|
|
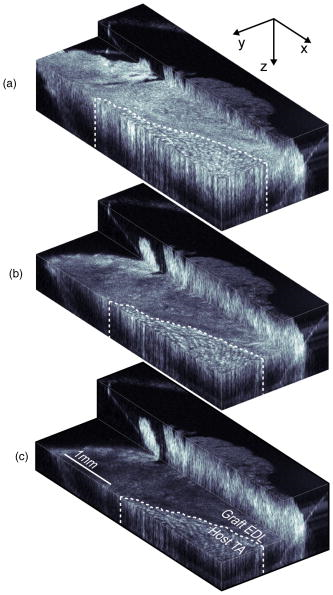
1.IntroductionMuscle tissue from experimental animal models is routinely studied to understand the cause, disease progression, and efficacy of treatment of human Duchenne muscular dystrophy. This common form of muscular dystrophy, occurring in approximately 1 in 3500 male births worldwide,1 is characterized by the progressive degeneration and loss of skeletal muscle, leading to death from respiratory or cardiac failure.2 Myofiber (muscle fiber) necrosis (cell death) normally results in new muscle formation to repair the damaged tissue, but in Duchenne muscular dystrophy repeated cycles of necrosis over time result in failed regeneration with the progressive loss of myofibers and replacement with fibrous and fatty connective tissue.3 The mouse whole-muscle autograft provides a large mass of tissue with distinctive histological features of muscle necrosis, inflammation, and regeneration (representing events in human disease): this model is well characterized, highly predictable, and reproducible between animals.4 The model also provides a convenient in vivo bioassay for assessing pharmacological interventions designed to reduce inflammation associated with necrosis5, 6 (as also occurs in muscular dystrophy). Such drug interventions have been shown to ameliorate skeletal muscle necrosis and pathology in the X-chromosome-linked muscular dystrophy mouse experimental animal model for human Duchenne muscular dystrophy,3, 6, 7 and in patients with the disease.2, 3, 8 Histology is conventionally used to study the morphology of skeletal muscle. Typically (e.g., for whole-muscle grafts), the muscle tissue is removed from the animal, processed, and cut into transverse sections that are stained to identify and measure the amount of healthy and necrotic muscle tissue, the extent of inflammation, and the formation (density and size) of myotubes (multinucleated new muscle fibers) within the regenerating region.9 However, the full histological process is laborious and time consuming and not amenable to examining large tissue volumes. It requires sacrifice of the animal for tissue harvesting, tissue processing, and preservation; sectioning and staining; and microscopic examination of many images. It would be preferable to be able to perform detailed 3D analysis on large volumes of tissue in situ without the need for muscle biopsy or animal sacrifice, and subsequent sectioning, staining, and imaging. Furthermore, longitudinal studies require the sacrifice of multiple animals for each time point, preventing the observation of sequential development of the same tissue within an individual animal. In vivo imaging, without the need for sacrifice, would enable repeated observation on an individual animal (optimal experimental scenario) and decrease the number of animals necessary per experiment (this is ethically desirable and has the added advantage of reduced cost). The opportunity to track the same tissue over time would greatly reduce the complication of biological variation between animals (at each time point) that is a considerable problem in conventional sampling. Such an increase in the efficiency and throughput of tissue characterization would signify a major shift in current experimental technique with wide application to animal studies and potential human tissue analysis. Biomedical imaging modalities that could potentially be used to characterize skeletal muscle pathology, ex vivo and in vivo, include ultrasonography, magnetic resonance imaging (MRI), confocal and multiphoton microscopy, and optical coherence tomography. Ultrasonography10 has been used clinically to probe the gross structure of human muscle tissue with large penetration depth, but is unable to resolve individual myofibers with an average diameter of . MRI has been used for in vivo studies of muscle in whole live animals,11, 12 but requires an exogenous contrast agent for localized detection of muscle pathology,13, 14 and its resolution14 of is insufficient to image the individual myofibers. Confocal and multiphoton microscopy, with their superior resolution, can resolve individual myofibers in vivo,15 but are limited in penetration depths to a few hundred micrometers, and routinely depend on the detection of fluorescent signals from exogenous contrast agents. Optical coherence tomography (OCT) is an emerging imaging modality capable of real-time, noninvasive, cross-sectional imaging of thick biological tissue approaching micrometer resolution.16 OCT uses near-IR light to perform optical ranging analogous to ultrasound B-mode imaging, but with resolution of two orders of magnitude greater. Typically, cross-sectional images are generated, which represent the spatially localized intensity of backscattered light with a penetration depth of up to in tissue.17 A third scan dimension can be added to enable 3D imaging of samples. OCT has been used to investigate a wide range of biological tissues and structures,17 but its use in the study of muscle tissue has been limited. Recently, Pasquesi reported the first study directed specifically toward skeletal muscle tissue and muscular dystrophy. This study18 used polarization-sensitive OCT to measure changes in the birefringence of skeletal muscle of the mouse model for Duchenne muscular dystrophy, and correlated these changes with myofiber damage due to dystrophy. Other previous studies with OCT have involved muscle tissue only incidentally; e.g., measuring the refractive index of cardiac muscle in humans,19 and determining the birefringence of animal skeletal20, 21, 22, 23, 24 and cardiac25, 26 muscle using polarization-sensitive OCT. The purpose of this study is to determine if 3D-OCT is capable of imaging large volumes of ex vivo unstained mouse skeletal muscle in sufficient detail to be able to differentiate healthy and pathological tissue. Such capability is a precursor to eventual noninvasive in vivo assessment of muscular dystrophy in mouse models. We compare, for the first time, 3D-OCT images of whole-muscle autografts with conventional histology, to identify damaged and regenerating muscle adjacent to healthy undamaged myofibers. In the remainder of this paper, we describe the imaging method, the anatomy of the whole-muscle autograft, and the surgical procedure. We present coregistered OCT images and conventional histology of the autografted muscle tissue, as well as 3D-OCT volume reconstructions, and discuss the results and prospects for in vivo imaging. 2.Methods2.1.OCTWe performed 3D-OCT imaging using a 3D-scanning optical fiber interferometer, as shown in Fig. 1a . The input light is split by a fiber optic coupler to reference and sample arms, and a circulator is used to enable dual-balanced detection to reduce optical intensity noise.27 The broadband light source is based on a semiconductor optical amplifier (JDS Uniphase, Milpitas, California) with a center wavelength of and a full-width-at-half-maximum bandwidth of . Fig. 1(a) Schematic of the fiber-optic OCT system: BBS, broadband source; PD, photodetector; VOA, variable optical attenuator; FD-ODL, frequency-domain optical delay line; and PC, polarization controller. (b) Whole-muscle autograft sample orientation. The arrow indicates the alignment of myofibers: EDL, extensor digitorum longus and TA, tibialis anterior. (c) En face OCT image plane indicating direction of inflammatory response (arrows). 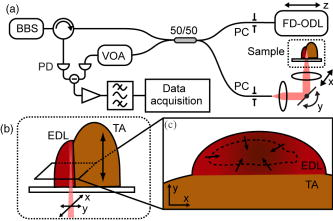 Measurement of the sample in the axial direction (along the beam) was performed by scanning the path length of the reference arm, using a frequency-domain optical delay line,28 with a measured resolution in air of . Transverse scanning in a single dimension was performed by a galvanometer mirror and objective lens setup with a beam numerical aperture of 0.07, measured in-focus transverse resolution ( intensity beam diameter) of and calculated axial Rayleigh range of . The dual-balanced photodetected difference signal was bandpass filtered and logarithmically demodulated before analog-to-digital conversion (PCI-6111E, National Instruments, Austin, Texas). The OCT system had a measured detection sensitivity of with incident on the sample. Two-dimensional (2D) OCT images were acquired in , and the third dimension was obtained by sequential cross-sectional scans at intervals along the axis obtained by linear translation of the objective lens. The 3D data sets consisting of images were acquired in and measured . En face OCT images at various sample depths were reconstructed from the 3D data set. 2.2.Animal Model and Whole-Muscle Autograft SurgeryThree female C57BL/10ScSn -old mice were obtained from the Animal Resources Centre, Murdoch, Western Australia. The mice were housed and treated according to the Western Australian Prevention of Cruelties to Animals Act (1920) and the Australian National Health and Medical Research Council guidelines, under protocols approved by the University of Western Australia Animal Ethics Committee. For whole-muscle autograft surgery, the mice were anaesthetized with 1.5% (v/v) Rodia halothane (Merial, Parramatta, Australia), , and and transplantation of the whole intact extensor digitorum longus (EDL) muscle was performed on one or both legs, described in detail elsewhere.9, 29 Briefly, the EDL muscle with both tendons attached was removed from the anatomical bed of the hindlimb of the mouse and transplanted onto the anterior surface of the tibialis anterior (TA) muscle of the same leg. The tendons of the EDL were sutured onto the host TA muscle, the skin closed, and the mice allowed to recover. Five days after the surgery, the mice were anaesthetized and sacrificed via cervical dislocation, and the entire TA muscle with attached EDL muscle was excised and placed in phosphate-buffered saline (PBS) on ice for transportation for ex vivo 3D-OCT imaging. 2.3.OCT Imaging ProtocolAll muscle samples were imaged within of animal sacrifice. A number of imaging geometries were investigated on the first two samples, and the optimum protocol, used on subsequent samples, is described in the following. The sample was bisected along the midregion to expose the transverse face of both the host TA and graft EDL, and then placed on an imaging window and bathed in PBS to prevent drying, and to reduce the refractive-index mismatch by displacing any air gaps between the glass plate and sample. The muscle sample was oriented with its myofibers parallel to the optical axis and its transverse face parallel to the en face imaging plane, as shown in Fig. 1b, and full-volume 3D-OCT scans were obtained. The image axial depth was scaled by 1.4, the approximate refractive index of muscle tissue.30 This sample alignment was preferred as it enabled the gross muscle tissue morphology and individual myofibers of the transverse plane of the EDL and TA to be displayed over the large area of the en face OCT image plane, for comparison with conventional transverse histology. 2.4.Histological Processing and Image CoregistrationImmediately following OCT imaging, samples were fixed in 4% (w/v) paraformaldehyde (pH 7.6) for , transferred to 70% (v/v) ethanol followed by automated standard paraffin preparation (Shandon Citadel 1000, Thermo Fisher Scientific, Inc., Waltham, Massachussets). Transverse -thick sections were cut on a microtome and collected on glass slides, and stained with hematoxylin and eosin (H&E) for light microscopy (DM RBE, Leica Microsystems GmbH, Wetzlar, Germany) and digital image capture (DXM 1200F, Nikon Corporation, Ontario, Canada). Nonoverlapping images of the grafts were obtained with an automated microscope translation stage, and images were tiled and stitched together using commercial software (ImagePro Plus 4.5.1, Media Cybernetics Inc., Silver Spring, Maryland) on a personal computer. Coregistration of OCT and light microscopy images was performed by matching for depth and visually correlating structural features of interest. 3.ResultsAll en face images taken from 3D-OCT data sets obtained using the protocol described in Sec. 2.3 were found to be consistent in appearance. The typical results presented in this section were taken from one mouse. Figure 2 shows a representative en face OCT image and the corresponding H&E-stained histological image of the transverse face of the EDL and TA tissues through the midregion of the muscle sample at a depth of . A magnified image (right) shows the graft boundary (dashed line) and nearby healthy myofibers (H) of the healthy host TA tissue. A mean filter was applied to this image to reduce speckle noise. The kernel size was chosen to roughly match the OCT in-focus transverse resolution. As a result of surgery, the blood and nerve supply to the EDL muscle is severed, causing the grafted tissue to undergo necrosis, subsequent inflammation, and regeneration.5 The histology in Fig. 2b shows inflammatory cells (I) infiltrating the grafted EDL: this is associated with the revascularization of the graft in part from the host TA. In the peripheral zone of inflammation, the necrotic tissue is phagocytozed, although some myofibers survive at the periphery (S), and myoblasts (precursor muscle cells) are activated and fuse to form myotubes.5 The centrally located area of persisting necrotic tissue (N) remaining within the EDL is termed the necrotic core, and constitutes approximately 50% of the entire area of the graft in these samples (at ). Fig. 2(a) En face OCT image and (b) corresponding histology of a transverse section of the autografted muscle sample showing the grafted EDL overlying the host TA muscle, separated by a dashed line added for clarity. A magnified view (right) shows the graft boundary, with healthy myofibers (H); surviving myofibers (S); and inflammatory cells (I). 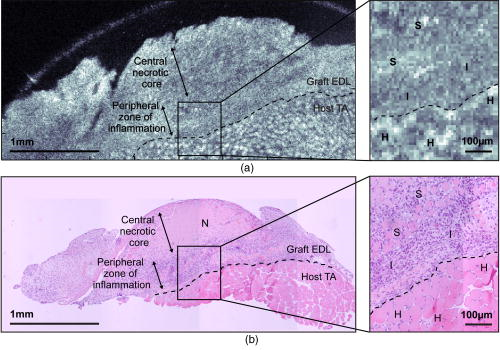 In the OCT image in Fig. 2a, the grafted EDL tissue is readily distinguished from the host TA tissue, meeting at a clearly defined boundary (dashed line). The TA tissue displays a regular pattern of circular features, in marked contrast to the EDL graft, which lacks any such prominent features, instead exhibiting a more uniform texture corrupted by the speckle noise typical of OCT images.16 The magnified image (right) highlights the distinction between the two muscle types, and the circular features of the OCT image in Fig. 2(a, right) are of a size and shape that correlate well with the healthy myofibers (H) of the corresponding histology [Fig. 2(b, right)]. In the entire 3D-OCT data set, we observed the grafted EDL tissue to be clearly distinguished from the host TA tissue with a distinct boundary between the two. Three representative en face OCT images at different depths within the whole-muscle autograft are presented in Fig. 3 , along with coregistered histology. At all depths, the OCT images [Figs. 3a, 3c, 3e] correspond well in overall structure with the histology [Figs. 3b, 3d, 3f]. The pattern of circular structures in the TA tissue persists throughout the 3D-OCT data set, with a shape and size similar to that expected for healthy myofibers. In the grafted EDL tissue, however, a similar pattern is lacking throughout. The tissue generally appears more uniform in texture, although there are sparsely distributed small regions of high intensity, which resemble the circular features found in the TA tissue. Fig. 3(a), (c), and (e) En face OCT images, and (b), (d), and (f) the corresponding histology of transverse sections of the autografted muscle sample at depths of (a) and (b) 100, (c) and (d) 250, and (e) and (f) . Surviving myofibers (S). The magnified view (insets) show the graft boundary. 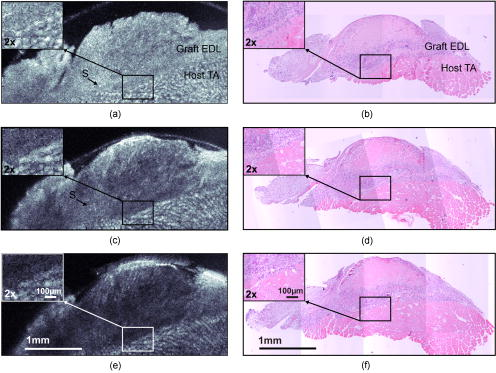 The contrast between the two muscle tissue types is more apparent, particularly at the graft boundary (see Fig. 3 insets), as the image depth increases. Another difference between the grafted EDL and host TA tissue is that the average signal intensity, particularly at the graft boundary as seen in Fig. 3a, 3c, 3d (insets), is attenuated to a greater extent in the grafted EDL tissue than in the neighboring region of TA tissue at the same depth. In the histological sections, the necrotic core within the EDL graft is obvious in all sections, and constitutes approximately 50% of the entire area of the graft. However, this feature is not evident in the corresponding OCT images. The average myofiber diameter of this region is smaller than those of the host TA muscle. The 3D-OCT cutaway reconstructions of the autografted muscle sample reveal the transverse face of the graft EDL tissue and host TA tissue at three depths into the sample in Fig. 4 . The reconstructed en face plane of the 3D-OCT data set is by far the most informative section for revealing the muscle morphology. The and image views in Fig. 4 show far fewer readily identifiable features. 4.DiscussionThis preliminary investigation has demonstrated that 3D-OCT is a promising tool for examining the morphology of muscle pathology and identifying individual muscle fibers. Previous studies involving muscle tissue using conventional B-scan oriented OCT have presented images that do not show individual muscle fibers or reveal significant morphological detail of the muscle tissue.18, 20, 21, 24, 31 A possible exception is the study by Hsiung 32 of the hamster cheek pouch at ultrahigh resolution ( at ), wherein, within the superficial and deep muscle layers , structures resembling muscle fibers are observed, although the origin of these structures was not discussed. Similarly, Guo presented a B-scan OCT image of rabbit muscle in which structure is evident, but the nature of this structure was not discussed, nor was there comparison with histology.22 In working with excised muscle, we were able to optimally orientate the sample to match the largest scan area (en face) to the transverse face of the whole-muscle autograft, which is the orientation of most interest to experimental biologists and conventionally examined in histological sections. We have found the availability of 3D data indispensable in identifying gross and fine features in the autografted muscle sample. The availability of alternative 2D sectional views provided by the 3D data set, as well as the ability to visualize the muscle sample in cut-away 3D movies (not presented here), proved invaluable in orientating the samples and presenting the en face images shown here. By contrast, our early work (not reported) recording only 2D sections was less successful. En face OCT images at all depths were found to be significant for visualizing the transverse-face whole-muscle autograft structure and features of interest; in contrast, images from the or planes were less informative. The en face OCT images generated from the 3D-OCT data set reveal a distinct difference between the muscle structure of the grafted EDL and the host TA, which correlates well with the corresponding histology. We observed that the muscle tissue features were smaller in the histology than in the OCT images. This shrinkage is due to dehydration of the tissue during the histological process, as previously reported.33 The most noticeable feature of the OCT images, confirmed by histology, is the observation of healthy myofibers (H) in the TA muscle, and their general absence in the graft EDL [Figs. 2a, 3a, 3c, 3e] with a few exceptions. Surviving myofibers (S) were identified in OCT images of the EDL [Figs. 2a, 3a, 3c] (confirmed in the corresponding histological sections) by their distinctive high-intensity circular shape comparable with the appearance of the healthy myofibers of the TA, which differentiated them from the generally featureless and lower intensity surrounding regions containing necrotic tissue and inflammatory cells. The 3D-OCT volume enhanced identification of surviving myofibers, as they could be tracked in depth and seen in successive en face OCT images reconstructed along the axis [Figs. 3a and 3b]. By after transplantation, revascularization of the EDL muscle and infiltration of inflammatory cells into the grafted EDL tissue is well advanced. The inflammatory tissue in the en face OCT images of the EDL has an appearance similar to those features observed in the study by Cobb of wound healing in mouse skin, particularly the granulation tissue in the dermis.34 The presence of the inflammatory cells in the EDL with the breakdown of necrotic muscle tissue significantly increases the density of cell nuclei is this region, staining dark blue in the histology of Fig. 2b. These nuclei ( in diameter) are around an order of magnitude smaller than the resolution of the OCT system and therefore cannot be visualized. This is a plausible explanation for the more uniform appearance of the EDL muscle graft, which is especially obvious at the boundary of the two tissues, as seen in Fig. 3 (insets). The presence of inflammatory cells and subsequent increase in the density of nuclei in the EDL tissue, particularly at the graft boundary, should also affect the attenuation of the OCT signal with depth (A-scan profile). The expected relatively stronger scattering in the EDL is confirmed in Fig. 3 (insets), which shows that the average signal strength in the EDL tissue is lower than in the neighboring TA tissue, increasingly so at greater depth [Fig. 3c]. While the morphology evident in the en face OCT images presented here correlates well with the corresponding histology, several issues must be resolved for the technique to be a useful replacement for conventional histology. New regenerating myofibers, which are of particular interest in the study of inflammation and regeneration in muscle tissue, could not be readily identified in the OCT images. Typically, activated myoblasts have fused to form myotubes and new myofibers are visible at the edge of the graft by day 5, becoming evenly distributed throughout5, 9 the EDL around days 7 to 8. Discrimination in histology relies on resolving the nucleus and its position with respect to the myofiber, as myotubes and regenerated myofibers have centrally located nuclei, while original myofibers have peripherally located nuclei. Such resolution is beyond the ability of the OCT setup used in this experiment but may be attainable with ultrahigh-resolution OCT (Ref. 32) and by utilizing recently published resolution enhancement methods.35 The distinction between myofiber types may also be possible through examination of the variation in the A-scans, but more work is required to establish this. En face OCT images could not be used to readily identify the necrotic core within the grafted EDL tissue and, therefore, could not be used to assess the percentage of this necrotic tissue relative to the total graft area, an important measure that is used to assess the effectiveness of drugs aimed at reducing inflammation.5 Future in vivo 3D-OCT imaging of mouse skeletal muscle tissue must contend with the anatomical orientation of muscle tissue and the presence of overlying skin tissue. Beyond improvements to the resolution afforded by ultrahigh resolution32 and high speed,36, 37 there are other potential strategies to facilitate in vivo imaging. Optical clearing agents could potentially be used to increase the imaging depth of OCT through the skin and cutis to the underlying muscle tissue of interest.38 Less desirably, direct access to muscle could be made via surgical incision15 or needle incision.39, 40 5.ConclusionThe findings here demonstrate the promise of 3D-OCT as a tool for the study of the morphology of mouse skeletal muscle. In the whole-muscle autograft model of damage and inflammation, OCT clearly shows the boundary between the tissues of the grafted and host muscles. Further improvements to enable the distinction between muscle fiber types (original undamaged versus regenerated) and the detection of the necrotic core, however, are required. Our preliminary results suggest that OCT may have a future role in the morphological assessment of the pathology of mouse skeletal muscle with applications in the study of muscular dystrophy. AcknowledgmentsThe authors thank Marilyn Davies for technical assistance with both the whole-muscle autograft surgery and skeletal muscle histology, James Ridgley for his contribution to the histological digital imaging, Thea Shavlakadze for useful discussions on whole-muscle autograft biology, and Timothy Hillman for preparation of Fig. 1a. ReferencesA. E. H. Emery,
“The muscular dystrophies,”
Lancet, 359
(9307), 687
–695
(2002). https://doi.org/10.1016/S0140-6736(02)07815-7 0140-6736 Google Scholar
W. D. Biggar,
H. J. Klamut,
P. C. Demacio,
D. J. Stevens, and
P. N. Ray,
“Duchenne muscular dystrophy: current knowledge, treatment, and future prospects,”
Clin. Orthop. Relat. Res., 401 88
–106
(2002). 0009-921X Google Scholar
H. G. Radley,
A. D. Luca,
G. S. Lynch, and
M. D. Grounds,
“Duchenne muscular dystrophy: focus on pharmaceutical and nutritional interventions,”
Int. J. Biochem. Cell Biol., 39
(3), 469
–477
(2007). https://doi.org/10.1016/j.biocel.2006.09.009 1357-2725 Google Scholar
P. Roberts,
J. K. McGeachie, and
M. D. Grounds,
“The host environment determines strain-specific differences in the timing of skeletal muscle regeneration: cross-transplantation studies between SJL/J and BALB/c mice,”
J. Anat., 191
(4), 585
–594
(1997). https://doi.org/10.1046/j.1469-7580.1997.19140585.x 0021-8782 Google Scholar
M. D. Grounds,
M. Davies,
J. Torrisi,
T. Shavlakadze,
J. White, and
S. Hodgetts,
“Silencing activity by using Remicade or Enbrel blocks inflammation in whole-muscle grafts: an in vivo bioassay to assess the efficacy of anti-cytokine drugs in mice,”
Cell Tissue Res., 320
(3), 509
–515
(2005). 0302-766X Google Scholar
M. D. Grounds and
J. Torrisi,
“Anti- (Remicade®) therapy protects dystrophic skeletal muscle from necrosis,”
FASEB J., 18
(6), 676
–682
(2004). https://doi.org/10.1096/fj.03-1024com 0892-6638 Google Scholar
S. Hodgetts,
H. Radley,
M. Davies, and
M. D. Grounds,
“Reduced necrosis of dystrophic muscle by depletion of host neutrophils, or blocking function with Etanercept in mdx mice,”
Neuromuscul Disord., 16
(9–10), 591
–602
(2006). 0960-8966 Google Scholar
W. D. Biggar,
V. A. Harris,
L. Eliasoph, and
B. Alman,
“Long-term benefits of deflazacort treatment for boys with Duchenne muscular dystrophy in their second decade,”
Neuromuscul Disord., 16
(4), 249
–255
(2006). 0960-8966 Google Scholar
J. D. White,
A. Scaffidi,
M. Davies,
J. McGeachie,
M. A. Rudnicki, and
M. D. Grounds,
“Myotube formation is delayed but not prevented in MyoD-deficient skeletal muscle: Studies in regenerating whole-muscle grafts of adult mice,”
J. Histochem. Cytochem., 48
(11), 1531
–1543
(2000). 0022-1554 Google Scholar
M. Narici,
“Human skeletal muscle architecture studied in vivo by noninvasive imaging techniques: functional significance and applications,”
J. Electromyogr. Kinesiol., 9
(2), 97
–103
(1999). 1050-6411 Google Scholar
L. M. McIntosh,
R. E. Baker, and
J. E. Anderson,
“Magnetic resonance imaging of regenerating and dystrophic mouse muscle,”
Biochem. Cell Biol., 76
(2–3), 532
–541
(1998). https://doi.org/10.1139/bcb-76-2-3-532 0829-8211 Google Scholar
G. A. Walter,
L. Cordier,
D. Bloy, and
H. L. Sweeney,
“Noninvasive monitoring of gene correction in dystrophic muscle,”
Magn. Reson. Med., 54
(6), 1369
–1376
(2005). https://doi.org/10.1002/mrm.20721 0740-3194 Google Scholar
H. Amthor,
T. Egelhof,
I. McKinnell,
M. E. Ladd,
I. Janssen,
J. Weber,
H. Sinn,
H.-H. Schrenk,
M. Forsting,
T. Voit, and
V. Straub,
“Albumin targeting of damaged muscle fibres in the mdx mouse can be monitored by MRI,”
Neuromuscul Disord., 14
(12), 791
–796
(2004). 0960-8966 Google Scholar
V. Straub,
K. M. Donahue,
V. Allamand,
R. L. Davisson,
Y. R. Kim, and
K. P. Campbell,
“Contrast agent-enhanced magnetic resonance imaging of skeletal muscle damage in animal models of muscular dystrophy,”
Magn. Reson. Med., 44
(4), 655
–659
(2000). https://doi.org/10.1002/1522-2594(200010)44:4<655::AID-MRM22>3.0.CO;2-T 0740-3194 Google Scholar
M. Bartoli,
N. Bourg,
D. Stockholm,
F. Raynaud,
A. Delevacque,
Y. Han,
P. Borel,
K. Seddik,
N. Armande, and
I. Richard,
“A mouse model for monitoring calpain activity under physiological and pathological conditions,”
J. Biol. Chem., 281
(51), 39672
–39680
(2006). https://doi.org/10.1074/jbc.M608803200 0021-9258 Google Scholar
D. D. Sampson and
T. R. Hillman,
“Optical coherence tomography,”
Lasers and Current Optical Techniques in Biology, 481
–571 Royal Society of Chemistry, Cambridge
(2004). Google Scholar
Handbook of Optical Coherence Tomography, Marcel Dekker(2002). Google Scholar
J. J. Pasquesi,
S. C. Schlachter,
M. D. Boppart,
E. Chaney,
S. J. Kaufman, and
S. A. Boppart,
“In vivo detection of exercise-induced ultrastructural changes in genetically-altered murine skeletal muscle using polarization-sensitive optical coherence tomography,”
Opt. Express, 14
(4), 1547
–1556
(2006). https://doi.org/10.1364/OE.14.001547 1094-4087 Google Scholar
G. J. Tearney,
M. E. Brezinski,
J. F. Southern,
B. E. Bouma,
M. R. Hee, and
J. G. Fujimoto,
“Determination of the refractive index of highly scattering human tissue by optical coherence tomography,”
Opt. Lett., 20
(21), 2258
–2260
(1995). 0146-9592 Google Scholar
J. F. de Boer,
S. M. Srinivas,
B. H. Park,
T. H. Pham,
Z. Chen,
T. E. Milner, and
J. S. Nelson,
“Polarization effects in optical coherence tomography of various biological tissues,”
IEEE J. Sel. Top. Quantum Electron., 5
(4), 1200
–1204
(1999). https://doi.org/10.1109/2944.796347 1077-260X Google Scholar
J. F. de Boer,
T. E. Milner, and
J. S. Nelson,
“Determination of the depth-resolved Stokes parameters of light backscattered from turbid media by use of polarization-sensitive optical coherence tomography,”
Opt. Lett., 24
(5), 300
–302
(1999). 0146-9592 Google Scholar
S. G. Guo,
J. Zhang,
L. Wang,
J. S. Nelson, and
Z. Chen,
“Depth-resolved birefringence and differential optical axis orientation measurements with fiber-based polarization-sensitive optical coherence tomography,”
Opt. Lett., 29
(17), 2025
–2027
(2004). https://doi.org/10.1364/OL.29.002025 0146-9592 Google Scholar
B. H. Park,
M. C. Pierce,
B. Cense, and
J. F. de Boer,
“Jones matrix analysis for a polarization-sensitive optical coherence tomography system using fiber-optic components,”
Opt. Lett., 29
(21), 2512
–2514
(2004). https://doi.org/10.1364/OL.29.002512 0146-9592 Google Scholar
M. Yamanari,
S. Makita,
V. D. Madjarova,
T. Yatagai, and
Y. Yasuno,
“Fiber-based polarization-sensitive Fourier domain optical coherence tomography using B-scan-oriented polarization modulation method,”
Opt. Express, 14
(14), 6502
–6515
(2006). https://doi.org/10.1364/OE.14.006502 1094-4087 Google Scholar
M. J. Everett,
K. Schoenenberger,
B. W. J. Colston, and
L. B. Da Silva,
“Birefringence characterization of biological tissue by use of optical coherence tomography,”
Opt. Lett., 23
(3), 228
–230
(1998). 0146-9592 Google Scholar
K. Schoenenberger,
B. W. J. Colston,
D. J. Maitland,
L. B. Da Silva, and
M. J. Everett,
“Mapping of birefringence and thermal damage in tissue by use of polarization-sensitive optical coherence tomography,”
Appl. Opt., 37
(25), 6026
–6036
(1998). 0003-6935 Google Scholar
A. M. Rollins and
J. A. Izatt,
“Optimal interferometer designs for optical coherence tomography,”
Opt. Lett., 24
(21), 1484
–1486
(1999). 0146-9592 Google Scholar
A. V. Zvyagin,
E. D. J. Smith, and
D. D. Sampson,
“Delay and dispersion characteristics of a frequency-domain optical delay line for scanning interferometry,”
J. Opt. Soc. Am. A, 20
(2), 333
–342
(2003). https://doi.org/10.1364/JOSAA.20.000333 0740-3232 Google Scholar
M. D. Grounds and
J. K. McGeachie,
“Therapeutic uses of muscle and factors controlling efficiency of whole-muscle autograft regeneration,”
Composite Tissue Transplantation, 121
–137 R. G. Landes Bioscience Co., Texas
(1998). Google Scholar
A. M. Zysk,
S. G. Adie,
J. J. Armstrong,
M. S. Leigh,
A. Paduch,
D. D. Sampson,
F. T. Nguyen, and
S. A. Boppart,
“Needle-based refractive index measurement using low-coherence interferometry,”
Opt. Lett., 32
(4), 385
–387
(2007). https://doi.org/10.1364/OL.32.000385 0146-9592 Google Scholar
C.-W. Sun,
Y.-M. Wang,
L.-S. Lu,
C.-W. Lu,
I.-J. Hsu,
M.-T. Tsai,
C. C. Yang,
Y.-W. Kiang, and
C.-C. Wu,
“Myocardial tissue characterization based on a polarization-sensitive optical coherence tomography system with an ultrashort pulsed laser,”
J. Biomed. Opt., 11
(5), 054016
(2006). https://doi.org/10.1117/1.2363358 1083-3668 Google Scholar
P.-L. Hsiung,
P. R. Nambiar, and
J. G. Fujimoto,
“Effect of tissue preservation on imaging using ultrahigh resolution optical coherence tomography,”
J. Biomed. Opt., 10
(6), 064033
(2005). https://doi.org/10.1117/1.2147155 1083-3668 Google Scholar
E. M. Anger,
A. Unterhuber,
B. Hermann,
H. Sattmann,
C. Schubert,
J. E. Morgan,
A. Cowey,
P. K. Ahnelt, and
W. Drexler,
“Ultrahigh resolution optical coherence tomography of the monkey fovea. Identification of retinal sublayers by correlation with semithin histology sections,”
Exp. Eye Res., 78
(6), 1117
–1125
(2004). https://doi.org/10.1016/j.exer.2004.01.011 0014-4835 Google Scholar
M. J. Cobb,
Y. Chen,
R. A. Underwood,
M. L. Usui,
J. Olerud, and
X. Li,
“Noninvasive assessment of cutaneous wound healing using ultrahigh-resolution optical coherence tomography,”
J. Biomed. Opt., 11
(6), 064002
(2006). https://doi.org/10.1117/1.2388152 1083-3668 Google Scholar
T. S. Ralston,
D. L. Marks,
P. S. Carney, and
S. A. Boppart,
“Interferometric synthetic aperture microscopy,”
Nat. Phys., 3
(2), 129
–134
(2007). 1745-2473 Google Scholar
Y. Watanabe,
K. Yamada, and
M. Sato,
“Three-dimensional imaging by ultrahigh-speed axial-lateral parallel time domain optical coherence tomography,”
Opt. Express, 14
(12), 5201
–5209
(2006). https://doi.org/10.1364/OE.14.005201 1094-4087 Google Scholar
Y. Yasuno,
V. D. Madjarova,
S. Makita,
M. Akiba,
A. Morosawa,
C. Chong,
T. Sakai,
K.-P. Chan,
M. Itoh, and
T. Yatagai,
“Three-dimensional and high-speed swept-source optical coherence tomography for in vivo investigation of human anterior eye segments,”
Opt. Express, 13
(26), 10652
–10664
(2005). https://doi.org/10.1364/OPEX.13.010652 1094-4087 Google Scholar
R. Cicchi,
F. S. Pavone,
D. Massi, and
D. D. Sampson,
“Contrast and depth enhancement in two-photon microscopy of human skin ex vivo by use of optical clearing agents,”
Opt. Express, 13
(7), 2337
–2344
(2005). https://doi.org/10.1364/OPEX.13.002337 1094-4087 Google Scholar
X. Li,
C. Chudoba,
T. Ko,
C. Pitris, and
J. G. Fujimoto,
“Imaging needle for optical coherence tomography,”
Opt. Lett., 25
(20), 1520
–1522
(2000). https://doi.org/10.1364/OL.25.001520 0146-9592 Google Scholar
H. Li,
B. A. Standish,
A. Mariampillai,
N. R. Munce,
Y. Mao,
S. Chiu,
N. E. Marcon,
B. C. Wilson,
A. Vitkin, and
V. X. D. Yang,
“Feasibility of interstitial Doppler optical coherence tomography for in vivo detection of microvascular changes during photodynamic therapy,”
Lasers Surg. Med., 38
(8), 754
–761
(2006). https://doi.org/10.1002/lsm.20387 0196-8092 Google Scholar
|