|
|
|
Numerous areas of science employ Monte Carlo (MC) methods to simulate complex processes. Light propagation in random media, often referred to as photon migration, is an area of physics in which such methods are of great importance. Prime examples include radiative transfer in highly scattering materials such as biological tissues, clouds, and pharmaceuticals. There, MC simulation is generally considered the gold standard of modeling and is used to investigate complex systems and processes, to validate simpler models, as well as to evaluate data. Several authors have, for example, used MC to simulate photon migration in complex structures, such as the adult human head.1 The technique has also been used to understand the generation of fluorescence and Raman signals in random media.2, 3 Furthermore, the validity of the widely used diffusion approximation of radiative transport theory is typically tested by comparing its predictions with the outcome of MC simulations.4, 5, 6 When diffusion theory (or other approximative models) is invalid and cannot be used to evaluate experimental data, the direct use of MC simulations plays an important roll. Simpson evaluated integrating sphere measurements using a so-called MC inversion technique to determine the near-IR optical properties of human skin ex vivo.7 Bevilacqua employed MC to solve the inverse problem of deriving optical properties from spatially resolved spectroscopic data generated by systems used for intraoperative characterization of human brain.8 Similarly, for breast cancer diagnostics, Palmer and Ramanujam employed inverse MC to derive absorption and reduced scattering coefficients from point measurements of diffuse reflectance.9 Furthermore, Hayakawa 10 have shown that MC-based data evaluation can be used to extract optical properties of tissue heterogeneities (i.e., solve inverse two-region problems). In the case of time-domain photon migration, Alerstam have shown a general necessity for MC-based evaluation of experimental photon time-of-flight spectroscopy11, 12 (TOFS), and Svensson used this approach to extract in vivo optical and physiological characteristics of human prostate tissue from TOFS data.13 The main drawback of MC methods is the extensive computational burden. The general development of faster and faster computer processors is, of course, of great importance in making MC feasible for photon migration modeling, but other developments are equally important. Various variance reduction techniques are frequently used, the most fundamental being the one where absorption is modeled by reducing photon weights rather the by photon termination.14 Zolek 15 showed that MC simulations of photon migration in tissue can be made a factor 4 faster by employing certain approximations during calculations of logarithmic and trigonometric functions. A conceptually different, but extremely efficient, method to reduce the computational cost is to avoid the requirement for multiple simulations at different optical properties by rescaling single MC simulations.11, 12, 16, 17, 18 Rescaling with respect to different absorption coefficients is straightforward, while scaling with respect to scattering coefficients is possible only in certain simple geometries. This letter presents a technical approach that dramatically increases the speed of MC simulations of photon migration. A $110 programmable graphics processing unit (GPU; NVIDIA GeForce 8800GT) is used for parallel computing, and the fully parallelable character of photon migration simulations renders the approach particularly effective. We choose to exemplify the great value of the GPU for the field of biomedical optics by considering time-resolved MC simulations of photon migration in semi-infinite (half-space) geometry. Since the fundamentals of MC can be found elsewhere,14 we focus on issues particular to parallel computing with GPUs. Our work is based on NVIDIA’s Compute Unified Device Architecture (CUDA), a hardware and software architecture for general-purpose computing on graphics processing units19, 20 (GPGPU). Due to the great computational power of the GPU, the GPGPU method has proven valuable in various areas of science and technology.21 Although GPGPU for MC simulations of photon migration is neither described nor used in the scientific literature, it has been suggested.22 The approach is, on the other hand, used in various other fields relying on MC methods.20, 23 The modern GPU is a highly data-parallel processor, optimized to provide very high floating point arithmetic throughput for problems suitable to solve with a single program multiple data (SPMD) model. On a GPU, the SPMD model works by launching thousands of threads running the same program (called the kernel) working on different data. The ability of the GPU to rapidly switch between threads in combination with the high number of threads ensures the hardware is busy at all times. This ability effectively hides memory latency, and in combination with the several layers of very high bandwidth memory available in modern GPUs also improves GPU performance. CUDA programs are based on the C programming language with certain extensions to utilize the parallelism of the GPU. These extensions also provide very fast implementations of standard mathematical functions such as trigonometric functions, floating point divisions, logarithms, etc. The code defining the kernel looks almost like an ordinary scalar C function. The only major difference is that some effort must be made to optimize the function to work as efficiently as possible in a parallel environment, e.g., to minimize divergence among the threads and synchronize thread memory read/writes. For more detailed information on CUDA, see to the CUDA Programming Guide.20 The MC implementation used in this paper is a modified version of a previously described code for simulation of photon migration in semi-infinite, homogenously scattering, and nonabsorbing media.11 Briefly, photons are launched into the medium at the origin, perpendicular to the surface. Photon transport is simulated until the photon escapes the medium or the total time of flight (TOF) exceeds a predefined maximum value, . If the photon exits the medium within the area of a fictive detector (in this paper, between 9.5 and from the origin), the TOF histogram time bin corresponding to the photon TOF is incremented by one. To make maximum use of the parallel computational ability of the GPU, a new photon is immediately launched when a photon is terminated. Otherwise, photons that take many steps before being terminated would significantly delay the launch of new photons. Using this method of launching photons, it is not computationally efficient to define an exact number of photons to be simulated. Instead, defining a certain number of photon steps per thread is beneficial. As long as this number is large enough (so that a large number of full photon paths can be simulated by all those steps), this approach will not influence the simulation results. As all current CUDA-enabled devices are optimized for floating point precision, this was the precision used in our code (the new NVIDIA GTX-200 architecture does, however, support double, , floating point precision). As this differs from most CPU MC implementations, the results from our precision code were compared to results from our white Monte Carlo (WMC) implementation, where the simulation is performed in double precision.11 One very important aspect of running MC simulations in parallel is the choice of method of random number generation. Running many concurrent threads using the same random number generator (RNG) would most likely result in many threads performing exactly the same computations if the generator was seeded with a timestamp, as is commonly done in scalar MC programs. Even if the RNG is perfectly seeded, one may run into the problem of using the same sequence of random numbers if the number of parallel processes is large, the number of required random numbers is large, and the period of the RNG is short. To overcome this problem one can use a RNG with an extreme period, such as the Mersenne Twister24 and make sure that the different threads are properly and differently seeded. However, in the NVIDIA GPU architecture, the available memory per thread is limited, making use of the relatively memory-hungry Mersenne Twister RNG less preferable. Instead we have employed the multiply-with-carry (MWC) RNG, featuring a period of about . The beauty of this approach is that different RNG parameters can be chosen for each individual thread so that they all generate unique sequences of random numbers while requiring a minimum amount of memory. If a longer period or better randomness is required, the MWC RNG can be easily be extended to a lag- MWC or a KISS RNG (at the cost of an increased need of memory and computation). A good overview of the MWC RNG and its derivatives can be found in Ref. 25 To compare CPU and GPU speed, the code developed for the CUDA implementation was carefully converted to a CPU equivalent (including the MWC RNG) to perform exactly the same computations as the CUDA code (e.g., both using the same random number sequences and single precision). Note, however, that as the two different implementations does not use exactly the same functions to approximate, for example, trigonometric functions, they can never yield exactly the same result (although they should be statistically equivalent). Simulations were performed for a semi-infinite material defined by , , , and . Photons were tracked for , and the source-detector separation was set to . The GPU code launched 26,880 simultaneous threads performing 500,000 photon steps each, while the CPU sequentially ran 26,880 independent “virtual threads” (each comprising 500,000 photon steps). The GPU used was a low-cost ($110 at the time of writing) NVIDIA 8800GT, while the CPU was a Intel Pentium 4 HT . The results of these two simulations and a third reference MC simulation (double precision WMC) can be seen in Fig. 1 . Fig. 1TOF histogram of three different MC simulation methods. WMC (simulated using double precision) is used for reference. The WMC results comprise 2,133,796 detected photons (building the database took using all four cores of an Intel Core 2 Quad ). The GPU and CPU results (both simulated using single precision), which should be statistically equivalent, both comprise about 326,000 detected photons out of about full photon paths simulated. The simulation times were for the GPU (a NVIDIA 8800GT) and 8513s for the CPU (an Intel Pentium 4 HT ), i.e., the GPU proves to be faster. Comparing the CPU and GPU results to the WMC result validates the MC code for both implementation and confirms that single precision is sufficient as long as proper care is taken during calculations. 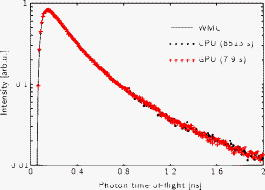 The CPU and GPU codes yield very similar result, both simulating about full photon paths. Although the two implementations perform the same task, the GPU implementation is about faster than the conventional CPU implementation. Both the CPU and GPU results agree well with the reference WMC simulation. This confirms that single-precision floating point calculations are sufficient as long as proper care is taken, e.g., normalization of the direction vector after each direction change to avoid the accumulation of small numerical errors. The massive speedup of GPGPU versus traditional CPU MC simulations should be of great interest to the field of biomedical optics. With the results presented in this paper, MC takes a huge leap toward being a viable option in many applications of photon migration modeling. This includes, for example, its use for data evaluation in geometries not eligible for the scalable WMC method (e.g., heterogenous media or media featuring complex boundaries). While there are no fundamental architectural limitations restricting the addition of, e.g., complex heterogeneities/boundaries and more advanced photon scoring methods, note that such additions will influence performance (depending on MC implementation and memory access patterns). The interest in fast MC for biomedical applications is reflected in previous work on parallel computing. Kirkby and Delpy used a 24-computer network for simulations of light transport in tissue,26 and Colasanti used a CRAY parallel processor computer with 128 processors.27 The use of multiple computers (or processors) is, however, both inconvenient and expensive, especially if such systems are to reach the extreme speed of a GPU. This is reflected by the fact that numerous areas of computational science are starting to use20, 21 programmable GPUs. Yet another interesting aspect of GPGPU is the rapid development of parallel processors for graphics processing. While the present work presents a speedup using a relatively low end card, the current NVIDIA-generation high-end cards can provide significantly higher performance. A few such cards in parallel can, for example, provide a 10-fold increase in computing power (providing an estimated speedup). Also keep in mind the high-end cards from ATI/AMD, as well as the future release of the Intel Larrabee architecture. In conclusion, we presented a GPGPU as a technical approach to MC simulation of photon migration, providing a massive speedup over traditional CPU code. This tremendous reduction in simulation time should make MC an attractive tool in many applications involving photon migration. Finally, we would like to invite everyone interested in GPGPU MC simulation of photon migration to download current and future versions of our CUDA MC code from our webpage: http://www.atomic.physics.lu.se/biophotonics/ AcknowledgmentsThe authors gratefully acknowledge financial support from the Swedish Research Council and from EC UB Nano. We also acknowledge the work on random number generation using CUDA by Steven Gratton generously made available at http://www.ast.cam.ac.uk/~stg20/. ReferencesD. A. Boas,
J. P. Culver,
J. J. Stott, and
A. K. Dunn,
“Three dimensional Monte Carlo code for photon migration through complex heterogeneous media including the adult human head,”
Opt. Express, 10
(3), 159
–170
(2002). 1094-4087 Google Scholar
K. Vishwanath,
B. Pogue, and
M. A. Mycek,
“Quantitative fluorescence lifetime spectroscopy in turbid media: comparison of theoretical, experimental and computational methods,”
Phys. Med. Biol., 47
(18), 3387
–3405
(2002). https://doi.org/10.1088/0031-9155/47/18/308 0031-9155 Google Scholar
N. Everall,
T. Hahn,
P. Matousek,
A. W. Parker, and
M. Towrie,
“Photon migration in Raman spectroscopy,”
Appl. Spectrosc., 58
(5), 591
–597
(2004). https://doi.org/10.1366/000370204774103426 0003-7028 Google Scholar
S. T. Flock,
M. S. Patterson,
B. C. Wilson, and
D. R. Wyman,
“Monte Carlo modeling of light-propagation in highly scattering tissues: I model predictions and comparison with diffusion-theory,”
IEEE Trans. Biomed. Eng., 36
(12), 1162
–1168
(1989). https://doi.org/10.1109/TBME.1989.1173624 0018-9294 Google Scholar
M. S. Patterson,
B. Chance, and
B. C. Wilson,
“Time resolved reflectance and transmittance for the noninvasive measurement of tissue optical-properties,”
Appl. Opt., 28
(12), 2331
–2336
(1989). 0003-6935 Google Scholar
F. Martelli,
D. Contini,
A. Taddeucci, and
G. Zaccanti,
“Photon migration through a turbid slab described by a model based on diffusion approximation: II. Comparison with Monte Carlo results,”
Appl. Opt., 36
(19), 4600
–4612
(1997). https://doi.org/10.1364/AO.36.004600 0003-6935 Google Scholar
C. R. Simpson,
M. Kohl,
M. Essenpreis, and
M. Cope,
“Near-infrared optical properties of ex vivo human skin and subcutaneous tissues measured using the Monte Carlo inversion technique,”
Phys. Med. Biol., 43
(9), 2465
–2478
(1998). https://doi.org/10.1088/0031-9155/43/9/003 0031-9155 Google Scholar
F. Bevilacqua,
D. Piguet,
P. Marquet,
J. D. Gross,
B. J. Tromberg, and
C. Depeursinge,
“In vivo local determination of tissue optical properties: applications to human brain,”
Appl. Opt., 38
(22), 4939
–4950
(1999). https://doi.org/10.1364/AO.38.004939 0003-6935 Google Scholar
G. M. Palmer and
N. Ramanujam,
“Monte Carlo-based inverse model for calculating tissue optical properties. Part i: theory and validation on synthetic phantoms,”
Appl. Opt., 45
(5), 1062
–1071
(2006). https://doi.org/10.1364/AO.45.001062 0003-6935 Google Scholar
C. K. Hayakawa,
J. Spanier,
F. Bevilacqua,
A. K. Dunn,
J. S. You,
B. J. Tromberg, and
V. Venugopalan,
“Perturbation Monte Carlo methods to solve inverse photon migration problems in heterogeneous tissues,”
Opt. Lett., 26
(17), 1335
–1337
(2001). https://doi.org/10.1364/OL.26.001335 0146-9592 Google Scholar
E. Alerstam,
S. Andersson-Engels, and
T. Svensson,
“White Monte Carlo for time-resolved photon migration,”
J. Biomed. Opt., 13
(4), 041304
(2008). https://doi.org/10.1117/1.2950319 1083-3668 Google Scholar
E. Alerstam,
S. Andersson-Engels, and
T. Svensson,
“Improved accuracy in time-resolved diffuse reflectance spectroscopy,”
Opt. Express, 16
(14), 10434
–10448
(2008). https://doi.org/10.1364/OE.16.010434 1094-4087 Google Scholar
T. Svensson,
E. Alerstam,
M. Einarsdóttír,
K. Svanberg, and
S. Andersson-Engels,
“Towards accuracte in vivo spectroscopy of the human prostate,”
J. Biophoton., 1
(3), 200
–203
(2008). Google Scholar
L. H. Wang,
S. L. Jacques, and
L. Q. Zheng,
“MCML Monte Carlo modeling of light transport in multilayered tissues,”
Comput. Methods Programs Biomed., 47
(2), 131
–146
(1995). https://doi.org/10.1016/0169-2607(95)01640-F 0169-2607 Google Scholar
N. Zolek,
A. Liebert, and
R. Maniewski,
“Optimization of the Monte Carlo code for modeling of photon migration in tissue,”
Comput. Methods Programs Biomed., 84
(1), 50
–57
(2006). 0169-2607 Google Scholar
R. Graaff,
M. H. Koelink,
F. F. M. Demul,
W. G. Zijlstra,
A. C. M. Dassel, and
J. G. Aarnoudse,
“Condensed Monte Carlo simulations for the description of light transport,”
Appl. Opt., 32
(4), 426
–434
(1993). 0003-6935 Google Scholar
A. Pifferi,
P. Taroni,
G. Valentini, and
S. Andersson-Engels,
“Real-time method for fitting time-resolved reflectance and transmittance measurements with a Monte Carlo model,”
Appl. Opt., 37
(13), 2774
–2780
(1998). https://doi.org/10.1364/AO.37.002774 0003-6935 Google Scholar
A. Kienle and
M. S. Patterson,
“Determination of the optical properties of turbid media from a single Monte Carlo simulation,”
Phys. Med. Biol., 41
(10), 2221
–2227
(1996). https://doi.org/10.1088/0031-9155/41/10/026 0031-9155 Google Scholar
T. R. Halfhill,
“Parallel processing with CUDA,”
(2008) Google Scholar
D. Blythe,
“Rise of the graphics processor,”
Proc. IEEE, 96
(5), 761
–778
(2008). 0018-9219 Google Scholar
K. Gossage, Google Scholar
S. Tomov,
M. McGuigan,
R. Bennett,
G. Smith, and
J. Spiletic,
“Benchmarking and implementation of probability-based simulations on programmable graphics cards,”
Comput. Graph., 29
(1), 71
–80
(2005). 0097-8930 Google Scholar
M. Matsumoto and
T. Nishimura,
“Mersenne twister: a 623-dimensionally equidistributed uniform pseudorandom number generator,”
ACM Trans. Model. Comput. Simul., 8
(1), 3
–30
(1998). https://doi.org/10.1145/272991.272995 1049-3301 Google Scholar
G. Marsaglia,
“Random number generators,”
J. Mod. Appl. Stat. Meth., 2
(1), 2
–13
(2003). Google Scholar
D. R. Kirkby and
D. T. Delpy,
“Parallel operation of Monte Carlo simulations on a diverse network of computers,”
Phys. Med. Biol., 42
(6), 1203
–1208
(1997). https://doi.org/10.1088/0031-9155/42/6/016 0031-9155 Google Scholar
A. Colasanti,
G. Guida,
A. Kisslinger,
R. Liuzzi,
M. Quarto,
P. Riccio,
G. Roberti, and
F. Villani,
“Multiple processor version of a Monte Carlo code for photon transport in turbid media,”
Comput. Phys. Commun., 132
(1–2), 84
–93
(2000). https://doi.org/10.1016/S0010-4655(00)00138-7 0010-4655 Google Scholar
|

