|
|
1.IntroductionHemoglobins (Hb) can reversibly bind to dioxygen or other small ligands, and their main purpose is to transport oxygen to local tissues through the vascular system. The iron atom is the ligand binding site of the protein and is found in the heme prosthetic group. After binding of the iron atom to the proximal histidine residue, there is one remaining axial coordination site on the distal side available for ligand binding. The binding site is termed penta-coordinated if accessible for exogenous ligand binding, or hexa-coordinated if an amino acid side chain is connected to the site through an H-bridge. The main difference between the two forms is that biomolecular reactions are fast and straightforward in traditional penta-coordinate globins, whereas ligand binding to hexa-coordinated globins involves a competition between the exogenous ligand and intramolecular coordination by the local amino acid (histidine) side chain. Thus, hexa-coordination is considered an alternative mechanism for regulating ligand binding to the heme, and new interest in the function structure relationship of heme proteins has begun.1, 2 A milestone within the heme-research was set in the year 2000 when Burmester discovered a new mammalian globin predominantly expressed in the brain and therefore termed neuroglobin (NGB).3 Surprisingly, neuroglobin is the first mammal globin found to be in a hexa-coordinated low spin form both in the deoxy and oxy form, giving rise to many studies and suggestions on the still unknown function of the protein. 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21 The proposals are manifold: the globin might have a myoglobin-like function in the metabolism, and it may protect neuronal cells from hypoxic-ischemic insults.18 Also, it might function as an oxidative stress-responsive sensor protein, or participate in a signal transduction pathway that modulates activities of regulatory proteins in response to changes in , , and concentrations. There is also the alternative of neuroglobin sustaining scavenging and or reactive oxygen species (ROS) detoxification. It has also been found that oxygenation and carbonylation are linked to the redox state of the cell via intramolecular disulphide bridge formation/dissociation.10 However, the observations were made in vitro and no data are so far available about the physiological function of this disulfide bridge in vivo.20 In the course of our previous studies on the -binding of neuroglobin proteins,5, 22 we discovered that the oxidation of neuroglobin in cell lysates obtained from E. coli cells overexpressing neuroglobin is very slow in contrast to the fast autoxidation rate reported for these proteins.23 Several in vitro experiments on the cell lysates strongly indicate the presence of an enzymatic system capable of reducing the ferric form of neuroglobin, and similar effects were observed in eukaryotic cells (data published elsewhere24). For many of the biological relevant ligand-binding functions ascribed to globins, the reduction of the ferric globin is postulated to play an important role, although little or no information is available on the identity and in vivo working of the enzymatic systems that facilitate this reduction.25 Our findings on the reduction of neuroglobin in E. coli as well as eukaryotic cells prompted us to investigate the possibility of observing the redox reaction in living cells, preferably on a single cell level, and thus probe directly the influence of changing external conditions, such as pH or oxygen levels, on the oxidation state of the globin. To enable in vivo measurements on the single cell level, we combined Raman spectroscopy with a microfluidic system and an optical trap. Recently, we demonstrated how the combination of a micro-Raman spectrometer with a microfluidic system and optical tweezers can be used to study the oxygenation cycle of hemoglobin in optically trapped single red blood cells.26 Resonance Raman spectroscopy is an excellent tool to investigate heme proteins. The resonance effect is strong enough to investigate the heme group without interference by surrounding media or other components of the sample, and therefore enabling whole cell studies.27 Microfluidic systems are miniature structures of channels and reservoirs that give control over the diffusion of substances and chemical reactions in a confined space. They have become important tools in biomedicine and biochemistry, since they can mimic in vivo conditions in an in vitro environment.28, 29, 30 Such flow systems can easily be integrated into existing experimental setups due to their small sizes. Because of the flow, the cells under investigation need to be immobilized. Optical tweezers are ideal tools to avoid cell damage while guaranteeing stable immobilization of cells.31 The method is based on the fact that the photon momentum of light can be transferred onto small objects. A high gradient field close to a tightly focused Gaussian laser beam creates a force strong enough to trap micron-sized dielectric particles in three dimensions. Since the introduction of optical tweezers, they have become a significant tool in biochemistry. 31, 32, 33, 34 Given the fact that the previously described approach of combining a microresonance Raman setup with optical tweezers and a microfluidic system holds promise for investigations on diverse heme-protein-containing cells,26, 35 this method presents a possible strategy for tacking the earlier mentioned observation of the redox processes in cells (over)expressing NGB. Ideally, the method should be used to observe NGB in eukaryotic cells. However, the protein is found in low concentrations in brain tissue3 and can therefore not be directly used for the experiment, since the blood hemoglobin corrupts the experimental results even for perfused brains. Furthermore, the culture of nerve cell lines is not straightforward and gives low yield. In this work, we therefore use E. coli bacteria overexpressing wild-type (wt) human NGB and its E7Leu mutant (His at position E7 substituted by Leu) to test the applicability and modifications of the microresonance Raman method. The choice of the system is also interesting in view of the earlier observation of the presence of an enzymatic system. The main adjustment to the technique is that the system now is equipped with a gas-tight flow cell coupled to a continuous pump. This gives the opportunity to vary the oxygen concentration in the buffer flushed through the gas-tight cell, which triggers oxygen release or binding of the heme. As mentioned before, this method has previously proven to be feasible for studies of the oxygenation cycle of single red blood cells containing 33% of Hb and having a size of . The question we try to answer in this work is if it will hold to study single E. coli bacteria being about 100 times smaller with approximately 10 times less protein concentration. Therefore, part of the current study is dedicated to investigating the sensitivity of the setup, effects of laser power and optical trapping, and the possibility to visualize responses of the oxygenation state of the globin to environmental changes. The advantages and disadvantages of the technique are discussed. Finally, the current observations are linked to our earlier findings. 2.Materials and Methods2.1.SetupFor this study, a home-built microscope equipped with a microfluidic system and optical tweezers were combined with a Raman spectrometer, as shown in Fig. 1 . The flow cell used consisted of a flat Plexiglas ring ( , ) that was tightly sealed with vacuum grease between two cover glasses. The cell was flushed by a continuous pump connected to a switch leading to two buffer-containing vessels purged with air or , respectively. This system enabled us to vary the aerobic conditions in the flow cell while simultaneously monitoring spectral changes. A Kr-ion laser tuned to was used as an excitation source for the Raman measurements. The excitation and backscattering collection system consisted of an inverted microscope equipped with a water-immersion objective . The laser light was guided into the microscope by a dichroic mirror (DM1), reflecting while transmitting the rest of the visible light. The Raman backscattered light was guided into a XY Dilor Raman spectrometer equipped (gratin ) by a second dichroic mirror (DM2), and thereafter passing a objective to match the ratio of the spectrometer entrance . The spectra were recorded by a liquid-nitrogen-cooled charge-coupled device (CCD) camera. Since the image of the laser focus in the sample is enlarged by the added optics, the variable slit in front of the resolving stage had to be open to pass the entire image, giving a calculated spectral resolution of . However, comparing spectra taken in the microconfiguration with the macroconfiguration (spectral resolution ), no loss of spectral information could be observed. Hence, the calculated spectral resolution of might be overestimated. Fig. 1Schematic of the setup. A Kr-ion laser tuned to is used for Raman excitation. The excitation and backscattering collection system consists of an inverted microscope equipped with a water-immersion objective . DM1 leads the laser line into the microscope. The Raman backscattered light is guided into a Dilor XY800 Raman spectrometer by a second dichroic mirror (DM2), passing a objective to match the ratio of the spectrometer entrance. The trapping laser is guided by DM3 through the microscope objective. A 1:5 beam expander is mounted in the trapping laser beam to assure stable trapping and to bring together the foci from the different lasers. The sample is monitored by a web camera. The microfluidic system, a Plexiglas ring ( , ) with an in- and outlet sealed between two cover glasses, is flushed with buffer by a continuous pump, equipped with a switch and connected to buffer containers purged with air or . 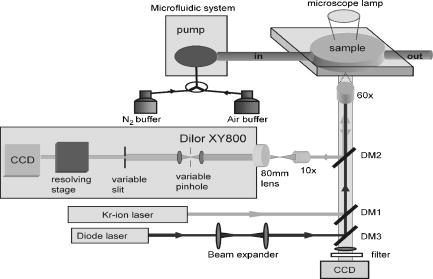 The trapping laser (a diode laser emitting at ) was guided by a third dichroic mirror (DM3) through the microscope objective used for the Raman setup and for the monitoring of the sample. Stable trapping was accomplished by overfilling the entrance aperture of the microscope objective. Therefore, a 1:5 beam expander was mounted in the light path of the trapping laser. The beam expander was also adjusted to get the focal point of the 413.1-nm beam and the 830-nm trapping laser to coincide. Safe visual observation/monitoring of the transmission microscopic image of the sample under white light illumination was accomplished using a CCD chip from a web camera coupled to a PC. 2.2.SampleThe growing of E. coli cells containing the expression plasmid encoding wt NGB or its E7Leu mutant has been described previously.7 Briefly, NGB cDNA cloned in pET3a were mutated using the site-directed mutagenesis method (Stratagene), whereby the E7His was mutated to a Leucine (E7LeuNGB). The wt and mutant NGB expression plasmids (cDNA of NGB in pET3a) were transformed into E. coli, strain BL21(DE3)pLysS. The cells were grown at in TB medium (1.2% bactotryptone, 2.4 % yeast extract, 0.4 % glycerol, 72-mM potassium phosphate buffer pH 7.5) containing ampicilin, chloramphenicol, and 1-mM d-amino-levulinic acid. The culture was induced at by the addition of isopropyl-1thio-D-galactopyranoside of the final concentration of , and expression was continued overnight.5 The fresh cells were washed and put in a suspension of the washing buffer (5-mM glucose, 25-mM Tris pH 8, and 10-mM EDTA). A drop of the cell suspension was put in the flow cell directly before the measurements started. The cell suspension was either taken directly from the batch (E7 LeuNGB) or diluted between to times (NGB), depending on the experiment. 3.ResultsThe first question addressed in this work was if it is possible to monitor single bacteria. In a previous study we showed that the microresonance Raman setup works very well to study the oxygenation cycle of hemoglobin in erythrocytes. However, red blood cells are large [typically 80 to 96 ] and contain on average hemoglobin.36 Considering that human hemoglobin contains four heme groups, the total heme concentration in erythrocytes is around . E. coli cells are much smaller [typically 1 ]37 and the E. coli cells overexpressing NGB used in these experiments have, on average, globin (and hence heme) concentrations of around 2 to . In a first step, we therefore compare the quality of resonance Raman spectra from different numbers of trapped E. coli cells overexpressing wt NGB. The concentration of cells was varied by altering the dilution from and to . A drop of of the diluted bacteria suspension was put directly on a cover glass. The number of cells in the optical trap is determined from the optical microscopy image prior to switching to the Raman mode. Similarly, the cells were checked for lysis before and after illumination. Since the E. coli bacteria are known to lyse very fast after cell death, this can be used as a measure to determine whether the cells are still alive. The results are depicted in Fig. 2 , showing that the resolution of the setup is adequate to record resonance Raman spectra from a single, optically trapped bacterium. The signal increases with the number of trapped cells and is considerably lower when the optical trap is off. Remarkably, if only one cell is trapped, the signal decreases while the noise increases. This is due to the fact that the area of a single trapped bacterium is smaller than the focused laser spot. Hence, also light reflected from the glass/water interface can be recollected. This is not the case when the whole measurement volume is filled with a dense population of cells. The band at , the iron oxidation state marker, is in all spectra the most dominant feature. When the optical trap and the flow are off, it is the only band detected, even with a high density of cells in the suspension (data not shown). The same is true if only a single or a few cells are trapped while a flow is applied through the flow cell. Using the optical trap to assemble a high density of cells within the Raman focus, the signal-to-noise ratio (SNR) of the spectra can be increased substantially (Fig. 2). Even though to 20 cells should be sufficient to fill the laser spot in the focal plane, the signal continues to increase for a larger number of cells, mostly because of the contributions from other (out of focus) planes, as the focus of the laser beam is elongated along the direction. During the initial test measurements, it was also found that only fresh cell suspensions gave a good SNR in the Raman spectra. This was clearly linked to the increasing cell lysis upon aging of the suspension. Furthermore, cell lysis is possibly enhanced under laser illumination and the level of cell disruption may in addition depend on the type of protein that is overexpressed in the cells, as is discussed later. Fig. 2Stacked spectra retrieved from different numbers of trapped E. coli bacteria overexpressing wt NGB. The setup can resolve spectra at the single cell level where the SNR increases with the number of cells trapped. The integration time was and the power, measured prior to the microscope objective, was . The number of cells in the optical trap is determined from the optical microscopy image prior to switching to the Raman mode. 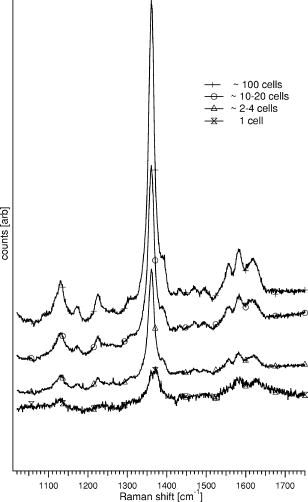 Secondly, we conducted an experiment where resonance Raman spectra of as-harvested E. coli cells were compared with spectroscopic data obtained earlier on the system and on the purified proteins. Previous studies, many of them involving some of the present coauthors, have led to a clear spectroscopic identification of neuroglobin and some of its mutants. 4, 5, 7, 16, 20, 22, 38 Furthermore, both wt and E7Leu NGB are fully folded in the cytoplasm of the E. coli cells and are as such extracted from the cell lysate upon purification.7 We can therefore validate in detail the information obtained in our setup to probe possible corruption of the data by laser-induced processes. Figure 3 (upper trace) shows the resonance Raman spectrum from E. coli cells overexpressing wt NGB. In this case, the washed cell suspension was diluted 50 times with buffer. In an earlier work, we showed that during protein expression in E. coli, the metabolism of the cells gradually shifts from aerobic to anaerobic respiration, since all available is consumed by the increasing population of cells.5 Nitrate reductase is then induced, causing nitrate to be reduced to nitrite and further to nitric oxide . The E. coli cells under study are grown under these conditions. Using a combination of absorption spectroscopy and electron paramagnetic resonance (EPR), we proved earlier that wt NGB overexpressed in E. coli cell cultures favors the conformation, whereby only a small fraction of the protein binds .5, 22 The present resonance Raman spectra taken from a group of optically trapped E. coli cells overexpressing wt NGB (upper trace, Fig. 3) confirm that the globin is in a reduced state, where no oxygen is bound to the heme, since the oxidation state marker is found at . The band is found at , confirming that the heme is in a hexa-coordinated low spin form.4, 38 Furthermore, the measured spectrum agrees well with earlier resonance Raman studies on the purified NGB, revealing that the globin is in its bis-histidine coordinated form in the deoxy ferrous state.4 The acquisition time in the experiment was set to per spectrum. In our previous absorption and EPR study, we showed that the nitrosyl ferrous form of E7Leu NGB is readily observed in E. coli cells overexpressing this mutant, since the competition with the distal histidine has now disappeared.5, 22 Figure 3 (bottom trace) shows the resonance Raman spectrum of the trapped E. coli cells overexpressing E7Leu NGB. Fewer and broader bands are observed than in the previous case and a high fluorescence background is present. Also, the cell suspension is considerably less diluted and more cells had to be trapped to get a sufficiently resolved Raman spectrum. This parallels the somewhat lower protein expression in this case . Furthermore, the band at and the band at indicate that the heme is in the five-coordinated ferrous high-spin form as could be easily confirmed from a comparison with the resonance Raman spectrum of E7Leu NGB reduced with dithionite (not shown). This observation contrasts the EPR and absorption findings for these cell suspensions. This indicates that, besides the decreased stability of the E. coli cells against lysis (apparent from fluorescence background and visual cell lysis on microscope images), the local laser power is causing the bond to break (photochemistry). In contrast, the Fe-His bond is known to be far more stable against photodissociation. This experiment thus shows that photoinduced cell degradation and photodissociation can put a limit on the applicability of the technique and has to be monitored closely to avoid corruption in the interpretation of results. Fig. 3Upper stacked spectrum shows typical ferrous Raman bands from 200 to of wt NGB overexpressed in E. coli cells. The stacked spectrum below shows the same experiment for the E7Leu NGB. The integration time was set to at a power of prior to the microscope objective. 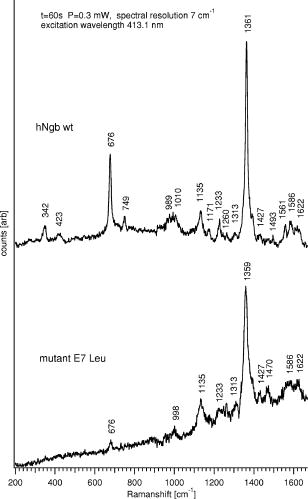 In a third experiment, the influence of the cell environment on the globin was investigated. In our previous study, it was shown how the oxygenation process of a single erythrocyte under changing oxygen concentrations could be followed in time.26 In the next step, we wanted to investigate if the environmentally induced changes of the oxidation/oxygenation state of the NGB variants could be monitored in E. coli cells. Therefore, the E. coli bacteria overexpressing wt NGB were exposed to a flow of air-purged buffer. Time series of spectra were taken to see how the bacteria react to environmental changes (Fig. 4 ). The power of the Raman excitation line at into the microscope objective was measured to be , whereas the power of the trapping laser was . The optical trap assembled the cells at the focal point of the collection spot and kept the cells in place while the flow was applied. The integration time was set to , and the spectra were recorded subsequently with no time delay in between, to register spectral changes over time. It was possible to work with the same sample for without any photoinduced degradation of the sample or cell lysis. Approximately 20 cells were in the optical trap. The flow of the air-purged buffer ( , flow-cell dimensions: , ) was put on and off twice, while the subsequent shift of the band in the resonance Raman spectra was recorded. When the cells are flushed with air-purged buffer, the band shifts from 1361 to . identifies uniquely the reduced deoxy ferrous form of the globin, whereby agrees with both the oxy and the met form.4 The transfer between the forms is represented in Fig. 4 by the change in the ratio between the intensity values at 1361 and . Fig. 4Time series of the resonance Raman spectra of wt NGB overexpressed in E. coli cells. Raman laser power is and the trapping laser power is measured prior to the MO. Integration time is , and spectra are taken subsequently with no time delay. The graph shows the ratio between the band at 1361 and , respectively, giving a measure of the oxygen binding to the wt NGB. The inset shows the position of the band for the oxy and the deoxy state from which the ratio was taken. 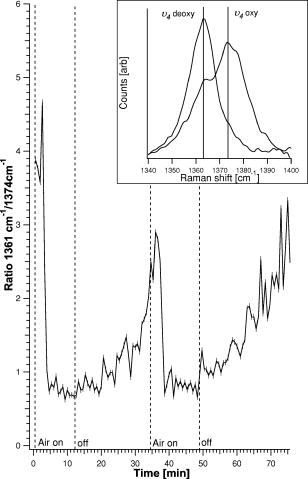 The time span measured for the trapped cells to go from a clear deoxy state to a mixture between the deoxy and oxy form was around . When the flow was turned off, it took approximately for the cells to return to the reduced state again. From the graph, it can be seen that the cells are striving to be in the reduced state, and even when flushed with oxygen a pure oxy state is never observed in the spectra (data not shown). It is intriguing that wt NGB returns to a completely reduced state, although there should remain some oxygen in the flow cell, and the autoxidation rates are known to be very high from in vitro studies on purified NGB.7 Note that recorded time series of spectra of the E. coli did not reveal any photochemistry at the applied power. The experiment was repeated for the E. coli cells overexpressing the E7Leu NGB mutant, giving a quite different result, as can be seen in Fig. 5 . The results are worth presenting, since they show that great care has to be taken to avoid misinterpretation of the results retrieved with the applied technique. Fig. 5Time series of stacked resonance Raman spectrum of NGBE7Leu overexpressed in E. coli cells. The first spectrum taken is shown at the bottom of the graph. Raman laser power is and the trapping laser power is measured prior to the microscope objective. The integration time is and spectra are taken in sequence with no time delay in-between. The flow starts after the second spectrum. An initial oxygenation is followed by a photoinduced degradation of the sample. 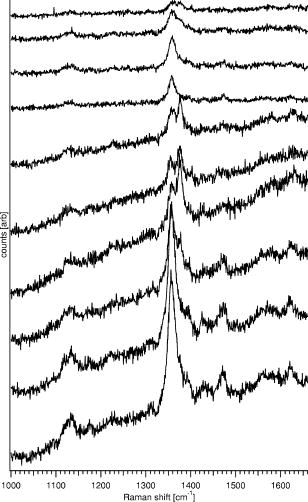 The recorded spectra are taken from about 100 trapped E. coli cells in a very dense cell suspension, i.e., the cell suspension was not diluted from the beginning. The high density of cells in the trap was achieved by carefully collecting bacteria while scanning the sample. In this case, a flow of air-purged buffer was started after the recording of the second spectrum. The laser power was . An immediate increase of the band at can be observed in Fig. 5. After three more spectra, this band disappears again for two spectra and then increases again. The disappearance of the is accompanied by an increase in the signal-to-noise ratio. As mentioned before, the concentration of bacteria in the flow cell is very high to get a good signal; hence due to the flow, neighboring cells can be dragged into the focus of the trapping beam and give rise to the unexpected flipside shift of the band. These fluctuations could also be observed on the microscope image. However, the decrease of the signal and the shift of the peak to imply that the sample is degrading quickly, despite of the access of fresh cells. This leads to the conclusion that when oxygen is abundant in the flow cell, the photoinduced degradation of the sample may even be accelerated. The observation that the Raman signal decreases significantly while the fluorescent background disappears over time is unexpected. As mentioned earlier, the E. coli bacteria overexpressing E7Leu NGB are extremely receptive to lysis, and only very fresh samples can be used in the experiments. Hence, the most likely explanation for the loss in signal, both Raman and the fluorescent background, is the ongoing lysis of the cells, which might be accelerated by the Raman-laser illumination, since the E. coli absorb significantly at . Due to lysis, the content of the bacteria will diffuse into the flow cell instead of being assembled in the trapping focus. Further, it is obvious that the E. coli overexpressing the E7Leu NGB are more delicate and susceptible to photoinduced chemistry than E. coli overexpressing wt NGB. 4.DiscussionIn a previous study, we presented the possibility of monitoring the oxygenation cycle of hemoglobin within single functional erythrocytes.26 A double microscope configuration, equipped with two separate microscope objectives, was used to separate the optical trap from the Raman spectrometer. The reason for this was that the spectrometer was initially equipped with an upright microscope, and hence it was necessary to fine tune the optical conditions of each process individually to enable the integration of the microfluidic system. The method showed potential, but the oxygenation cycle was triggered by the addition of sodium dithionite, which resulted in a protein denaturation after one and a half cycles.26, 27 In the present study, the spectrometer did not have a microscope integrated to start with, and hence an inverted microscope could be added and the same objective could be used for optical trapping and the recording of Raman spectra. Furthermore, the flow through the gas-tight microfluidic system was generated by a continuous pump. This gives the possibility to switch between aerobic or anaerobic conditions, resembling physiological circumstances more than adding sodium dithionite into an open channel system. The time series presented in Fig. 4 shows that the state of the art equipment provides conditions that keep the cells functional over long periods of time, and hence it allows for the reversible monitoring of the oxygenation cycle of heme proteins. The microfluidic system provides an ideal system to vary simultaneously or separately different aspects such as gas and salt concentrations and pH. In general, fluidic setups are required to be able to study continuously the influence of these varying circumstances, independently, whether one aims at a macroscopic or microscopic detection of the changes. Such a fluidic system implies fixation of the cells. The fixation of the cells using optical tweezers avoids cell damage that is known to occur when cells are immobilized to poly-lysine covered glass slides.27 It allows, in combination with an optical microscope, for the selection and fixation of well-defined numbers of cells for the experiment, which is exactly the ultimate aim. The choice for a microscopic detection method is in this case evident. In comparison with absorption spectra, the resonance Raman spectra of globins are in principle richer in information. Moreover, the focusing of the laser light in resonance Raman is more optimal than the focusing obtained in a microabsorption setup, where the large range of wavelengths needed for the experiment limits the focus characteristics. Therefore, the combined resonance Raman setup has in principle clear advantages over other techniques when observation of globin molecules in cells is targeted. Here, it is demonstrated that the system is indeed suitable to study E. coli bacteria overexpressing NGB in vivo on the singe-cell level, but the signal-to-noise ratio is low (Fig. 2). When the flow is started, the optical trap has to be applied not only to keep the cells in place but also to gather a high density of cells in the focus of the Raman beam to get good quality spectra. It has been demonstrated previously that optical trapping at does not harm E. coli bacteria, and therefore it is an ideal immobilization tool, especially in combination with a microfluidic system.33 The performance of the setup is limited by different factors. If the cell concentration in the suspension is too high, new cells can be dragged into the optical trap easily, corrupting the data in the time series (Fig. 5). Higher concentrations may, however, be necessary for cells with low protein expression needing a larger amount of cells in the trap to obtain a good quality spectrum. Furthermore, laser-induced cell lysis can put a limit to the applicability of the technique, and the trapped cells should be monitored by optical microscopy before and after the experiment. The approach can give insight on a cellular level, as is evident from the experiments on E. coli cells overexpressing wt NGB (Fig. 4). As stated in the introduction, the current study was triggered by the discovery that an enzymatic system is present in E. coli and in eukaryotic cells that is capable of reducing NGB.23 If we can develop a method that allows detection of the heme state of NGB in cells, we open the way of studying the behavior of the full NGB and enzyme system under varying conditions in a cell. To test the feasibility of using a microresonance Raman setup for these purposes, we focus in this study on the analysis of E. coli cells overexpressing wt NGB. In our in vitro studies, we found that the lysates of E. coli cells can reduce ferric wt NGB.24 After the breaking of E. coli cells overexpressing wt NGB, an immediate formation of oxygenated wt NGB was observed due to the sudden presence of oxygen, but wt NGB was soon after found to return to the reduced state, despite the continuous exposure to air and despite the high autoxidation rate of NGB.7 Different experiments involving fractionation of the lysate, addition of tripsine or cofactors, showed unambiguously that the reducing agent is an enzyme. The microresonance Raman experiments confirm that wt NGB is in a ferrous bis-histidine coordinated state in the E. coli cells grown under anaerobic circumstances, as was observed earlier5, 22 (Fig. 3). Flushing with an oxygen-rich buffer induces the formation of oxygenated (or even oxidized) NGB, as could be determined from the shift of the band (Fig. 4). However, based on the high autoxidation rate of NGB,7 one would expect at least partial oxidation of the NGB molecules in the cell. In contrast, when the oxygen flush is stopped, NGB is again reduced to the initial ferrous form. A full shift from the oxy form back to the deoxy form within after the stop of flow might be partially explained by the fact that the cells are consuming dioxygen at a high rate, but it cannot explain the lack of autoxidation of the protein under oxygen-rich conditions. The latter parallels the in vitro observations and shows that we have in principle a tool of measuring the coordination and oxidation state of globins.24 However, to differentiate between the oxygenated and oxidized form of wt NGB, higher resolution resonance Raman spectra need to be taken, since the most prominent and bands do not allow for a differentiation between these forms. This implies an increase in measuring time and thus increases the time between spectra in a time series. Furthermore, the results can be corrupted by the technique itself, as became apparent from comparing the results on E. coli cells overexpressing wt NGB with those of the bacteria overexpressing E7 LeuNGB. Despite the fact that both proteins are not naturally abundant in the E. coli cell and may thus influence the host cell, and that in both cases the E. coli bacteria are found to be receptive to lysis, the lysis effect is considerably more pronounced for the E. coli cells overexpressing the E7Leu mutant than the wt globin. In the former case, even fresh samples show a high fluorescent background in the resonance Raman spectra, and many expected Raman bands cannot be resolved. The observation indicates that the cells are decomposing and the released DNA as well as the photodegradation of the heme give rise to the increased fluorescence. Furthermore, it is not possible to record spectral time series of these cells because of protein and cell degradation (Fig. 5). Our earlier absorption studies revealed that in E. coli cells overexpressing the NGB E7Leu mutant, the globin is found to be predominantly in a -ligated ferrous form.5, 38 The local laser power needed for the present in vivo resonance Raman experiment is shown to dissociate the bond (Fig. 3). This will cause a sudden release of in the cell, which may well contribute to the higher receptivity to protein and cell degradation in comparison to the cells overexpressing the wt NGB, which is predominantly in the ferrous form. Furthermore, the -ligated ferrous forms of globins are known to absorb stronger at than the bis-histidine coordinated ferrous form of heme proteins.22 This may induce a higher local heating of the cell, which again can contribute to the increased susceptibility to lysis of the cell. One should also note that the quantum efficiency for -ligated globins is in general lower than for or -ligated globins.39 The current fast photodissociation is thus undoubtedly related to overall protein degradation due to lysis, and the formation of the band at is thus most probably related to the formation of ferric heme. Apart from the susceptibility of the E7Leu mutant toward photoinduced chemistry, the high density of cells necessary for measurements causes problems. Fresh, close cells can be dragged into the optical trap, which can lead to a misinterpretation of the oxygenation state of the cells, especially when the flow is applied. 5.ConclusionsIn conclusion, we show that a microresonance Raman spectrometer combined with optical tweezers and a microfluidic system allows for the reversible monitoring of oxygenation and redox cycles of heme proteins on a microscopic level in bacterial cells. This allows us to study not only the NGB system presented here under varying exposure of the E. coli cells to buffer purged with relevant ligands, but it also opens the way to study bacterial, Archaea, or eukaryotic globins in their native cells in the future. However, the present study also shows that photoinduced damage and low protein concentrations may put a serious limit to the method. These factors are system-bound and it is at all times necessary to monitor the validity of the data by checking the cells via optical microscopy and comparing the start and end point measurements with (macroscopic) absorption experiments. AcknowledgmentsThis work was supported by the European Commission Sixth Framework Programme through the project ATOM-3D (contract number 508952), from the European Science Foundation EUROCORES Programme, SPANAS, through funds from the Swedish Research Council, and from the European Commission Sixth Framework Programme. Further acknowledgement goes to the Fund for Scientific Research-Flanders (FWO) for support through grant G.0468.03 and the postdoctoral fellowships granted to S.D. and W.W.. For technical assistance, we would like to thank Heinrich Riedl and Paul Casteels. ReferencesT. Egawa and
S. R. Yeh,
“Structural and functional properties of hemoglobins from unicellular organisms as revealed by resonance Raman spectroscopy,”
J. Inorg. Biochem., 99 72
–96
(2005). 0162-0134 Google Scholar
R. E. Weber and
N. Vinogradov,
“Nonvertebrate hemoglobins: functions and molecular adaptions,”
Physiol. Rev., 81 569
–628
(2001). 0031-9333 Google Scholar
T. Burmester,
B. Welch,
S. Reinhardt, and
T. Hankeln, Nature (London), 407 520
–523
(2000). 0028-0836 Google Scholar
M. Couture,
T. Burmester,
T. Hankeln, and
D. L. Rousseau,
“The heme environment of mouse neuroglobin: evidence for the presence of two conformations of the heme pocket,”
J. Biol. Chem., 276 36377
–36382
(2001). 0021-9258 Google Scholar
S. Van Doorslaer,
S. Dewilde,
L. Kiger,
S. V. Nistor,
E. Goovaerts,
M. C. Marden, and
L. Moens,
“Nitric oxide binding properties of neuroglobin,”
J. Biol. Chem., 278 4919
–4925
(2003). 0021-9258 Google Scholar
J. T. Trent,
A. N. Hvitved, and
M. S. Hargrove,
“A model for ligand binding to hexacoordinate hemoglobins,”
Biochemistry, 40 6155
–6163
(2001). 0006-2960 Google Scholar
S. Dewilde,
L. Kiger,
T. Burmester,
T. Hankeln,
V. Baudin-Creuza,
T. Aerts,
M. Marden,
R. Caubergs, and
L. Moens,
“Biochemical characterization and ligand binding properties of neuroglobin, a novel member of the globin family,”
J. Biol. Chem., 276 38949
–38955
(2001). 0021-9258 Google Scholar
J. M. Kriegl,
A. J. Bhattacharyya,
K. Nienhaus,
P. Deng,
O. Minkow, and
G. U. Nienhaus,
“Ligand binding and protein dynamics in neuroglobin,”
Proc. Natl. Acad. Sci. U.S.A., 99 7992
–7997
(2002). 0027-8424 Google Scholar
D. Hamdane,
L. Kiger,
S. Dewilde,
B. N. Green,
A. Pesce,
J. Uzan,
T. Burmester,
T. Hankeln,
M. Bolognesi,
L. Moens, and
M. Marden,
“The redox state of the cell regulates the ligand binding affinity of human neuroglobin and cytoglobin,”
J. Biol. Chem., 278 51713
–51721
(2003). 0021-9258 Google Scholar
E. Vinck,
S. Van Doorslaer,
S. Dewilde, and
L. Moens,
“Structural change of the heme pocket due to disulfide bridge formation is significantly larger for neuroglobin than for cytoglobin,”
J. Am. Chem. Soc., 126 4516
–4517
(2004). 0002-7863 Google Scholar
J. Uzan,
S. Dewilde,
T. Burmester,
T. Hankeln,
L. Moens,
D. Hamdane,
M. Marden, and
L. Kiger,
“Neuroglobin and other hexacoordinated hemoglobins show a weak temperature dependence of oxygen binding,”
Biophys. J., 87 1
–7
(2004). https://doi.org/10.1529/biophysj.103.030601 0006-3495 Google Scholar
K. Nienhaus,
J. M. Kriegl, and
G. U. Nienhaus,
“Structural dynamics in the active site of murine neuroglobin and its effects on ligand binding,”
J. Biol. Chem., 279 22944
–22952
(2004). 0021-9258 Google Scholar
S. V. Nistor,
E. Goovaerts,
S. Van Doorslaer,
S. Dewilde, and
L. Moens,
“EPR-spectroscopic evidence for a dominant His-FEIII-His coordination in ferric neuroglobin,”
Chem. Phys. Lett., 361 355
–361
(2002). 0009-2614 Google Scholar
A. Fago,
C. Hundahl,
S. Dewilde,
K. Gilany,
L. Moens, and
R. E. Weber,
“Allosteric regulation and temperature dependence of oxygen binding in human neuroglobin and cytoglobin,”
J. Biol. Chem.,
(2004). 0021-9258 Google Scholar
J. T. Trent,
R. A. Watts, and
M. S. Hargrove,
“Human neuroglobin, a hexacoordinate hemoglobin that reversibly binds oxygen,”
J. Biol. Chem., 276 30106
–30110
(2001). 0021-9258 Google Scholar
H. Sawai,
M. Makino,
Y. Mizutani,
T. Ohta,
H. Sugimoto,
T. Uno,
N. Kawada,
N. Yoshizawa,
T. Kitagawa, and
Y. Shiro,
“Structural characterization of the proximal and distal histidine environment of cytoglobin and neuroglobin,”
Biochemistry, 44 13257
–13265
(2005). 0006-2960 Google Scholar
Y. Sun,
K. Jin,
X. O. Mao,
Y. Zhu, and
D. A. Greenberg,
“Neuroglobin is up-regulated by and protects neurons from hypoxic-ischemic injury,”
Proc. Natl. Acad. Sci. U.S.A., 98 15306
–15311
(2001). 0027-8424 Google Scholar
S. Herold,
A. Fago,
R. E. Weber,
S. Dewilde, and
L. Moens,
“Reactivity studies of the Fe(III) and Fe(II)NO forms of human neuroglobin reveal a potential role against oxidative stress,”
J. Biol. Chem., 279 22841
–22847
(2004). 0021-9258 Google Scholar
E. Geuens,
S. Dewilde,
D. Hoogewijs,
A. Pesce,
K. Nienhaus,
G. U. Nienhaus,
J. Olson,
J. Vanfleteren,
M. Bolognesi, and
L. Moens,
“Nerve globins in invertebrates,”
IUBMB Life, 56 653
–656
(2004). 1521-6543 Google Scholar
S. Van Doorslaer,
V. E. F. Trandafir,
I. Ioanitescu,
S. Dewilde, and
L. Moens,
“Tracing the structure-function relationship of neuroglobin and cytoglobin using resonance Raman and electron paramagnetic resonance spectroscopy,”
IUBMB Life, 56 665
–670
(2004). 1521-6543 Google Scholar
A. Pesce,
M. Bolognesi,
A. Bocedi,
P. Ascenzi,
S. Dewilde,
L. Moens,
T. Hankeln, and
T. Burmester,
“Neuroglobin and cytoglobin: Fresh blood for the vertebrate globin family,”
EMBO Rep., 3 1146
–1151
(2002). 1469-221X Google Scholar
F. Trandafir,
S. Van Doorslaer,
S. Dewilde, and
L. Moens,
“Temperature dependence of NO binding modes in human neuroglobin,”
Biochim. Biophys. Acta, 1702 153
–161
(2004). 0006-3002 Google Scholar
F. Trandafir,
S. Dewilde,
L. Moens, and
S. Van Doorslaer,
“NO-binding properties and (non-)enzymatic reduction of human neuroglobin,”
FEBS J., 272
(Suppl. 1),
(2005). 1742-464X Google Scholar
F. Trandafir,
D. Hoogewijs,
F. Altieri,
P. Rivetti di Val Cervo,
K. Ramser,
W. Wenseelers,
S. Dewilde,
S. Van Doorslaer,
J. Vanfleteren, and
L. Moens,
“Neuro- and cytoglobin as potential enzymes or substrates,”
Gene, 0378-1119 Google Scholar
M. Brunori,
“Nitric oxide moves myoglobin centre stage,”
Trends Biochem. Sci., 26 209
–210
(2001). 0167-7640 Google Scholar
K. Ramser,
J. Enger,
M. Goksör,
D. Hanstorp,
K. Logg, and
M. Käll,
“A microfluidic system enabling Raman measurements of the oxygenation cycle in single optically trapped red blood cells,”
Lab Chip, 5 431
–436
(2004). https://doi.org/10.1039/b416749j 1473-0197 Google Scholar
K. Ramser,
E. J. Bjerneld,
C. Fant, and
M. Käll,
“Importance of substrate and photoinduced effects in Raman spectroscopy of single functional erythrocytes,”
J. Biomed. Opt., 8 173
–178
(2003). https://doi.org/10.1117/1.1559730 1083-3668 Google Scholar
G. M. Walker,
H. C. Zeringue, and
D. J. Beebe,
“Microenvironment design considerations for cellular scale studies,”
Lab Chip, 4 91
–97
(2004). 1473-0197 Google Scholar
D. J. Beebe,
G. A. Mensing, and
G. M. Walker,
“Physics and applications of microfluidics in biology,”
Annu. Rev. Biomed. Eng., 4 261
–286
(2002). https://doi.org/10.1146/annurev.bioeng.4.112601.125916 1523-9829 Google Scholar
T. Fujii,
“PDMS-based microfluidic devices for biomedical applications,”
Microelectron. Eng., 61-62 907
–914
(2002). 0167-9317 Google Scholar
M. J. Lang and
S. Block,
“Resource Letter: LBOT-1: Laser-based optical tweezers,”
Am. J. Phys., 71 201
–212
(2003). https://doi.org/10.1119/1.1532323 0002-9505 Google Scholar
A. Ashkin,
“History of optical trapping and manipulation of small-neutral particle, atoms, and molecules,”
IEEE J. Sel. Top. Quantum Electron., 6 841
–856
(2000). https://doi.org/10.1109/2944.902132 1077-260X Google Scholar
K. C. Neuman,
E. H. Chadd,
G. F. Liou,
K. Bergman, and
S. Block,
“Characterization of photodamage to Escherichia coli in optical traps,”
Biophys. J., 77 2856
–2863
(1999). 0006-3495 Google Scholar
C. Kuyper and
D. T. Chiu,
“Optical trapping: a versatile technique for biomanipulation,”
Appl. Spectrosc., 56 300A
–312A
(2002). https://doi.org/10.1366/00037020260377652 0003-7028 Google Scholar
K. Ramser,
K. Logg,
M. Goksör,
J. Enger,
M. Käll, and
D. Hanstorp,
“Resonance Raman spectroscopy of optically trapped functional erythrocytes,”
J. Biomed. Opt., 9
(3), 593
–600
(2004). https://doi.org/10.1117/1.1689336 1083-3668 Google Scholar
G. B. Nash and
H. J. Meiselman,
“Red cell and ghost viscoelasticity. Effect of hemoglobin concentration and in vivo aging,”
Biophys. J., 43 63
–73
(1983). 0006-3495 Google Scholar
T. E. Shehata and
A. G. Marr,
“Effect of temperature on the size of Escherichia coli cells,”
J. Bacteriol., 124 857
–862
(1975). 0021-9193 Google Scholar
T. Uno,
D. Ryu,
H. Tsutsumi,
Y. Tomisugi,
Y. Ishikawa,
A. J. Wilkinson,
H. Sato, and
T. Hayashi,
“Residues in the distal heme pocket of neuroglobin: implications for the multiple binding steps,”
J. Biol. Chem., 279 5886
–5893
(2004). 0021-9258 Google Scholar
W. A. Saffran and
Q. H. Gibson,
“Photodissociation of ligands from heme and heme proteins. Effect of temperature and organic phosphate,”
J. Biol. Chem., 252 7955
–7958
(1977). 0021-9258 Google Scholar
|

