“Local nanomechanical motion of the cell wall of Saccharomyces cerevisiae,”
Science, 305
(5687), 1147
–1150
(2004). https://doi.org/10.1126/science.1097640 0036-8075 Google Scholar
F. Julicher and
J. Prost,
“Spontaneous oscillations of collective molecular motors,”
Phys. Rev. Lett., 78
(23), 4510
–4513
(1997). https://doi.org/10.1103/PhysRevLett.78.4510 0031-9007 Google Scholar
H. Akaike,
“New look at statistical-model identification,”
IEEE Trans. Autom. Control, AC19
(6), 716
–723
(1974). https://doi.org/10.1109/TAC.1974.1100705 0018-9286 Google Scholar
K. P. Burnham and
D. R. Anderson, Model Selection and Multimodel Inference: A Practical-Theoretical Approach, Springer, New York
(2002). Google Scholar
X. Hong,
P. M. Sharkey, and
K. Warwick,
“Automatic nonlinear predictive model-construction algorithm using forward regression and the PRESS statistic,”
IEE Proc.: Control Theory Appl., 150
(3), 245
–254
(2003). https://doi.org/10.1049/ip-cta:20030311 1350-2379 Google Scholar
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||

