|
|
1.IntroductionColorectal cancer is the second leading cause of cancer-related death in the United States. In 2007, over 153,760 new cases were diagnosed and over 52,180 deaths resulted from colorectal cancer.1 As these cancers have a long development phase from inception to cancer,2 theoretically all these cancers are preventable with accurate universal screening. Screening colonoscopy substantially reduces the risk of colorectal cancer in long-term follow-up.3, 4 The identification and removal of polyps has reduced the rate of mortality from colon cancer.5 However, the decrease in long-term incidence of colorectal cancer varies greatly in these patients. Reasons for these differences have been attributed to variations in study populations, colonoscopy technique,6 and the presence of hard-to-detect cancer precursor lesions.7 Colonic mucosa contains numerous pits, the crypts of Lieberkühn, with roundish openings arranged in a regular pattern on the surface of normal colorectal mucosa.8 Prior studies have shown that shapes of colonic crypts change with disease state and show characteristic patterns.9, 10, 11 It has also been shown that crypt patterns and histopathological diagnosis are well correlated.11, 12, 13, 14, 15 For example, aberrant crypt foci (ACF) were first described by Bird 16 in 1987 in mice given carcinogens. Aberrant colonic crypts have larger lumens than normal crypts and have a thickened epithelium that stains darker with dyes compared to surrounding crypts.17, 18 The lumens of the aberrant crypts often appear slit-shaped rather than circular. Aberrant crypts usually occur in groups of four or more (hence, ACF) and are readily identified on the mucosal surface of the colon by microscopy. There is compelling evidence that ACF are biomarkers for colorectal carcinoma.19, 20, 21, 22, 23 ACF are more numerous in patients with colon cancer.19, 20, 21 It is also known that the orientation of crypts changes with the progression of the disease.24, 25, 26, 27, 28 Normal colonic crypts are oriented parallel to each other. Aberrant crypts and crypts in cancerous tissue are oriented less parallel to each other. High-resolution endoscopic methods have been demonstrated to visualize colonic crypts in humans, including high-magnification chromoendoscopy, high-magnification narrow band imaging, and endoscopic confocal microscopy. Chromoendoscopy is a technique that utilizes tissue stains, such as methylene blue, applied topically to the gastrointestinal mucosa to better characterize lesions.29, 30 Narrow band imaging (NBI) may be regarded as the optical analog of chromoendoscopy, without the use of staining agents, which is accomplished by narrowing the bandwidth of spectral transmittance of the red/green/blue optical filters used in the sequential-frame imaging method for videoendoscopic imaging.31, 32, 33 Both chromoendoscopy and NBI can be used in conjunction with magnification endoscopy, which uses optical systems to magnify endoscopic images, allowing one to see the minute colonic surface structures.33, 34, 35 Although these high-magnification methods can visualize the morphology of the crypt openings at the mucosal surface, they cannot visualize the three-dimensional (3-D) structure of the crypts. Endoscopic confocal microscopy produces high-resolution images of the gastrointestinal epithelium and is also sensitive to fluorescence. Furthermore, confocal imaging is capable of sectioning in depth and can therefore visualize 3-D structures. In vivo confocal fluorescence microscopy has been demonstrated in the upper36, 37 and lower37, 38, 39 gastrointestinal (GI) tracts. However, the depth of crypts range from ,26 while the in-depth observation range of endoscopic confocal microscopy is limited to a few hundred micrometers.36, 38 Therefore, this method cannot always visualize the entire depth of crypts. Endoscopic optical coherence tomography (EOCT) provides subsurface, high-resolution real-time imaging of GI mucosa.40, 41, 42 EOCT provides depth sectioning to from the mucosal surface, overcoming the limitations of magnification endoscopy and endoscopic confocal microscopy for visualizing the 3-D structure of crypts. Furthermore, in vivo EOCT imaging of colon polyps has shown good correlation with the pathology observed in these polyps after their removal.43 3-D EOCT imaging of the colon in an in vivo animal model (rabbit) has recently been demonstrated, including a discussion of crypt visualization and basic manual quantification of crypt size.44 3-D EOCT imaging of human colonic crypts has not been demonstrated to date, but the potential is clear.44, 45 It has been shown that image analysis can quantify colonic crypt structures and improve interpretation of crypt patterns. Quantification of crypt structures in histopathological24, 25, 46, 47, 48, 49 and chromoendoscopic28, 50 images has been recently demonstrated. In this work, we demonstrate that colonic crypt structures can be imaged and quantified by automated algorithms using 3-D optical coherence tomography and by microscopy with methylene blue staining. 2.Methods and MaterialsUnder a protocol approved by the Institutional Review Board of University Hospitals Case Medical Center, we studied 79 samples of resected fresh colon tissues obtained from colectomies (59 patients, including 31 female, old) using a microscope-integrated bench-top OCT scanner described elsewhere.51 The time-domain system utilized a high-power broadband source centered around with a bandwidth of . Axial and transverse resolution was (FWHM coherence length in tissue) and ( spot diameter), respectively. With optical power of incident on the sample, measured sensitivity was . Images were acquired at 8 frames per second with 500 axial lines (A-scans) per image. Data included 14 normal samples (from 12 patients), 44 samples of normal-appearing tissue adjacent to cancer (NA) (from 32 patients), and 21 malignant samples (from 18 patients). Cancer was diagnosed by routine clinical pathology, and tissue was classified as “normal” or “normal adjacent to cancer” by the surgeon. The clinical target for high-resolution colon imaging is to detect abnormal variation of crypt structure in fields of apparently normal colonic tissues where premalignancy may be suspected but not detectable by conventional endoscopy. While malignant colonic tissue is readily detectable by endoscopy and therefore not a clinical diagnostic target for OCT imaging nor for high-magnification chromoendoscopy, we include images of malignant samples in our descriptive results in order to demonstrate that our observations match what is already known about the architecture of malignant colon. We excluded the malignant samples from the data set for automatic quantification of colonic crypt morphology. 2.1.Sample Preparation and Image AcquisitionPreparation of colon tissue was meant to simulate approximately in vivo imaging with high-magnification chromoendoscopy. First, the surface of the freshly resected colon specimen was washed using 10% acetylcysteine, a mucolytic agent, and washed by water to remove the coating mucous and to prepare the tissue for dye staining. Subsequently, tissue was stained with 10% methylene blue to enhance the contrast of crypts under the microscope. After dye staining, several microscopic images were taken under different magnifications using a commercial CCD camera (PixelLink model PL-A642). 3-D OCT image volumes were obtained in the same region of interest (ROI) and registered with the microscopic images. For each 3-D OCT volume, 9 times frame averaging was used to reduce the speckle noise. 2.2.Microscopic Image and OCT En Face View SegmentationEach 2-D color microscopic image was transformed to a gray-level image. Then, uneven illumination was corrected by morphological opening with a large structure element,52 and background noise was removed using a median filter. Subsequently, the morphological gradient image was calculated in order to capture the edges of the crypts.53 A modified gradient image was created by thresholding using a threshold value calculated by Otsu’s method.54 In the modified gradient image, gradient intensities lower than the threshold were set to zero, and intensities higher than threshold were unchanged. The crypt edges produce the strongest gradient values, so this method prevents oversegmentation by removing gradients representing structures other than the crypt edges. Watershed segmentation is subject to oversegmentation because it is sensitive to every local minimum and maximum. This can be avoided by providing foreground markers, which are locations known to be inside of crypt lumens, and background markers, which are locations known to be outside of crypt lumens. These were automatically computed by morphological reconstruction.55, 56 Watershed transformation using immersion simulation57 was executed on the superimposed foreground and background markers and modified gradient image to segment the crypts within the ROI. Because strands of connective tissue within intervening nonpitted mucosa (indicated by white arrows in Figs. 3 and 4) have similar contrast characteristics as crypts, they can lead to inclusion of “false crypts” among the set of segmented crypts in an ROI. Therefore, crypts having a ratio of major axis length to minor axis length larger than three were rejected as false crypts. Finally, the crypts touching the edges of the ROI were removed prior to subsequent morphological feature extraction. Fig. 3Segmentation results of representative NA sample: (A) the microscopic image with methylene blue and (B) corresponding OCT volume of a sample of NA colonic tissue; (C) the crypt segmented within the 2-D micrograph and (D) the crypts segmented in 3-D within the OCT volume. The crypts touching the edges of the ROI were removed. White arrows indicate intervening nonpitted mucosa. Yellow arrows indicate missed crypts. (Color online only.) 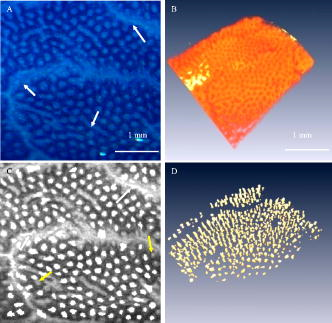 Fig. 4Segmentation results of representative ACF sample: (A) the microscopic image with methylene blue and (B) corresponding OCT volume of a sample of colonic tissue including an ACF; (C) the crypts segmented within the 2-D micrograph and (D) the crypts segmented in 3-D within the OCT volume. The crypts touching the edges of the ROI were removed. White arrows indicate intervening nonpitted mucosa. Yellow arrows indicate missed crypts. (Color online only.)  In the OCT volume data, each en face plane is analogous to a gray-level microscopic image. Therefore, we adapted the 2-D microscopic image segmentation described above to segment the crypts within each en face plane of the 3-D OCT volumes, resulting in segmented crypts in three dimensions, which could be visualized using volume rendering. To validate the automated segmentation results, one representative microscopic image and one representative OCT volume were manually segmented, and the manual segmentation results were quantitatively compared with the marker-controlled watershed segmentation results. The contours of crypts within one micrograph of normal colonic tissue were manually traced and compared with the contours resulting from marker-controlled watershed segmentation of the same image. In the same way, the contours of the crypts within each en face plane of one OCT volume were manually traced and compared with the contours resulting from marker-controlled watershed segmentation of the same volume. The precision ratio (PR) and the match ratio (MR)58 between the manually segmented contours and the marker-controlled watershed segmented contours were calculated to quantify the comparisons. The PR is defined as , where is the number of pixels that differ between the manually determined contour and the marker-controlled watershed segmented contour and is the number of pixels in the manual contour. The MR is defined as , where and denote the areas enclosed by manually segmented contours and the areas covered by marker-controlled watershed segmented contours, respectively. 2.3.Morphological Feature ExtractionWithin the ROI of the 2-D microscopic images, the following features of each segmented crypt were extracted: the area, the major axis length, minor axis length, and eccentricity of the best-fitting ellipse, and the solidity. Solidity is the ratio of the area of the crypt to the area of its convex hull and is a measure of the degree to which the crypt is concave or convex. In addition, the density of crypts within the ROI was calculated as the number of crypts divided by the area of the ROI. The 3-D OCT image volumes were visualized by volume rendering and were used to quantify crypt orientation. In order to quantify the crypt orientation, it is useful to extract the “skeleton” of each crypt, the curve representing the center of the lumen of the crypt in three dimensions. The conventional method for skeleton extraction, morphological thinning via distance transformation, led to unwanted branches59 instead of a single curve, which complicated quantification of the orientation (Fig. 7). Therefore, we developed a method to estimate the skeleton of each crypt by linking the centroids of each crypt in the ROI at each en face plane of the segmented 3-D OCT volume. Fig. 7Results of skeleton extraction from 3-D segmented crypts using the conventional method, morphological thinning via distance transformation, and our centroid-linking method. Skeletons extracted from each crypt within a 3-D OCT volume of a representative sample of NA using (A) the conventional method and (B) our method, respectively. Skeletons extracted from each crypt within a 3-D OCT volume of a sample including an ACF using (C) the conventional method and (D) our method, respectively.  Skeleton estimation by centroid-linking was carried out in two steps. First, the centroids of the segmented crypts in each en face plane of the OCT volume were calculated. Second, the centroids of each crypt were linked with the centroids of the same crypts in adjacent en face planes, resulting in strings of coordinates representing the skeletons of each crypt. This linking step was initiated from a plane below the surface of the tissue, approximately the location of the focus of the scanner. This initial plane was selected using the volume rendering to include as many crypts as possible. Then, for each crypt identified in the initial plane, the linking proceeded deep to the initial plane until either the crypt terminated or the data ended. Then the linking was carried out from the initial plane up to the tissue surface. Because of noise in the images, occasionally crypts were not segmented in single en face planes, resulting in “missing layers.” Also, occasionally false crypts were identified due to oversegmentation. In order to link discontinuous crypts caused by missing layers, if an overlying crypt was not found in the plane adjacent to the current plane, subsequent planes were searched until an overlaying crypt was found. This crypt centroid was linked and skeleton coordinates were assigned to missing layers by linear interpolation. If an overlaying crypt was not found within a maximum gap of , the crypt was assumed to be terminated. False crypts were only problematic if they occurred in the initial plane or if they overlaid a true crypt. False crypts in the initial plane were mitigated, because any crypt of in extent was removed from analysis. If more than one overlaid segmented crypt was found in adjacent planes, then the centroids of the crypts having the largest ratio of overlaid area were linked. The other crypt was assumed to be false. Crypt orientation was quantified from the skeletons by measuring the straightness of each crypt skeleton and how parallel they were to each other. In order to quantify the straightness of each crypt, each crypt skeleton was fit to a line by calculating the coefficients of the first principal component of the skeleton. The goodness of fit, assessed by the coefficient of determination (also referred to as the value), was taken as our measure of the straightness of that crypt. The value for the fit indicates the fraction of the variation in the skeleton of crypt that is explained by the line (i.e., the closer that value is to 1, the straighter the crypt is). In order to quantify how parallel the crypts in a ROI were to each other, the direction vector of the line fitted to the skeleton of each crypt was compared to the mean direction vector of all of the crypts in the ROI. The comparison was quantified by calculating the dot product of each crypt vector and the mean vector, resulting in a value representing the parallelism. If this value is close to 1, the crypts within an ROI are more parallel to each other. Within an ROI, the straightness and parallelism measures were calculated to represent crypt orientation in the region. Running MATLAB version 7.4 on a PC with a CPU and RAM, the entire image analysis algorithm ran in including of preprocessing, of segmentation, and of morphological feature extraction for one micrograph. It ran in including of preprocessing, of segmentation, and of morphological feature extraction, for a 3-D OCT volume (599 slices). 3.Results3.1.Descriptive ObservationsObserved characteristics of normal colonic tissue under the microscope and OCT include round, small crypts that are uniformly distributed,8 straight, and oriented parallel to each other. Observed characteristics of ACF under the microscope and OCT are larger and more often elongated crypts compared with normal surrounding crypts17, 18 and crypt orientation that is less straight and less parallel. These observations are consistent with previously reported observations;8, 24, 25, 26 however, to our knowledge the 3-D orientations of colonic crypts have not been quantified previously.25, 26 Within the 14 samples of normal colonic tissues, no ACF were found. Within the 44 samples of apparently normal tissue adjacent to cancer, four ACF were found. ACF are an endoscopic diagnosis, not a histopathological diagnosis, based on observation of the surface appearance of the tissue. Here, the observation of ACF was based on the documented typical appearance of ACF and confirmed by a GI pathologist. Within the 21 samples of malignant tissue, two types of cancer were observed, moderately differentiated adenocarcinoma and mucinous adenocarcinoma arising from a tubulovillous adenoma. In the 19 samples of moderately differentiated adenocarcinoma, dramatically distorted crypt patterns were observed. In the two samples of moderately differentiated mucinous adenocarcinoma arising from a tubulovillous adenoma, small, oblique, tubulovillous structures were observed. Examples of each of these mucosa types are shown in Fig. 1 . Figure 1A is a micrograph of normal colonic mucosa with methylene blue staining; Fig. 1 is an OCT cross-sectional image, taken at the location indicated by the yellow line in Fig. 1A. Similarly, Figs. 1B and 1 demonstrate an ACF found in a sample of apparently normal tissue adjacent to a tumor. Figures 1C and 1 demonstrate moderately differentiated adenocarcinoma, and Figs. 1D and 1 demonstrate moderately differentiated mucinous adenocarcinoma arising from a tubulovillous adenoma. Fig. 1Examples of the appearance of normal colonic mucosa (A and ), ACF (B and ), moderately differentiated adenocarcinoma (C and ), and mucinous adenocarcinoma arising from a tubulovillous adenoma (D and ). The first column shows micrographs of the tissue samples under methylene blue staining. The second column shows cross-sectional OCT images of the same samples at the locations indicated by the yellow lines in the micrographs. (Color online only.) 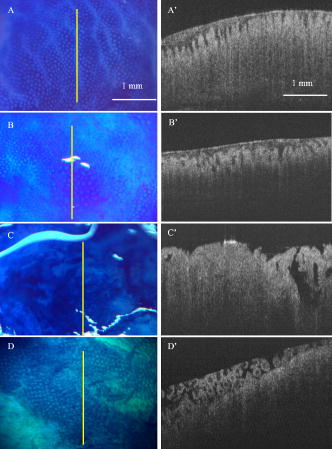 3.2.Quantitative ResultsImages of a typical aberrant crypt focus appear in Fig. 2 . Figure 2A is the micrograph with methylene blue staining; Fig. 2B is the corresponding OCT B-scan image at the location indicated by the yellow line. Figure 2C is the maximum intensity projection of the 3-D OCT volume. Figures 2D, 2E, 2F, 2G, 2H, 2I are the OCT en face views at depths of 129, 258, 430, 602, 774, and from the surface of the tissue, respectively. En face planes were filtered with a median kernel to smooth the contours of segmented crypt boundaries. Figures 3 and 4 show segmentation results of representative NA and ACF samples. Figure 3A shows the microscopic image with methylene blue of a sample of NA colonic tissue, while Fig. 3B shows the corresponding OCT volume. Figure 3C shows the crypts segmented within the 2-D micrograph; Fig. 3D shows the crypts segmented in three dimensions within the OCT volume. Similarly, Fig. 4 shows microscopic and OCT images of an ACF as well as the crypts segmented in two and three dimensions. Fig. 2Images of a typical aberrant crypt focus: (A) the micrograph with methylene blue staining; (B) the corresponding OCT B-scan image at the yellow line location; (C) the maximum intensity projection of the 3-D OCT volume; and (D) the OCT en face views at depth of 129 (E) 258, (F) 430, (G) 602, (H) 774, and (I) from the surface of the tissue, respectively. En face planes were filtered with a median kernel to reduce speckle noise. (Color online only.) 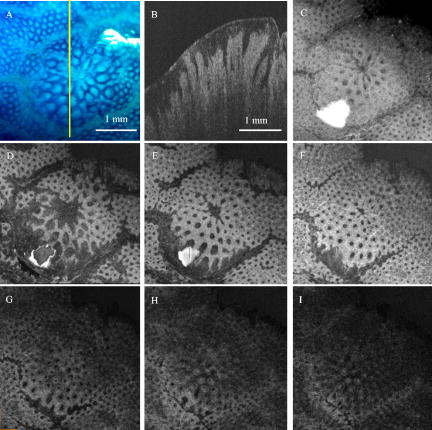 The results of the segmentation validation experiments are shown in Figs. 5 and 6 . Figure 5 shows the results of the manual and marker-controlled watershed segmentation of one microscopic image of a normal colon. Figure 5A is the microscopic image. The yellow contours in Fig. 5B are the manually traced contours of each crypt. Figure 5C shows the contours of crypts segmented automatically. Figure 6 shows the results of the manual and marker-controlled watershed segmentation on one OCT volume. Figure 6A shows the volume rendering of the OCT image volume of a sample including an aberrant crypt focus. Figure 6B shows the volume rendering overlaid with the manually segmented crypts, while Fig. 6C shows the volume rendering overlaid with the automatically segmented crypts. Only selected crypts were manually segmented, and these were quantitatively compared with the marker-controlled watershed segmentation of the same selected crypts. Comparison between the manual and automatic segmentation of this OCT image volume at various depths from the surface of tissue resulted in the PR and MR values shown in Table 1 . Fig. 5Results of the manual and marker-controlled watershed segmentation of one microscopic image of a normal colon: (A) the microscopic image, (B) the yellow contours of crypts traced manually, and (C) the contours of crypts segmented automatically. (Color online only.)  Fig. 6Results of manual and marker-controlled watershed segmentation of one OCT volume: (A) volume rendering of the OCT image volume of a sample including an aberrant crypt focus, (B) the volume rendering overlaid with the manually segmented selected crypts, and (C) the volume rendering overlaid with the automatically segmented crypts.  Table 1Comparison between the manuual and automic segmentation of the micrograph shown in Fig. 5 and the OCT image shown in Fig. 6 at various depths from the surface of tissue.
Figure 7 shows the results of skeleton extraction from 3-D segmented crypts using the conventional method, morphological thinning via distance transformation, and our centroid-linking method. Using both methods, a skeleton of each segmented crypt was extracted. Figures 7A, 7B demonstrate the skeletons extracted from each crypt within a 3-D OCT volume of a representative sample of NA tissue using the conventional method and our method, respectively. Similarly, Figs. 7C, 7D demonstrate the skeletons extracted from each crypt within a 3D OCT volume of a sample including an ACF. Skeletons of individual crypts can be clearly observed in the figure insets (eight times magnified). Note that the unwanted branches on the skeletons extracted by morphological thinning are not present on the skeletons extracted by centroid-linking. Also, the skeletons extracted by centroid-linking do not include any false crypts shorter than . To compare the suitability of the skeletons extracted by both methods for the purpose of quantifying the straightness and parallelism of the segmented crypts, the metrics were computed from a representative sample of NA tissue and a sample including ACF. These results are summarized in Table 2 . To evaluate the effect of noise reduction by nine-times frame averaging and median filtering on the measurement of crypt orientation, straightness and parallelism of the segmented crypts were computed from the same two tissue samples. These results are summarized in Table 3 . Table 2Calculated straightness and parallelism values of a representative sample of NA tissue and a sample including ACF, from skeletons extracted by morphological thinning via distance transformation and by centroid-linking.
Table 3Straightness (S) and parallelism (P) values of a representative sample of NA tissue and a sample including ACF from skeletons extracted by centroid-linking using single frames and nine-times frame averaging, with and without 3×3 median filtering.
The six morphological features were calculated within the segmented ROI of the 14 normal colonic tissue samples, the tissue samples, and the 4 ACF. The dominant value of each feature in one sample was taken to be the peak of the distribution of values within that sample. The first six columns within Table 4 show the mean and standard deviation of the dominant values of the six morphological features of all samples of three types. The first two rows within Fig. 8 display the histograms of each morphological feature, separated into the three sample types. Table 5 shows the correlation coefficients for each pair of the six morphological features. Because some features were correlated, the first two principal components (PCs) of the six features were calculated. The mean and standard deviations of the first two PCs are shown in the last two columns within Table 4. Also, the histograms of the first two PCs are displayed in the third row of Fig. 8. Fig. 8Histogram of each morphological feature and the first two PCs of those features within the segmented ROI of the 14 normal colonic tissues, colonic tissues, and 4 ACF within colonic tissues. 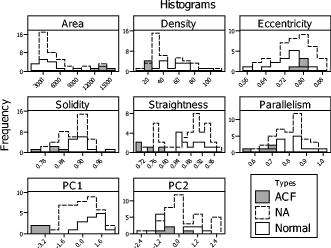 Table 4Mean and standard deviation of the dominant values of the morphological features and first two PCs of those six features within the segmented ROI of the 14 normal colonic tissues, 40NA colonic tissues, and 4 ACF within colonic tissues.
Table 5Correlation coefficients between each morphological feature.
4.Discussion and ConclusionFrom our methylene blue-stained micrographs, samples of normal mucosa and normal-appearing mucosa adjacent to cancer showed predominantly round crypts distributed uniformly, while ACF stained dark and had crypts with large, irregular lumens. Previous studies using high-magnification chromoendoscopy have reported equivalent observations.11, 60, 61 From the micrographs, topology of the tissue surface and crypt orientation are not readily apparent. However, because it records 3-D image data with depth resolution, OCT provides visualization of these features. This difference is illustrated clearly in Fig. 2, where the crypt openings of the ACF can be clearly visualized in the micrograph (A), but the OCT cross section (B) and volume rendering (C) demonstrate more clearly the topology of the slightly raised ACF and the less-parallel orientation of the crypts. From the OCT data, we observed that normal crypts were generally very straight and parallel to each other. Crypts within ACF were observed to be somewhat less straight and significantly less parallel to each other compared with the orientation of normal crypts. Crypts within moderately differentiated adenocarcinoma showed oblique orientation which was different from the orientation of normal and ACF crypts, which were more perpendicular to the tissue surface. Although there was variability from sample to sample, generally contrast in the OCT volumes was sufficient to visualize and segment crypts to a depth of at least . However, we observed that crypts were observable deeper in fresher samples, up to in depth. This gives us confidence that measurement of crypt orientation by OCT is feasible in vivo. Figure 2 displays OCT en face images of an ACF at various depths down to from the tissue surface. Two previous studies observed crypt orientation (without quantification) from histopathology slides, and our observations are consistent with those findings.18, 25 It should be noted, however, that our observations were obtained from images of intact, unfixed tissue. These observations confirm that the observed characteristics of colonic crypts reported here are consistent with known crypt morphology, suggesting that the analysis methods described here will be applicable to high-magnification chromoendoscopy and endoscopic OCT images obtained in patients. Previously, several demonstrations of quantitative analysis of colonic tissue have been reported from histological sections. Image-feature quantification methods based on texture analysis,24, 50, 62, 63 fractals,48 or frequency analysis28 have been shown, but these methods do not directly quantify colonic crypts morphology. Mophometric methods of analyzing histological sections have also been demonstrated, 25, 46, 47, 48, 49, 63, 64 including automated segmentation of cellular and crypt features.47 However, the image properties of photographs of histological sections are vastly different than those of chromoendoscopy and OCT, so those methods are not directly applicable to chromoendoscopy and OCT data. A few demonstrations of quantification of crypt morphology have been reported using high-magnification chromoendoscopy,11, 65, 66, 67 but to our knowledge, automated segmentation of colonic crypts from chromoendoscopic or OCT imaging has not been previously reported. Our methods reported here demonstrate the feasibility of automatically quantifying crypt morphometry from 2-D micrographs with methylene blue staining and from 3-D OCT volumes. This is significant, because these methods are applicable to the high-potential endoscopic microscopy techniques of high-magnification chromoendoscopy and EOCT. The marker-based watershed segmentation method presented here achieved accurate segmentation of colonic crypts from 2-D micrographs with methylene blue stain as well as from 3-D OCT volumes. It can be observed in Figs. 3 and 4 that crypts were generally accurately segmented, few false crypts were included, and few crypts were missed. To validate the accuracy of automatic segmentation, we manually segmented one representative microscopic image and one representative OCT volume to compare with automatic segmentation. One representative image was considered adequate, because the accuracy of the segmentation did not depend on the shape of the crypts but on image contrast and resolution, which did not vary with tissue type. To quantify segmentation accuracy, we compared the manual and automated segmentation results using the PR and MR,58 as summarized in Table 1. Rejection of false crypts using a 3:1 ratio of major to minor axes was generally successful; however, some of these connective tissue artifacts (having a ratio of ) were retained. For OCT data, these remaining false crypts were usually rejected during the centroid-linking process. Automated segmentation of the micrographs with methylene blue achieved a PR and MR of , which is considered acceptable as indicating good agreement between the manual and automatic segmentation.58 Segmentation of the en face OCT image also achieved high values of PR and MR. Best agreement with manual segmentation was achieved at the plane of OCT volume corresponding to the focus of the OCT scanner, which is the location of best transverse image resolution ( below the surface of the tissue). This is indicated by both the high mean values and low standard deviations of PR and MR at this depth. Shallower than , the accuracy of the OCT segmentation was generally better than that of the micrograph. Deeper than , however, the accuracy decreases steadily. From the results summarized in Table 1, we observe that the accuracy of the automated segmentation of OCT images is influenced by two major factors, the transverse resolution, which peaks at the focal plane, and the image SNR, which decreases monotonically from the tissue surface into the tissue. We also observe that, at least near the surface, segmentation of OCT slices result in better PR and MR compared with segmentation of the corresponding crypts in the micrograph. This is likely due to the less-distinct crypt lumen borders within the micrographs as compared to the OCT slices. Also, OCT images are unaffected by uneven staining. As shown in Fig. 7, the conventional skeleton extraction method using 3-D morphological thinning via distance transformation resulted in many branches and artifacts, which are undesirable for quantifying orientation of crypts. The centroid-linking skeleton extraction method demonstrated here resulted in crypt skeletons without branches and also rejected artifactual false crypts. Table 2 shows that centroid-linking resulted in much narrower distributions of measured straightness and parallelism of crypts, as well as a clearer separation between normal and ACF tissues, as compared with the measurements of the skeletons extracted by 3-D morphological thinning. Table 3 shows that although noise reduction by nine times of frame averaging and median filtering slightly improves separation between NA and ACF tissues from crypt orientation measures, the improvement is not significant. This is because skeleton extraction by centroid-linking does not strongly depend on contour roughness of the segmented crypt boundaries. From this we are encouraged that the method does not require frame averaging, which reduces effective imaging rate, and is amenable to in vivo EOCT application. We have shown that the six morphological features of crypts summarized in Table 4 can be automatically quantified from micrographs with methelyne blue and/or OCT images. Our measurements of the area and density of crypts within normal tissue and NA tissues are in agreement with previous studies.26, 49 To our knowledge, eccentricity, solidity, straightness, and parallelism of crypts have not previously been quantified. The purpose of this work is to demonstrate crypt morphology quantification methods, not to prove that these methods can be used to classify tissue types. However, we can make some observations from the distributions of feature values represented by the histograms shown in Fig. 8. The distributions representing normal samples and NA samples appear generally very similar. However, NA may have broader distributions, for example, of area, solidity, parallelism, and eccentricity. The distribution of values representing ACF appear to have different means than normal/NA, except in the case of eccentricity. The distributions of area, density, straightness, and parallelism, and to a lesser extent solidity, show strong potential for separation of ACF from normal/NA tissues. The distributions of eccentricity for the three types of tissue appear to be similar to each other and broad compared to distributions of other parameters. This may be due to lack of control for the obliqueness of the image slices compared to the orientation of the crypts and the tissue surface topology. A more accurate measure of crypt eccentricity could be achieved by making use of 3-D crypt orientation and surface topology information from the OCT data. Some crypt morphology features were highly correlated. In particular, the mean luminal area and density of crypts within an ROI are highly correlated, with a correlation coefficient of as shown in Table 5. The straightness and parallelism of crypts within an ROI are also correlated, with a correlation coefficient of 0.49. It is intuitive and expected that these metrics should be correlated. Because of the correlation among these features, principal-component analysis was applied to these six features. The first principal component accounted for 56% variance of the six measured features. Together, the first two principal components accounted for 62% variance. From Fig. 8, we can observe that the distributions of the first PC for normal and NA tissues appear similar, with the NA distribution possibly being broader. Furthermore, the distribution of the first PC for ACF tissues appears to be significantly different than normal and NA tissues. The distributions of the second principal component do not appear to be particularly informative. This is not surprising, as the second PC does not account for much additional variance in the data. Because of the small sample size of ACF (four samples), no generalizable conclusion may be drawn, but these observations indicate that quantified morphological features of colonic crypts may have the potential to aid tissue classification for screening for early cancer biomarkers such as ACF. The observation that ACF occurred within NA tissue but not within normal tissue is consistent with the observation that the distributions of morphological feature values for NA tissue are broader than distributions for normal tissue and are shifted toward the ACF distributions. These observations taken together are consistent with the so called field effect, which refers to the idea that colon cancer arises in a wide field of mucosa that has previously undergone molecular changes.60, 68, 69 These observations suggest that subtle changes in crypt morphology may accompany molecular changes and that tissues progressing toward disease may be detectible by high-resolution endoscopic imaging. The methods described here are likely to be useful for analysis of data acquired in vivo with little modification. In high-magnification chromoendoscopy, the resolution is similar to our benchtop images; however, the image quality is expected to vary more due to less-consistent removal of mucus and staining. From subjective experience, we are confident that the shapes of crypts observable in vivo are not different than we observed in the data presented here. Similarly, image quality of endoscopic OCT data is expected to be less consistent. However, the morphological features we analyzed (parallelism and straightness) extracted from 3-D OCT volumes are rotation invariant, so absolute tissue orientation or gross orientation variations should not affect local orientation quantification of crypts in vivo. For both technologies, automated image-quality feedback to the operator would help to make the quality of images acquired in vivo more consistent. Also, for both technologies, real-time analysis would be most useful. We believe that this can be achieved by programming the algorithms using parallel processing techniques. In conclusion, we have shown that an automated image analysis algorithm can quantify colonic crypt morphology from high-resolution images approximating high-magnification chromoendoscopy and EOCT. These computerized algorithms can provide quantitative and objective measures, they are suitable for specific and repetitive image reading tasks, and they can potentially reduce inter- and intraobserver variability when human readers evaluate such images. This work can enable studies to determine the clinical utility of high-magnification chromoendoscopy and EOCT as well as studies to evaluate colonic crypt morphology as a biomarker for colonic disease progression. AcknowledgmentsThe authors acknowledge the contributions of David L. Wilson, Ph.D., Theresa P. Pretlow, Ph.D., Gerard Isenberg, M.D., Jeffry Katz, M.D., Metini Janyasupab, Christine Lemyre, Lateefa Russell, Wendi Barrett, and Sunny McClellan Morton. This work was supported in part by the National Institutes of Health (Grants No. CA114276 and No. CA110943). ReferencesA. Jemal,
R. Siegel,
E. Ward,
T. Murray,
J. Xu, and
M. J. Thun,
“Cancer statistics, 2007,”
Ca-Cancer J. Clin., 57 43
–66
(2007). 0007-9235 Google Scholar
S. Jones,
W. Chen,
G. Parmigiani,
F. Diehl,
N. Beerenwinkel,
T. Antal,
A. Traulsen,
M. A. Nowak,
C. Siegel,
V. E. Velculescu,
K. W. Kinzler,
B. Vogelstein,
J. Willis, and
S. D. Markowitz,
“Comparative lesion sequencing provides insights into tumor evolution,”
Proc. Natl. Acad. Sci. U.S.A., 105
(11), 4283
–4288
(2008). 0027-8424 Google Scholar
S. J. Winawer,
A. G. Zauber,
M. N. Ho,
M. J. O’Brien,
L. S. Gottlieb,
S. S. Sternberg,
J. D. Waye,
M. Schapiro,
J. H. Bond,
J. F. Panish,
F. Ackroyd,
M. Shike,
R. C. Kurtz,
L. Hornsby-Lewis,
H. Gerdes,
E. T. Stewart, and
T. N. P. S. Workgroup,
“Prevention of colorectal cancer by colonoscopic polypectomy,”
N. Engl. J. Med., 329
(27), 1977
–1981
(1993). https://doi.org/10.1056/NEJM199312303292701 0028-4793 Google Scholar
F. Citarda,
G. Tomaselli,
R. Capocaccia,
S. Barcherini,
M. Crespi, and
T. I. M. S. Group,
“Efficacy in standard clinical practice of colonoscopic polypectomy in reducing colorectal cancer incidence,”
Gut, 48 812
–815
(2001). 0017-5749 Google Scholar
S. Winawer,
A. G. Zauber,
M. N. Ho,
M. J. O’Brien,
L. S. Gottlieb,
S. S. Sternberg,
J. D. Waye,
M. Schapiro,
J. H. Bond,
J. F. Panish, F. Ackroyd,
M. Shike,
R. C. Kurtz,
L. Hornsby-Lewis,
H. Gerdes, and
E. T. Stewart,
“Prevention of colorectal cancer by colonoscopic polypectomy.,”
N. Engl. J. Med., 329
(27), 1977
–1981
(1993). https://doi.org/10.1056/NEJM199312303292701 0028-4793 Google Scholar
R. L. Barclay,
J. J. Vicari,
A. S. Doughty,
J. F. Johanson, and
R. L. Greenlaw,
“Colonoscopic withdrawal times and adenoma detection during screening colonoscopy,”
N. Engl. J. Med., 355
(24), 2533
–2541
(2006). 0028-4793 Google Scholar
R. M. Soetikno,
T. K. Kaltenbach,
R. V. Rouse,
W. Park,
A. Maheshwari,
T. Sato,
S. Matsui, and
S. Friedland,
“Prevalence of nonpolypoid (flat and depressed) colorectal neoplasms in asymptomatic and symptomatic adults,”
JAMA, J. Am. Med. Assoc., 299
(9), 1027
–1035
(2008). 0098-7484 Google Scholar
S. S. Poulsen,
K. C. Christensen,
M. Petri, and
S. Jarnum,
“Stereomicroscopic examination of stained rectal biopsies,”
Scand. J. Gastroenterol., 13 605
–608
(1978). 0036-5521 Google Scholar
S. Tada,
M. Iida,
T. Yao,
T. Matsumoto,
M. Tsuneyoshi, and
M. Fujishima,
“Stereomicroscopic examination of surface morphology in colorectal epithelial tumors,”
Hum. Pathol., 24 1243
–1252
(1993). 0046-8177 Google Scholar
S. Kudo,
S. Tamura,
T. Nakajima,
H. Yamano,
H. Kusaka, and
H. Watanabe,
“Diagnosis of colorectal tumorous lesions by magnifying endoscopy,”
Gastrointest. Endosc., 44 8
–14
(1996). 0016-5107 Google Scholar
S. Kudo,
C. A. Rubio,
C. R. Teixeira,
H. Kashida, and
E. Kogure,
“Pit pattern in colorectal neoplasia: endoscopic magnifying view,”
Endoscopy, 33 367
–373
(2001). 0013-726X Google Scholar
A. Iso,
S. Shimizu,
M. Tada, and
K. Kawai,
“Usefulness of real-time band enhancement in differential dianosis of colorectal tumors,”
Dig. Endosc., 7 397
–400
(1995). 0915-5635 Google Scholar
E. Jaramillo,
M. Watanabe,
R. Befrits,
E. Ponce,
C. Rubio, and
P. Slezak,
“Small, flat colorectal neoplasms in long-standing ulcerative colitis detected by high-resolution electronic video endoscope,”
Gastrointest. Endosc., 44 15
–22
(1996). 0016-5107 Google Scholar
T. Matsumoto,
F. Kuroki,
M. Mizuno,
S. Nakamura, and
M. Iida,
“Application of magnifying chromoscopy for the assessment of severity in patients with mild to moderate ulcerative colitis,”
Gastrointest. Endosc., 46 400
–405
(1997). 0016-5107 Google Scholar
S. Y. Tung,
C. S. Wu, and
M. Y. Su,
“Magnifying colonoscopy in differentiating neoplastic from non-neoplastic colorectal lesions,”
Am. J. Gastroenterol., 96 2628
–2632
(2001). 0002-9270 Google Scholar
R. P. Bird,
“Observation and quantification of aberrant crypts in the murine colon treated with a colon carcinogen: Preliminary findings,”
Cancer Lett., 37
(2), 147
–151
(1987). 0304-3835 Google Scholar
L. Roncucci,
D. Stamp,
A. Meline,
J. B. Cullen, and
W. R. Burce,
“Identification and quantification of aberrant crypt foci and microadenomas in the human colon,”
Hum. Pathol., 22 287
–294
(1991). 0046-8177 Google Scholar
C. D. Gregorio,
L. Losi,
R. Fante,
S. Modica,
M. Ghidoni,
M. Pedroni,
M. G. Tamassia,
L. Gafa,
P. de Leon, and
L. Roncucci,
“Histology of aberrant crypt foci in the human colon,”
Histopathology, 30
(4), 328
–334
(1997). 0309-0167 Google Scholar
T. P. Pretlow,
B. J. Barrow,
W. S. Ashton,
M. A. O’Riordan,
T. G. Pretlow,
J. A. Jurcisek, and
T. A. Stellato,
“Aberrant crypts: putative preneoplastic foci in human colonic mucosa,”
Cancer Res., 51 1564
–1567
(1991). 0008-5472 Google Scholar
L. Roncucci,
S. Modica,
M. Pedroni,
M. G. Tamassia,
M. Ghidoni,
L. Losi,
R. Fante,
C. Di Gregorio,
A. Manenti,
L. Gafa, and
M. Ponz De Leon,
“Aberrant crypt foci in patients with colorectal cancer,”
Br. J. Cancer, 77 2343
–2348
(1998). 0007-0920 Google Scholar
T. Takayama,
S. Katsuki,
Y. Takahashi,
M. Ohi,
S. Nojiri,
S. Sakamaki,
J. Kato,
K. Kogawa,
H. Miyake, and
Y. Niitsu,
“Aberrant crypt foci of the colon as precursors of adenoma and cancer,”
N. Engl. J. Med., 339 1277
–1284
(1998). 0028-4793 Google Scholar
E. McLellan,
A. Medline, and
R. P. Bird,
“Sequential analyses of the growth and morphological characteristics of aberrant crypt foci: putative preneoplastic lesions,”
Cancer Res., 51 5270
–5274
(1991). 0008-5472 Google Scholar
E. McLellan and
R. P. Bird,
“Effect of disulfiram on 1,2-dimethylhydrazine- and azoxymethane-induced aberrant crypt foci,”
Carcinogenesis, 12 969
–972
(1991). 0143-3334 Google Scholar
A. N. Esgiar,
R. N. G. Naguib,
B. S. Sharif,
M. K. Bennet, and
A. Murray,
“Microscopic image analysis for quantitative measurement and feature identification of normal and cancerous colonic mucosa,”
IEEE Trans. Inf. Technol. Biomed., 2
(3), 197
–203
(1998). 1089-7771 Google Scholar
H. Furukawa,
S. Asai,
T. Moriyasu,
H. Ono, and
T. Saiga,
“Three-dimensional reconstruction of the mucosa from sequential sections of biopsy specimens for understanding diseasae progress in ulcerative colitis,”
Dig. Endosc., 12 317
–321
(2000). 0915-5635 Google Scholar
S. Tamura,
Y. Furuya,
T. Tadokoro,
Y. Higashidani,
Y. Yokoyama,
K. Araki, and
S. Onishi,
“Pit pattern and three-dimensional configuration of isolated crypts from the patients with colorectal neoplasm,”
J. Gastroenterol., 37
(10), 798
–806
(2002). 0944-1174 Google Scholar
A. Ohta,
K. Tominaga, and
Y. Sakai,
“Efficacy of magnifying colonoscopy for the diagnosis of colorectal neoplasia: comparison with histopathological findings,”
Dig. Endosc., 16 308
–314
(2004). 0915-5635 Google Scholar
M. Haefner,
L. Brunauer,
H. Payer,
R. Resch,
F. Wrba,
A. Gangl,
A. Vecsei, and
A. Uhl,
“Pit pattern classification of zoom-endoscopical colon images using DCT and FFT,”
159
–164
(2007). Google Scholar
M. B. Kimmey,
R. W. Martin,
R. C. Gaggitt,
K. Y. Wang,
D. W. Franklin, and
F. E. Silverstein,
“Histologic correlates of gastrointestinal ultrasound images,”
Gastroenterology, 96 433
–441
(1989). 0016-5085 Google Scholar
D. P. Hurlstone,
S. S. Cross,
R. Slater, and
S. Brown,
“Detecting diminutive colorectal lesions at colonoscopy: A randomised controlled trial of pan-colonic versus targeted chromoscopy,”
Gut, 53 376
–380
(2004). 0017-5749 Google Scholar
K. Gono,
T. Obi,
M. Yamaguchi,
N. Ohyama,
H. Machida,
Y. Sano,
S. Yoshida,
Y. Hamamoto, and
T. Endo,
“Appearance of enhanced tissue features in narrow-band endoscopic imaging,”
J. Biomed. Opt., 9 568
–577
(2004). https://doi.org/10.1117/1.1695563 1083-3668 Google Scholar
J. E. East,
N. Suzuki,
M. Stavrinidis,
T. Guenther,
H. J. W. Thomas, and
B. P. Saunders,
“Narrow band imaging for colonoscopic surveillance in hereditary non-polyposis colorectal cancer,”
Gut, 57
(1), 65
–70
(2008). 0017-5749 Google Scholar
J. E. East,
N. Suzuki,
A. von Herbay, and
B. P. Saunders,
“Narrow band imaging with magnification for dysplasia detection and pit pattern assessment in ulcerative colitis surveillance: A case with multiple dysplasia associated lesions or masses,”
Gut, 55
(10), 1432
–1435
(2006). 0017-5749 Google Scholar
M. V. J. Sivak, Gastroenterologic Endoscopy, W. B Saunders Company, New York
(2000). Google Scholar
D. P. Hurlstone,
S. S. Cross,
I. Adam,
A. J. Shorthouse,
S. Brown,
D. S. Sanders, and
A. J. Lobo,
“Efficacy of high magnification chromoscopic colonoscopy for the diagnosis of neoplasia in flat and depressed lesions of the colorectum: A prospective analysis,”
Gut, 53
(2), 284
–290
(2004). 0017-5749 Google Scholar
H. Inoue,
S. Kudo, and
A. Shiokawa,
“Technology insight: Laser-scanning confocal microscopy and endocytoscopy for cellular observation of gastrointestinal tract,”
Nat. Clin. Pract. Gastroenterol. Hepatol., 2
(1), 31
–37
(2005). 1743-4378 Google Scholar
A. Polglase,
W. McLaren,
S. A. Skinner,
R. Kiesslich,
M. F. Neurath, and
P. Delaney,
“A fluorescence confocal endomicroscope for in vivo microscopy of the upper- and lower-GI tract,”
Gastrointest. Endosc., 62
(5), 686
–695
(2005). 0016-5107 Google Scholar
R. Kiesslich,
J. Burg,
M. Vieth,
J. Gnaendiger,
M. Enders,
P. Delaney,
A. Polglase,
W. McLaren,
D. Janell,
S. Thomas,
B. Nafe,
P. R. Galle, and
M. F. Neurath,
“Confocal laser endoscopy for diagnosing intraepithelial neoplasias and colorectal cancer in vivo,”
Gastroenterology, 127
(3), 706
–713
(2004). https://doi.org/10.1053/j.gastro.2004.06.050 0016-5085 Google Scholar
R. Kiesslich,
A. Hoffman,
A. Goetz,
S. Biesterfeld,
M. Vieth,
P. R. Galle, and
M. F. Neurath,
“In vivo diagnosis of collagenous colitis by confocal endomicoscopy,”
Gut, 55 591
–592
(2006). 0017-5749 Google Scholar
G. J. Tearny,
M. E. Brezinski,
B. E. Bouma,
S. A. Boppart,
C. Pitris,
J. F. Southern, and
J. G. Fujimoto,
“In vivo endoscopic optical biopsy with optical coherence tomography,”
Science, 276 2037
–2039
(1997). https://doi.org/10.1126/science.276.5321.2037 0036-8075 Google Scholar
A. M. Rollins,
R. Ung-arunyawee,
A. Chak,
R. C. K. Wong,
K. Kobayashi, M. V. Sivak Jr., J. A. Izatt,
“Real-time in vivo-imaging of human gastrointestinal ultrastructure by use of endoscopic optical coherence tomography with a novel efficient interferometer design,”
Opt. Lett., 24
(19), 1358
–1360
(1999). 0146-9592 Google Scholar
B. E. Bouma,
G. J. Tearney,
C. C. Compton, and
N. S. Nishi,
“High-resolution imaging of the human esophagus and stomach in vivo using optical coherence tomography,”
Gastrointest. Endosc., 51
(4), 467
–474
(2000). 0016-5107 Google Scholar
P. R. Pfau, M. V. Sivak Jr., A. Chak,
M. Kinnard,
R. C. Wong,
G. A. Isenberg,
J. A. Izatt,
A. M. Rollins, and
V. Westphal,
“Criteria for the diagnosis of dysplasia by endoscopic optical coherence tomography,”
Gastrointest. Endosc., 58
(2), 196
–202
(2003). 0016-5107 Google Scholar
D. C. Adler,
Y. Chen,
R. Huber,
J. Schmitt,
J. Connolly, and
J. G. Fujimoto,
“Three-dimensional endomicroscopy using optical coherence tomography,”
Nat. Photonics, 1 709
–716
(2007). 1749-4885 Google Scholar
S. H. Yun,
G. J. Tearney,
B. J. Vakoc,
M. Shishkov,
W. Y. Oh,
A. E. Desjardins,
M. J. Suter,
R. C. Chan,
J. A. Evans,
I. Jang,
N. S. Nishioka,
J. F. de Boer, and
B. E. Bouma,
“Comprehensive volumetric optical microscopy in vivo,”
Nat. Med., 12
(12), 1429
–1433
(2006). 1078-8956 Google Scholar
P. Schmitz-Moormann and
G. W. Himmelmann,
“Does quantitative histology of rectal biopsy improve the differential diagnosis of Crohn’s disease and ulcerative colitis in adults?,”
Pathol. Res. Pract., 183
(4), 481
–488
(1988). 0344-0338 Google Scholar
D. Thompson,
P. H. Bartels,
H. G. Bartels,
P. W. Hamilton, and
J. M. Sloan,
“Knowledge-guided segmentation of colorectal histopathologic imagery,”
Anal Quant Cytol. Histol., 15
(4), 236
–246
(1993). 0884-6812 Google Scholar
A. N. Esgiar,
R. N. G. Naguib,
B. S. Sharif,
M. K. Bennet, and
A. Murray,
“Fractal analysis in the detection of colonic cancer images,”
IEEE Trans. Inf. Technol. Biomed., 6
(1), 54
–58
(2002). 1089-7771 Google Scholar
P. W. Hamilton,
D. C. Allen,
P. C. H. Watt,
C. C. Patterson, and
J. D. Biggart,
“Classification of normal colorectal mucosa and adenocarcinoma by morphometry,”
Histopathology, 11 901
–911
(1987). 0309-0167 Google Scholar
M. P. Tjoa and
S. M. Krishnan,
“Feature extraction for the analysis of colon status from the endoscopic images,”
Biomed. Eng., 2
(9), 1
–17
(2003). 0006-2898 Google Scholar
Z. Hu and
A. M. Rollins,
“Quasi-telecentric optical design of a microscope-compatible OCT scanner,”
Opt. Express, 13
(17), 6407
–6415
(2005). https://doi.org/10.1364/OPEX.13.006407 1094-4087 Google Scholar
R. C. Gonzalez and
R. E. Woods,
“Digital image processing,”
Addison-Wesley, New York
(1992). Google Scholar
R. C. Gonzalez and
R. E. Woods,
“Digital image processing,”
Prentice Hall, New York
(2007). Google Scholar
N. Otsu,
“A threshold selection method from gray-level histograms,”
IEEE Trans. Syst. Man Cybern., SMC-9
(1), 62
–66
(1979). https://doi.org/10.1109/TSMC.1979.4310076 0018-9472 Google Scholar
L. Vincent,
“Morphological grayscale reconstruction in image analysis: applications and efficient algorithms,”
IEEE Trans. Image Process., 2
(2), 176
–201
(1993). https://doi.org/10.1109/83.217222 1057-7149 Google Scholar
P. Salembier,
“Morphological multiscale segmentation for image coding,”
Signal Process., 38 359
–386
(1994). https://doi.org/10.1016/0165-1684(94)90155-4 0165-1684 Google Scholar
L. Vincent and
S. Pierre,
“Watersheds in digital spaces: an efficient algorithm based on immersion simulations,”
IEEE Trans. Pattern Anal. Mach. Intell., 13
(6), 583
–598
(1991). https://doi.org/10.1109/34.87344 0162-8828 Google Scholar
Y. Huang and
D. Chen,
“Watershed segmentation for breast tumor in 2-D sonography,”
Ultrasound Med. Biol., 30
(5), 625
–632
(2004). https://doi.org/10.1016/j.ultrasmedbio.2003.12.001 0301-5629 Google Scholar
D. Reniers and
A. Telea,
“Skeleton-based hierarchical shape segmentation,”
(2007). Google Scholar
R. E. Rudolph,
J. A. Dominitz,
J. W. Lampe,
L. Levy,
P. Qu,
S. S. Li,
P. D. Lampe,
M. P. Bronner, and
J. D. Potter,
“Risk factors for colorectal cancer in relation to number and size of aberrant crypt foci in humans,”
Cancer Epidemiol. Biomarkers Prev., 14
(3), 605
–608
(2005). 1055-9965 Google Scholar
K. Seike,
K. Koda,
K. Odea,
C. Kosugi,
K. Shimizu,
M. Nishimura,
M. Shioiri,
S. Takano,
H. Ishikura, and
M. Miyazaki,
“Assessment of rectal aberrant crypt foci by standard chromoscopy and its predictive value for colonic advanced neoplasms,”
Aust. Occup. Ther. J., 101
(6), 1362
–1369
(2006). Google Scholar
A. N. Esgiar,
R. N. G. Naguib,
B. S. Sharif,
M. K. Bennet, and
A. Murray,
“Automated feature extraction and identification of colon carcinoma,”
Anal Quant Cytol. Histol., 20 297
–301
(1998). 0884-6812 Google Scholar
M. Haefner,
M. Liedlgruber,
A. Puespoek,
S. Thomas,
R. Schoefl,
F. Wrba,
A. Gangl, and
A. Uhl,
“Computer-assisted pit pattern analysis of colonic lesions,”
Gastrointest. Endosc., 63
(5), AB247
(2006). 0016-5107 Google Scholar
H. Yaegashi,
Y. Zhang,
F. Tezuka,
T. Takahashi, and
M. Fukumoto,
“Computer-assisted 3D mapping and morphometry of dysplastic zones in endoscopically resected colonic adenomas,”
J. Pathol., 191 143
–149
(2000). 0022-3417 Google Scholar
Q. Huang,
N. Fukami,
H. Kashida,
T. Takeuchi,
E. Kogure,
T. Kurahashi,
E. Stahl,
Y. Kudo,
H. Kimata, and
S. Kudo,
“Interobserver and intra observer consistency in the endoscopic assessment of colonic pit patterns,”
Gastrointest. Endosc., 60
(4), 520
–526
(2004). 0016-5107 Google Scholar
D. P. Hurlstone,
S. S. Cross,
I. Adam,
A. J. Shorthouse,
S. Brown,
D. S. Sander, and
A. J. Lobo,
“Endoscopic morphological anticipation of submucosal invasion in flat and depressed colorectal lesions: Clinical implications and subtype analysis of the kudo type V pit pattern using high-magnification-chromoscopic colonoscopy,”
Int. J. Colorectal Dis., 6
(5), 369
–375
(2004). 0179-1958 Google Scholar
D. P. Hurlstone,
M. Karajeh,
D. S. Sanders,
S. K. Drew, and
S. S. Cross,
“Rectal aberrant crypt foci indentification using high-magnification-chromoscopic colonoscopy: Biomarkers for flat and depressed neoplasia,”
Am. J. Gastroenterol., 100 1
–7
(2005). 0002-9270 Google Scholar
A. K. Gupta,
T. P. Pretlow, and
R. E. Schoen,
“Aberrant crypt foci: What we know and what we need to know,”
Nat. Clin. Pract. Gastroenterol. Hepatol., 5
(5), 526
–533
(2007). 1743-4378 Google Scholar
H. K. Roy,
Y. Liu,
R. K. Wali,
Y. L. Kim,
A. K. Kromine,
M. J. Goldberg, and
V. Backman,
“Four-dimensional elastic light-scattering fingerprints as preneoplastic markers in the rat model of colon carcinogenesis,”
Gastroenterology, 126
(4), 1071
–1081
(2004). https://doi.org/10.1053/j.gastro.2004.01.009 0016-5085 Google Scholar
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||

