|
|
1.IntroductionDespite a falling incidence rate of gastric cancer, it is still the fourth most common malignancy, and also the second leading cause of cancer deaths in humans, accounting for 600, 000 deaths worldwide.1, 2 If the tumor is detected early and treated before it has invaded the gastric wall, the survival rate of the patient will increase tremendously.2 However, early identification and demarcation of such lesions in the stomach can be very difficult using the conventional diagnostic method of a white-light endoscope, which heavily relies on the visual observation of gross morphological changes of tissue, leading to poor diagnostic accuracy. Excisional biopsy currently remains the standard approach for cancer diagnosis, but this method is invasive and impractical for screening high-risk patients who may have multiple suspicious lesions. Raman spectroscopy, which makes use of inelastic light scattering processes to capture “fingerprints” of specific molecular structures and conformations of a given tissue or disease state, has shown to be a promising optical diagnostic technique for identifying malignant tissues in various organs. 3, 4, 5, 6, 7, 8, 9, 10, 11, 12 To convert molecular differences subtly reflected in Raman spectra between different tissues types into valuable diagnostic information for clinicians, multivariate statistical techniques have been successfully deployed in developing effective diagnostic algorithms for Raman spectroscopic diagnosis of cancers.3, 4, 5, 6, 7 Due to the complexities of the biological tissues, principal component analysis (PCA), which is able to take into account the whole range of Raman spectral features of the tissue, has often been applied to simplify the computational complexities for the development of effective classifier algorithms [e.g., linear discriminant analysis (LDA), logistic regression] without compromising diagnostic accuracy. 3, 4, 5, 6, 7, 8, 9 However, PCA does not necessarily provide the physical meanings of component spectra for tissue classification.8 Very recently, the classification and regression tree (CART) technique,13 based on the recursive partitioning for generating discriminatory algorithms for classification of different subgroups in complex datasets,14 has received extensive attention in biomedical fields such as proteomics, genomics, and mass spectroscopy. 14, 15, 16, 17, 18, 19, 20 For instance, Luk 14 applied both neural networks and CART on liver cancer proteomes and found that both algorithms produced equally good predictive abilities. Garzotto 15 employed CART to identify prostate cancer from normal tissue with the sensitivity of 96.6%. Zhang 16 also made use of CART on mass spectral urine profiles to achieve the sensitivity of 93.3% and specificity of 87.0% for separating transitional cell carcinoma from normal bladder tissue. Despite these successful applications, to date, the CART technique has yet to be applied to Raman spectroscopy for elucidation of Raman spectra in tissue diagnosis. In this study, we explore the feasibility of applying the CART technique to develop effective diagnostic algorithms for differentiation of near-infrared (NIR) Raman spectra between normal and cancer tissue, and to further understand molecular changes reflected in Raman spectra of tissue associated with the onset of malignancy in the stomach. 2.Materials and Methods2.1.Raman InstrumentationThe instrument used for tissue Raman spectroscopic studies has been described in detail elsewhere.21 Briefly, this system consists of a diode laser, a transmissive imaging spectrograph with a Kaiser holographic grating, an NIR-optimized back-illuminated, deep-depletion charge-coupled device (CCD) detector (Princeton Instruments, Trenton, New Jersey), and a specially designed fiber optic Raman probe that can effectively eliminate interference from fiberoptic fluorescence and silica Raman signals. The laser is coupled to a core diameter fiber and the fiber is connected to the Raman probe for tissue excitation. Tissue NIR Raman signals collected by the probe are fed into the spectrograph and the holographic grating disperses the incoming light onto the liquid-nitrogen-cooled CCD detector controlled by a computer. The tissue Raman spectra are displayed on the computer screen in real time and can be saved for further analysis. The system acquired Raman spectra over the wavenumber range of , and each spectrum was acquired within with light irradiance of . The spectral resolution of the system is . All wavelength calibrated spectra were also corrected for the wavelength dependence of the system using a standard lamp (RS-10, EG and G Gamma Scientific, San Diego, California). 2.2.Gastric Tissue SamplesA total of 73 gastric tissue samples were collected from 53 patients (28 men and 25 women with a median age of ) who underwent endoscopy biopsies or gastrectomy operations with clinically suspicious lesions. All patients preoperatively signed an informed consent permitting the investigative use of the tissues, and this study was approved by the Ethics Committee of the National Healthcare Group (NHG) of Singapore. After biopsies or surgical resections, tissue samples were immediately sent to the laboratory for Raman measurements. After spectral measurements, the tissue samples were fixed in 10% formalin solution and then submitted back to the Hospital for histopathologic examinations. A gastrointestinal (GI) pathologist conducted pathologic examinations, and the results showed that among the 73 homogenous gastric tissue samples with clearly defined pathologies, 55 tissue specimens were normal, and 18 were cancer (adenocarcinoma). A total of 222 tissue Raman spectra were acquired from different sites of gastric tissues, in which 143 Raman spectra were from normal and 79 from cancer. Note that the gastric tissue samples were approximately in size, and the laser light with a beam size of was focused on the tissue surface to mimic the in-vivo clinical measurements. For the larger resection tissues, Raman spectra were acquired on two to five different sites of the same tissue samples, and the corresponding pathology examinations were also performed on the tissue sites measured to correlate with Raman spectra for tissue classification. Each tissue surface location measured was then marked and stained for pathology. After comparing with pathologic results, only those Raman spectra that were correctly acquired from the tissue surfaces were included in the data analysis. 2.3.Classification and Regression TreeClassification and regression tree (CART) is a statistical technique that selectively employs variables that are of the utmost importance from a large number of input variables in databases for binary discrimination. 13, 14, 15, 16, 17, 18, 19, 20 It is implemented by growing a tree structure with a root node containing all the objects that are then further divided into nodes by recursive binary splitting.14, 15 The split that gives the best reduction in impurity between the mother group and the daughter groups ( and ) at different nodes of the tree is sequentially selected in the construction of the CART. The maximization of change of impurity function at each node is defined as:20 where represents different variables for different values of from a total of variables; represents the best splitting value of when a maximum change of impurity function , is achieved.13, 18 , , and are the impurity functions belonging to the parent node , left child node , and right child node of the parent node, respectively. and are the probabilities of achieving left and right nodes, respectively. CART will search through all possible values of variables for the best splitter at the maximal . In this study, the Gini index14 is used to determine the impurity at each node, which forms the criterion for splitting.where is the number of different classes, is the total number of objects, and is the number of objects from class presented in the node. Generally, a tree is first grown to its maximal size until the terminal nodes are sufficiently small. However, the maximum tree size that is usually overfitted with noise could not generalize well for future datasets. Hence, the tree is usually gradually shrunk by pruning away terminal nodes that lead to the smallest decrease in accuracy. For each subtree , a complexity-misclassification cost function is generated:20 where is the resubstitution misclassification error of ; and and represent the number of terminal nodes and the cost of complexity per terminal node, respectively. During each successive pruning process, a smaller subtree [i.e., that minimizes ] with a smaller number of terminal nodes will be generated, but the cost of complexity will gradually increase. As a result, searching for an optimal tree size is equivalent to finding the correct , such that the information in the learning dataset is best fit rather than overfit or underfit.13Although only one variable would be selected as the best splitter at any node in a CART, there would always be a second best variable that could perform nearly as well as the best splitter. The second best variable(s) could be masked by the best splitter(s) and would not appear in the final CART tree. As such, to avoid masking the importance of any variables used in CART, the relative importance of each input variable is assessed based on its importance over all possible nodes and splits by a “variable ranking method.” The importance of a variable is defined as:13 with , which equals the maximal decrease in node impurity for the division of a parent node into daughter nodes and that are guided by a surrogate split . A surrogate split is defined by a surrogate variable. This variable is the second best variable, which follows the selected variable by giving the second best reduction in impurity of the mother group into the daughter groups. This maximal decrease in node impurity is summed for all the nodes of the optimal subtree to obtain the importance of a variable.In this study, a ten-fold cross-validation was chosen to select the optimal tree size.13 The learning dataset is randomly divided into ten subsets: one of the subsets is used as an independent testing dataset, while the other nine subsets are combined and used as training datasets. The tree growing and pruning procedure is repeated ten times with each time using a different subset as a testing dataset. For each tree size, the resubstitution and cross-validation error are calculated and averaged over all subsets. The misclassification cost obtained for each subtrees on the cross-validation subset is matched with the subtrees of the complete model learning dataset using the values. The optimal sized tree is selected to be within one standard error (SE) of the complexity-misclassification rate for the minimum complexity-misclassification rate.13, 20 2.4.Statistical AnalysisAmong the 222 Raman spectra of gastric tissues acquired, the Raman datasets were divided randomly into a model learning dataset (80% of total dataset) and a validation dataset (20% of total dataset) for CART analysis.15 An unpaired two-sided Student’s -test was first applied in the learning dataset to identify diagnostically significant prominent Raman peaks as input variables for the development of CART algorithms for binary-class classification. Equal misclassification costs were specified so that there was an unbiased cost associated with misclassifying a cancer case as a normal case, and vice versa. The prior probability of each class was defined as proportional to the sizes of the groups in the dataset. Thereafter, the predictive sensitivity and specificity of the resulting tree model were evaluated using both the model learning (80% of total dataset) and validation dataset (20% of total dataset). Note that for the assessment of diagnostic sensitivity and specificity of the Raman technique, histopathological results were regarded as the gold standard. 3.ResultsFigure 1 shows the mean Raman spectra of normal and cancer gastric tissue in the model learning dataset. Prominent Raman peaks are observed in both normal and cancer gastric tissue, which are located at around (C–C stretching of hydroxyproline), ( symmetric ring breathing of phenylalanine), (C–C stretching of phospholipids), ( asymmetric stretching of nucleic acids), (C–N stretching and N–H bending modes of amide III of proteins), , ( twisting of proteins and nucleic acids), ( bending of proteins and lipids), ( stretching of amide I of proteins), and ( stretching of phospholipids),6, 7, 8, 9, 10 respectively. The intensity differences between the two tissue types are remarkable. For example, cancer tissues show higher intensities at 1265, 1450, and , while lower at 875, 1004, 1100, and , compared to normal tissues. This suggests that there is an increase or decrease in the percentage of a certain type of biomolecule relative to the total Raman-active constituents in cancer tissue. There are also obvious changes of Raman peak positions and bandwidths in the ranges of , , and , which are related to the breathing mode of phenylalanine and C—C stretching of phospholipids, the amide III and amide I of proteins, twisting of proteins/nucleic acids, and stretching of phospholipids, respectively, for cancer tissue. The differences in Raman spectra between normal and cancer tissues demonstrate the utility of Raman spectroscopy for gastric cancer diagnosis. Fig. 1Mean Raman spectra of gastric tissues from (a) normal and (b) cancer in learning Raman dataset. 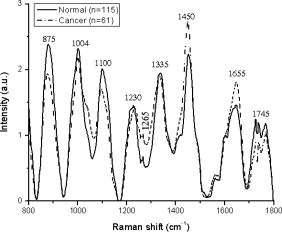 Table 1 lists the mean deviation (SD) of seven prominent Raman peaks for tissue classification. Overall, the intensity differences of these Raman peaks are statistically significant between normal and cancer tissues (unpaired two-sided Student’s -test, ). Based on the logistic regression analysis,22 the discrimination functions generated from each of these Raman peak intensities yield a sensitivity of 60 to 82% and the specificity of 60% to 75%, respectively, for identifying cancer from normal gastric tissues (Table 1). Table 1Statistical characteristics of diagnostically significant Raman peaks (unpaired two-sided Student’s t -test, p<0.05 ; 80% of total Raman dataset). Note that SD is standard deviation. The symbol * denotes a particular Raman peak intensity with cancer tissue being higher than normal tissue.
To further improve tissue classification, CART was subsequently employed to correlate all the diagnostically significant Raman peak intensities with tissue pathologies. Figures 2a and 2b show the relationship of complexity with respect to the misclassification cost and the number of terminal nodes for both cross-validated and resubstitution error after ten-fold cross-validation of the CART model learning dataset. The misclassification cost for the resubstitution error increases monotonically as the complexity increases with a corresponding decrease in terminal nodes. On the other hand, the misclassification cost for the cross-validated error increases at a slower rate compared to the resubstitution error. A local minimum misclassification cost of 0.1875 is found at a complexity of 0.00568 for the cross-validated error. Consequently, according to the cross-validated dataset, the optimal sized tree will be chosen at a complexity of 0.00852 with 13 terminal nodes, which is within one standard error (SE) of the complexity-misclassification cost for the local minimum complexity-misclassification cost. Fig. 2Dependence of complexity on (a) misclassification cost nodes for cross-validated error after ten-fold cross-validation, and resubstitution error, and on (b) number of terminal nodes for resubstitution error of the CART model learning dataset. The optimal sized tree was chosen to be at a complexity of 0.00852 with 13 terminal nodes within one standard error (SE) of the complexity-misclassification cost for the local minimum complexity-misclassification cost. 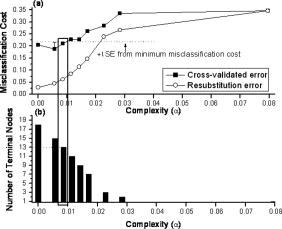 Figure 3 displays the CART analysis procedure in a classification model based on the model learning dataset (80% of total dataset). With the CART model, six diagnostically significant Raman peaks at 875, 1100, 1265, 1450, 1655, and are interlinked differently to build the following 13 subgroups (designated as either normal or cancer in the terminal subgroups): normal being groups 1, 3, 6, 7, 9, 11, and 13; cancer being groups 2, 4, 5, 8, 10, and 12. All these six significant Raman peak intensities are combined differently to build the seven normal and six cancer subgroups for best tissue classification. Fig. 3The optimal classification tree generated by the CART method after ten-fold cross-validation of the model learning dataset by utilizing six significant Raman peaks (875, 1100, 1265, 1450, 1655, and ). The binary classification tree is composed of 12 classifiers and 13 terminal subgroups. The decision-making process involves the evaluation of if-then rules of each node from top to bottom, which eventually reaches a terminal node with the designated class outcome, i.e., normal (N) or cancer (C). 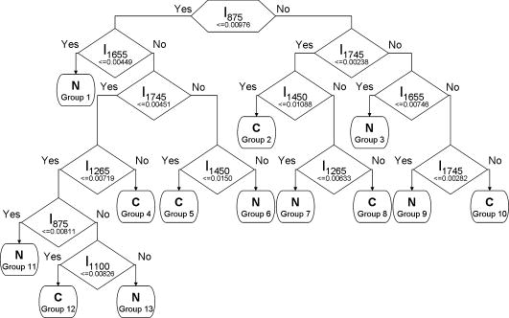 Table 2 tabulates the variable ranking of the Raman peaks and the total number of appearing times of different intensity features for generating the CART-based diagnostic model (Fig. 3). According to the variable ranking method,13 the most and least important Raman peaks are found to be located at 875 and , respectively. By assessing the final CART-based diagnostic model, the Raman peaks that appear the most and least number of times are located at 1745 and , respectively. Raman peaks located at 1655, 1265, 1100, and , which are in the order of descending variable rankings, appeared 2, 2, 1, and 2 times, respectively, in the final CART model. As a result, Raman peaks at 875, 1100, 1265, 1450, 1655, and are found to be most constructive toward building the final CART-based diagnostic model, and Raman peaks at 875 and appear to be the most important variables for tissue classification. Table 2The variable rankings of all the input Raman peak intensity features (n=7) computed by the CART algorithm, with the corresponding total number of times of the respective feature appearing in the final CART-based diagnostic model. Note that the symbol # denotes a particular Raman peak intensity with variable rankings (1 as the most important and 7 as the least important).
To evaluate the performance of the CART-based diagnostic algorithms for predicting the prospective cases (generalization), a randomly selected validation dataset (20% of total dataset) was used, in which six important Raman peaks (875, 1100, 1265, 1450, 1655, and ) were utilized as an input in the final CART-based diagnostic model. Table 3 summarizes the classification results of the two pathologic groups (normal versus cancer) for both the model learning dataset (after ten-fold cross-validation) (80% of total dataset), and the validation dataset (20% of total dataset). The sensitivity of 90.2% and specificity of 95.7% can be obtained for the model learning dataset, while a predictive sensitivity and specificity of 88.9 and 92.9% can be achieved for the independent validation dataset. The results show that CART-based diagnostic algorithms that utilize the most diagnostically important peaks of Raman spectra are powerful and robust for accurately predicting the tissue types in the prospective new cases. Table 3Classification results of Raman prediction of the two pathological groups with the model learning dataset (80% of total dataset) using the ten-fold cross-validation method, and the validation dataset (20% of total dataset) using a CART-based diagnostic algorithm.
4.DiscussionThe spectral analysis technique based on PCA-LDA has been widely practised in spectroscopy diagnosis of diseased tissue.3, 4, 5, 6, 7, 8 PCA is basically targeted to spectral data reduction rather than identification of biochemical components in tissue. The PCA-LDA model usually cannot interpret the physical meanings of the component spectra generated because the PCs that constitute most of the variance in the spectroscopic data are not necessary for the spectral parameters with the most diagnostic utility.3, 4, 8 In this study, a novel spectroscopy analysis method based on the classification and regression tree (CART) diagnostic model is explored to identify the distinctive Raman peak features for gastric tissue differentiation, and to relate to particular biochemical changes (e.g., vibrational modes of proteins, lipids, or nucleic acids) in tissue. We demonstrate that the CART-based diagnostic tree generated from the six significant Raman peaks (875, 1100, 1265, 1450, 1655, and ) can be used to correlate well with pathological classification of gastric tissues. We have identified the extent of significance of these prominent Raman peaks for the construction of the CART model (Table 2), and found that a Raman signal at [ (C—C) of hydroxyproline of collagen] and [ of phospholipids] are the most significant spectral features for gastric tissue diagnosis and characterization. However, the significance of all these Raman peak intensity features with respect to gastric tissue carcinogenesis has not been fully explored in the literature. In this work, the Raman peak intensity at has been found to decrease significantly with malignancy (Fig. 1), indicating a reduction in the percentage of collagen content relative to the total Raman-active components in cancer tissue. This observation is in agreement with the reports that cancerous cells proliferate, invade underlying layers, and express as a class of metalloproteases,23, 24 leading to a decrease in the amount of collagen level.25 In addition, the Raman peaks at 1265 and that are presumably ascribed to the amide III and amide I of proteins (in the -helix conformation of histones making up the chromatin)26 are also found to be important spectral features (Table 2). The significant increase of these two Raman intensity features indicates the elevated percentage of histones concentration with respect to the total Raman-active constituents in cancer. These findings accord with gastric cytologic studies of grading malignancy by the indication of nuclear hyperchromasia.27 On top of this, Raman peak intensities at 1100, 1450, and that represented mostly phospholipids at different molecular structures with different vibrational modes6, 7, 8, 9, 10 are also found to play pivotal roles toward tissue classification. Consistent with Raman studies on lung cancer diagnosis by Huang, 10 we also observed a lower percentage Raman signal of phospholipids (e.g., 1100 and ) in cancer gastric tissue. One notes that the important feature of Raman peaks at [ of phospholipids] appeared three times for the CART-based diagnostic model. This reveals that the significance of the variation in the concentration of phospholipids in gastric tissue and cells may be associated with malignant transformation. Hence, the CART technique may be a useful approach to identifying the origins of biochemical/biomolecular changes of Raman spectra for tissue carcinogenesis analysis. Applying the CART technique for classification of tissue Raman spectra, the predictive diagnostic sensitivity of 92.9% , specificity of 88.9% , and accuracy of 91.3% can be achieved for the independent validation dataset (Table 3). To compare with the CART method, we also performed the commonly practised PCA-LDA method using 80% of the same Raman dataset for training and 20% for testing for tissue classification, and a predictive sensitivity of 96.4% , specificity of 94.4% , and accuracy of 95.7% can be obtained for gastric cancer diagnosis. Further work based on PCA-LDA and CART techniques using 80% training/20% testing of the Raman dataset together with a five-fold cross-validation28 shows that the PCA-LDA approach yields a sensitivity of 92.4% , specificity of 94.4% , and accuracy of 93.7% ; whereas the CART method generates a sensitivity of 86.1% , specificity of 97.2% , and accuracy of 93.2% , respectively, for cancer detection. Hence, the CART-based diagnostic algorithms produce a similar level of diagnostic accuracy compared to the PCA-LDA model. These further reinforce that the CART-based diagnostic algorithms generated are robust and powerful for tissue diagnosis and characterization by Raman spectroscopy. Besides the ability for tissue classification, the CART diagnostic model could also provide a novel way for better understanding of the relationship between the disease-related biochemical changes of Raman spectra and tissue pathologies. We use the CART technique to evaluate how different Raman molecular information is correlated with tissue types by analyzing how the different Raman peaks are interlinked together to form different subgroups for best tissue classification. For instance, in Fig. 3, it is found that group 5 (cancer subgroup) has the highest probability of incidence, followed by group 3, which is a normal subgroup for both the model learning and validation datasets. In group 5, CART indicates that cancer gastric tissues are associated with a relative increase in Raman peak intensities at 1655 and , while a decrease at 875 and . These results, in fact, are consistent with reports on the decrease of Raman intensity ratio at associated with malignancies in the cervix and lung.10, 11 The CART model also shows that Raman peaks at 875 and representing collagen and phospholipids, respectively, appear to be significantly correlated with the Raman peaks at 1450 and for identifying the cancer subgroup (group 5). Conversely, the Raman peaks at 875, 1655, and utilized to construct a cancer subgroup (group 5) can be employed for identifying the normal subgroup (group 3). CART also indicates that compared to cancer tissue, normal gastric tissues tend to be related with high collagen contents (Raman peak at ) in extracellular matrix, high lipid contents (Raman peak at ) present in both extracellular matrix and cytoplasm, and a lower histones content (Raman peak at ) in the nucleus. Further investigation of other subgroups shows that heterogeneous molecular changes may occur in tissue, enabling cancer subgroups to be distinguished from normal. For example, collagen content typically decreases with malignancy, but there are quite a number of other cancer subgroups (groups 2, 8, and 10) associated with an increase in collagen content. These subgroups are accompanied by either less phospholipids or higher histones content, which could enable them to be distinguished from normal tissue. The prior CART-Raman analysis results indicate that most biochemical/biomolecular information from tissue and cells are essential for tissue discrimination, and the CART-based diagnostic model is able to partition different subgroups based on different compositions of Raman molecular information for separating gastric cancer from normal. Therefore, the CART-Raman technique may provide new insights into the biochemical/biomolecular changes associated with malignant transformation. In summary, the CART technique was first introduced and implemented to develop effective diagnostic algorithms for classification of Raman spectra between normal and cancer gastric tissues. This work shows that NIR Raman spectroscopy, in combination with powerful CART algorithms, has potential to provide an effective and accurate diagnostic means for cancer diagnosis in the gastric system. Further studies on a larger series of gastric tissues, in which the CART diagnostic algorithms are tested prospectively on new cases, are ongoing to reconfirm these preliminary findings. AcknowledgmentsThis research was supported by the Biomedical Research Council, the National Medical Research Council, the Academic Research Fund from Ministry of Education, and the Faculty Research Fund from National University of Singapore. ReferencesA. N. Milne,
R. Sitarz,
R. Carvalho,
F. Carniero, and
G. J. Offerhaus,
“Early onset gastric cancer: on the road to unraveling gastric carcinogenesis,”
Curr. Mol. Med., 7 15
–28
(2007). Google Scholar
K. G. Yeoh,
“How do we improve outcomes for gastric cancer?,”
J. Gastroenterol. Hepatol, 22 970
–972
(2007). 0815-9319 Google Scholar
M. G. Shim,
L. M. Song,
N. E. Marcon, and
B. C. Wilson,
“In vivo near-infrared Raman spectroscopy: demonstration of feasibility during clinical gastrointestinal endoscopy,”
Photochem. Photobiol., 72 146
–150
(2000). https://doi.org/10.1562/0031-8655(2000)072<0146:IVNIRS>2.0.CO;2 0031-8655 Google Scholar
T. C. Bakker Schut,
M. J. H. Witjes,
H. J. C. M. Sterenborg,
O. C. Speelman,
J. L. N. Roodenburg,
E. T. Marple,
H. A. Bruining, and
G. J. Puppels,
“In vivo detection of dysplastic tissue by Raman spectroscopy,”
Anal. Chem., 72 6010
–6018
(2000). https://doi.org/10.1021/ac000780u 0003-2700 Google Scholar
D. P. Lau,
Z. Huang,
H. Lui,
D. W. Anderson,
K. Berean,
M. D. Morrison,
L. Shen, and
H. Zeng,
“Raman spectroscopy for optical diagnosis in the larynx—preliminary findings,”
Lasers Surg. Med., 37
(3), 192
–200
(2005). https://doi.org/10.1002/lsm.20226 0196-8092 Google Scholar
A. Mahadevan-Jansen,
M. F. Mitchell,
N. Ramanujam,
A. Malpica,
S. Thomsen,
U. Utzinger, and
R. Richards-Kortum,
“Near-infrared Raman spectroscopy for in vitro detection of cervical precancers,”
Photochem. Photobiol., 68 123
–132
(1998). https://doi.org/10.1562/0031-8655(1998)068<0123:NIRSFV>2.3.CO;2 0031-8655 Google Scholar
N. Stone,
P. Stavroulaki,
C. Kendall,
M. Birchall, and
H. Barr,
“Raman spectroscopy for early detection of laryngeal malignancy: preliminary results,”
Laryngoscope, 110 1756
–1763
(2000). https://doi.org/10.1097/00005537-200010000-00037 0023-852X Google Scholar
Z. Huang,
H. Lui,
D. I. McLean,
M. Korbelik, and
H. Zeng,
“Raman spectroscopy in combination with near-infrared autofluorescence background enhances the in vivo assessment of malignant tissues,”
Photochem. Photobiol., 81
(5), 1219
–1226
(2005). https://doi.org/10.1562/2005-02-24-RA-449 0031-8655 Google Scholar
A. Robichaux-Viehoever,
E. Kanter,
H. Shappell,
D. Billheimer,
H. Jones, and
A. Mahadevan-Jansen,
“Characterization of Raman spectra measured in vivo for the detection of cervical dysplasia,”
Appl. Spectrosc., 61
(9), 986
–993
(2007). 0003-7028 Google Scholar
Z. Huang,
A. McWilliams,
H. Lui,
D. I. McLean,
S. Lam, and
H. Zeng,
“Near-infrared Raman spectroscopy for optical diagnosis of lung cancer,”
Int. J. Cancer, 107 1047
–1052
(2003). https://doi.org/10.1002/ijc.11500 0020-7136 Google Scholar
U. Utzinger,
D. L. Heintzelman,
A. Mahadevan-Jansen,
A. Malpica,
M. Follen, and
R. Richards-Kortum,
“Near-infrared Raman spectroscopy for in vivo detection of cervical precancers,”
Appl. Spectrosc., 55 955
–959
(2001). https://doi.org/10.1366/0003702011953018 0003-7028 Google Scholar
X. F. Ling,
Y. Z. Xu,
S. F. Weng,
W. H. Li,
X. Zhi,
R. M. Hammer,
W. G. Fateley,
F. Wang,
X. S. Zhou,
R. D. Soloway,
J. R. Ferraro, and
J. G. Wu,
“Investigation of normal and malignant tissue samples from the human stomach using Fourier Transform Raman spectroscopy,”
Appl. Spectrosc., 56
(2), 570
–573
(2002). https://doi.org/10.1366/0003702021955330 0003-7028 Google Scholar
L. Briman,
J. H. Friedman,
R. A. Olshen, and
C. J. Stone, Classification and Regression Trees,
(1984) Google Scholar
J. M. Luk,
B. Y. Lam,
N. P. Lee,
D. W. Ho,
P. C. Sham,
L. Chen,
J. Peng,
X. Leng,
P. J. Day, and
S. T. Fan,
“Artificial neural networks and decision tree model analysis of liver cancer proteomes,”
Biochem. Biophys. Res. Commun., 361
(1), 68
–73
(2007). 0006-291X Google Scholar
M. Garzotto,
T. M. Beer,
R. G. Hudson,
L. Peters,
Y. C. Hsieh,
E. Barrera,
T. Klein, and
M. Mori,
“Improved detection of prostate cancer using classification and regression tree analysis,”
J. Clin. Oncol., 23
(19), 4322
–4329
(2005). 0732-183X Google Scholar
Y. F. Zhang,
D. L. Wu,
M. Guan,
W. W. Liu,
Z. Wu,
Y. M. Chen,
W. Z. Zhang, and
Y. Lu,
“Tree analysis of mass spectral urine profiles discriminates transitional cell carcinoma of the bladder from noncancer patient,”
Clin. Biochem., 37 772
–779
(2004). 0009-9120 Google Scholar
K. R. Hess,
M. C. Abbruzzese,
R. Lenzi,
M. N. Raber, and
J. L. Abbruzzese,
“Classification and regression tree analysis of 1000 consecutive patients with unknown primary carcinoma,”
Clin. Cancer Res., 5 3403
–3410
(1999). 1078-0432 Google Scholar
I. Zlobec,
R. Steele,
N. Nigam, and
C. C. Compton,
“A predictive model of rectal tumor response to preoperative radiotherapy using classification and regression tree methods,”
Clin. Cancer Res., 11
(15), 5440
–5443
(2005). 1078-0432 Google Scholar
V. A. Valera,
B. A. Walter,
N. Yokoyama,
Y. Koyama,
T. Iiai,
H. Okamoto, and
K. Hatakeyama,
“Prognostic groups in colorectal carcinoma patients based on tumor cell proliferation and classification and regression tree (CART) survival analysis,”
Ann. Surg. Oncol., 14
(1), 34
–40
(2007). 1068-9265 Google Scholar
P. W. Stewart and
J. W. Stamm,
“Classification tree prediction models for dental caries from clinical, microbiological, and interview data,”
J. Dent. Res., 70
(9), 1239
–1251
(1991). 0022-0345 Google Scholar
Z. Huang,
H. Lui,
X. K. Chen,
A. Alajlan,
D. I. McLean, and
H. Zeng,
“Raman spectroscopy of in vivo cutaneous melanin,”
J. Biomed. Opt., 9
(6), 1198
–1205
(2004). https://doi.org/10.1117/1.1805553 1083-3668 Google Scholar
D. W. Hosmer and
S. Lemeshow, Applied Logistic Regression, John Wiley and Sons, New York
(1989). Google Scholar
S. Curran and
G. I. Murray,
“Matrix metalloproteinases in tumor invasion and metastasis,”
J. Pathol., 189 300
–308
(1999). 0022-3417 Google Scholar
C. Streuli,
“Extracellular matrix remodeling and cellular differentiation,”
Curr. Opin. Cell Biol., 11 634
–640
(1999). https://doi.org/10.1016/S0955-0674(99)00026-5 0955-0674 Google Scholar
G. I. Zonios,
R. M. Cothren,
J. T. Arendt,
J. Wu,
J. Van Dam,
J. M. Crawford,
R. Manoharan, and
M. S. Feld,
“Morphological model of human colon tissue fluorescence,”
IEEE Trans. Biomed. Eng., 43 113
–122
(1996). https://doi.org/10.1109/10.481980 0018-9294 Google Scholar
G. J. Thomas Jr., B. Prescott,
“Secondary structure of histones and DNA in chromatin,”
Science, 197
(4301), 385
–388
(1977). 0036-8075 Google Scholar
J. H. Hughes,
C. J. Leigh,
S. S. Raab,
S. Y. Hook,
M. B. Cohen, and
M. J. Suhrland,
“Cytologic criteria for the brush diagnosis of gastric adenocarcinoma,”
Cancer, 84
(5), 289
–294
(1998). 0008-543X Google Scholar
Y. Mao,
X. Zhao,
S. Wang, and
Y. Cheng,
“Urinary nucleosides based potential biomarker selection by support vector machines for bladder cancer recognition,”
Anal. Chim. Acta, 598
(1), 34
–40
(2007). 0003-2670 Google Scholar
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||

