|
|
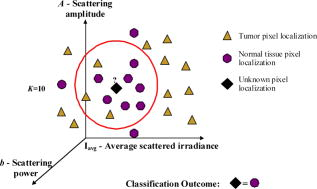
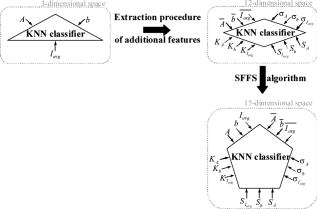
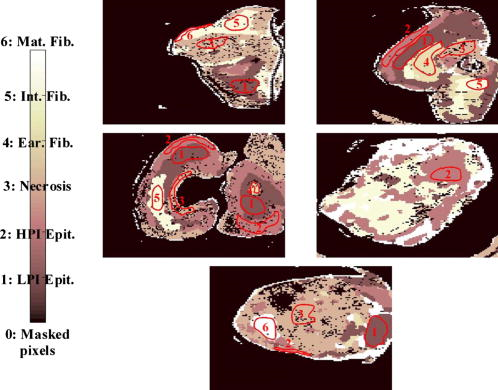
1.IntroductionSurgical microscopes are becoming more advanced all the time, and while there is great promise in their use, the problem still remains that the diffuse view seen by the surgeon does not allow analysis of the microscopic morphology of the tissue reliably. Many studies have looked at possible inclusion of color filtering,1, 2 polarization-based imaging,3, 4 or spectroscopic channel analysis5 in order to try and enhance the contrast at the margin between confirmed tumor and surrounding normal tissues. The creation of a tool that provides better delineation of the tissue at the microscopic level, with real-time viewing of the signal, in situ, would be a great asset, and therefore, it could gain clinical adoption readily. In this study, one approach to imaging tissue scatter spectra is used6 in conjunction with an automated classification approach to imaging. Scatter analysis of cells and tissues accomplished by angle-resolved or coherence-based methods has proved successful in the quantification the subcellular origin of certain features of tissue.7, 8, 9, 10, 11, 12 The measurement can be robust, and changes in scatter spectra are related to pathologic structures that occur in the tissue; and thus, measurement of this could provide a unique tool for guiding surgical resection if a way was developed to help the surgeon in data reduction and display in real time. A raster-scanning confocal reflectance imaging system to directly quantify tissue scatter in situ was previously designed and tested in tumor tissues.6 In addition, an attempt to establish a correlation between scatter changes and tissue morphologies was performed. The main conclusions of this study were that changes are subtle and the data are multiparametric. Therefore, an automated methodology to classify the encountered scatter changes according to their tissue subtypes is required before proceeding to clinical studies. An automated interpretation into what the signal means relative to the pathology has been designed here. The analysis was done in stages, with the first aim being the development of a methodology able to perform accurate tumor versus normal tissue discrimination, allowing reliable margin detection. However, the quantification of the scattering coefficient heterogeneity within tumor is also critical to treatment planning, because tumors can be extremely heterogeneous in terms of their fibrocystic and necrotic changes. Thus, the study here was done in these two stages and the automated identification process is described in detail for each. 2.Materials and Methods2.1.Tissue Scatter ImagingThe scatter imaging system consisted of a weakly confocal spectroscopic system having illumination and detection spot sizes smaller than one mean scattering length (typically, for tissue13), and a raster-scanning platform built using linear translation stages. This spot size was specifically chosen because it provides a scatter signal that does not have significant contributions from multiple scattering, making it essentially linearly dependent on the scatter coefficient. The instrument operates in the spectral waveband with a broadband fiber-coupled tungsten-halogen light source. An optical-fiber coupled to a CCD-based spectrometer was used for confocal spectroscopic detection, with a spectral resolution of . All spectral measurements were referenced to a Spectralon-based reflectance standard, which removes instrumental spectral response from the sample measurements and allows direct comparisons of the extracted scatter parameters across different samples. Average sample scanning time was , mainly due to the use of mechanical stages for raster scanning. Sufficient precautions were taken to prevent the sample from drying during the measurement sequence. A schematic and a more detailed description of the system can be found in a previous paper.6 In the presence of significant local absorption, for very small source-detector separations, the spectral reflectance can be estimated by an empirical relationship as follows: if the scattering and absorption coefficients are within the typical range found in tissue.14, 15 Parameter is the scattering amplitude, the scattering power, is the path length, is a constant proportional to the concentration of whole blood, and is the oxygen-saturation fraction. The extinction spectra of oxygenated and deoxygenated hemoglobin, and , were obtained from the Oregon Medical Laser Center database.16 Absorption contributions from other chromophores were assumed to be negligible in the waveband of interest (from ). Scattering parameters of interest for tissue morphology identification are and , along with the so-called average scattered irradiance, , which was calculated by integrating over a spectral range , which avoids the hemoglobin absorption peaks.6 The parameter is not entirely independent from and , but it provides a quick and direct estimate of average scatter without the need for an empirical model. Because, this parameter is not corrected for the effects of absorption in regions where higher than typical concentrations of blood are encountered,6 it could present subtle absorption-related features, in addition to the purely scatter-related features and , which could improve the accuracy of the discrimination algorithms. However, it should be noted that, in the absence of parameters and , alone has less value in terms of discriminating tissue types because this sensitivity to local absorption could lead to ambiguous situations where absorption artifacts are interpreted as scatter features.Human pancreatic tumor cells ASPC-1 were grown and injected subcutaneously in the flank region of male mice. Tumors were harvested seven weeks after injection when they measured in diameter and in thickness. Then they were dissected into thick sections and imaged using the mentioned scatter imaging system. In total, six tumor tissue sections harvested from four mice were imaged. After the measurement, the sample was routinely processed for histology evaluation by paraffin embedding, sectioning and H&E staining.6 The top full-view slide from each sample was used for pathologic analysis. A veterinary pathologist examined the H&E slides from each sample and identified several regions-of-interest corresponding to the observed tissue subtypes. These were classified under three major groups, namely, epithelium, fibrosis, and necrosis, with constituent subgroups: two distinct types of epithelial cells, according to the exhibited nucleus to cytoplasm ratio, were considered (low and high proliferation index) and regions exhibiting fibrosis were classified into early, intermediate, and mature fibrosis subgroups. Figure 1 shows an example of a pancreas tumor sample, where five regions of interest are shown overlaid on the scattered amplitude , scattering power , and average scattered irradiance images. Color bar scales of the scattering parameters are shown on the right of their corresponding images. Region 1 shows LPI tumor cells with less cellular density compared to HPI tumor cells found in region 2. Region 3 shows necrosis, and regions 4 and 5 show the early and intermediate stages of fibrosis, respectively. Apart from the tumor tissue samples mentioned above, -thick tissue sections of normal pancreas were harvested from three male mice and were imaged using the scatter imaging system. 2.2.Classification Methods2.2.1.-nearest neighborsThe -nearest neighbor (KNN) classifier17 consisted in the assignment of an unclassified vector using the closest vectors found in the training set. This approach can naturally deal with multiclass data while some of the more advanced classifiers, such as support vector machines (SVM),18 require the bridging of results from a combinatorial set of such classifiers to simulate multiclass parameters.19 Using the KNN classifier, both normal versus tumor sample segmentation and the discrimination of pathologically distinct tumor regions were performed using the same methodology. A schematic of the KNN process for classifying normal versus tumor tissue using three independent scatter-related parameters is depicted in Fig. 2 . The map was populated by points, each of which represents a tissue pixel with its own set of scattering parameters. Every pixel inside the predefined regions of interest was considered as a vector in a three-dimensional space (hereinafter called feature space), and it was plotted in the map using the three numerical values of its scattering parameters as the coordinate values. Because KNN is based on considering that similar data values should belong to the same class or region, the normal tissue or tumor in this case, then pixels with the most similar scattering parameters to an unclassified pixel were initially determined. This similarity was estimated in terms of the Euclidean distance where and are the two compared tissue pixel localizations, and , and stand for their scattering amplitude, scattering power, and average scattered irradiance, respectively. The unclassified pixel will be assigned to the most numerous tissue type (normal or tumor) among the closest neighbors. Therefore, this classifier has only one independent parameter, or the number of neighbors, to consider. In the proposed methodology, different values of the parameter are evaluated. Figure 3 shows in more detail how the KNN classifier works for a fixed value of parameter . The unknown black tissue pixel localization needs to be assigned either to tumor or normal tissue classes, depicted with triangles and circles, respectively. The first step is to determine the nearest ten neighbors as given by Eq. 2. The resulting ten pixel localizations are the ones surrounded by the red circle. As seven pixels out of ten are normal tissue and only three pixels are tumor, the outcome of the KNN classifier indicates that the unknown pixel corresponds to normal tissue.Fig. 2Schematic of the normal versus tumor discrimination by means of the KNN classifier, where the three measurement parameters are each considered a coordinate axis and the distance between points in this Cartesian space then defines how similar or different they are. 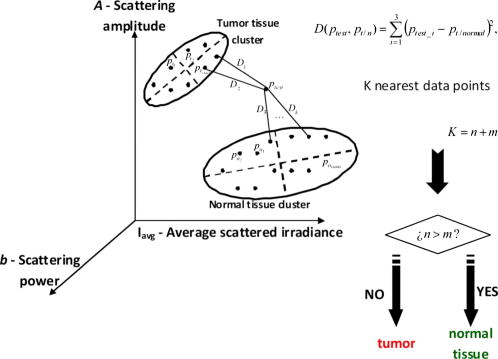 Because KNN is based on distances between sample points in the feature space, all parameters need to be normalized to prevent some features being more strongly weighted than others.19 Hence, all the three scattering parameters were unity normalized by the mean, before the classification process. Segmentation of pathologically distinct tumor regions by means of the KNN classifier was similar to the normal versus tumor classification described above, except for the fact that there were six different classes (low proliferation index and high proliferation index epithelium, necrosis, early, intermediate and mature fibrosis) and the unknown pixel was assigned to the most numerous class among these six. 2.2.2.Additional statistical data for the KNN classificationAs the number of features in a data set varies from 1 to , any classifier accuracy rises to a maximum and then falls back asymptotically.20 In the segmentation of the tumor regions, the six different tissue morphologies mentioned above have to be discriminated from a data set that lies in a three-dimensional space. Therefore, an additional feature extraction from the actual data set is required to decrease the discrimination error. In Ref. 19 a three-step automated breast-tissue classification methodology is proposed. The breast area is first segmented into fatty versus dense mammographic tissue, and the second step precisely consists of the extraction of morphological features by means of statistics calculations within the previously segmented breast areas. Because segmentation does not apply here (because that is the aim of the proposed methodology), a square spatial vicinity centered in each pixel location is defined. Then the first four statistical moments (mean, standard deviation, skewness, and kurtosis) of each scattering parameter are computed inside that vicinity region. Statistics are either concatenated with the fitted scattering parameters themselves or can be used on their own to form higher dimensional feature spaces. This procedure is graphically described in Fig. 4 . Initially, each data point lies in a three-dimensional space (the space). Then the statistical moments of each point are estimated inside a moving window centered in the pixel of interest. These moments on their own form a 12-dimensional space, whereas if they are concatenated with the three input parameters, each data point becomes a vector in a 15-dimensional space. The third and forth moments, the skewness, and the kurtosis, respectively, measure the bias and the probability of outlier occurrence, respectively, of each scattering parameter distribution inside the defined window. The behavior of the methodology in terms of the classification error is studied as a function of the number of neighbors but also as a function of the window size, where the latter is defined as the side length of the squared vicinity region. The study of KNN classification was completed with all the parameters for varying spatial window sizes, and an optimal window size was used for the final process. In addition, a further study on the capability of both the scattering parameters and their extracted statistics (mean, standard deviation, skewness, and kurtosis) to discriminate the different tumor regions was then performed. In this study, the sequential floating forward selection (SFFS) algorithm21 was used because it is widely applied to reduce the dimensionality (i.e., the number of features) of spectral data prior to interpretation.22, 23 When processing spectral data, the feature calculates mean values at each one of the spectral bands and the aim of SFFS in this case is to select the spectral bands that best discriminate among the subject classes, out of the total number initial bands, so . The discrimination among the classes, or class separability, can be calculated performing different statistical computations.22 The same fundamental is employed to sort the scattering parameters and their statistical values according to their subtype discrimination capability. In this way, the first feature selected by the algorithm will be the one with the greatest changes according to the pathology. As in Ref. 22, the Bhattacharya statistical distance was employed to measure these changes. Therefore, the difference in a scattering parameter , with , between two tissue subtypes, and , is given by where and are the mean and the variance matrix of for tissue subtype . Because there are six different tissue subtypes, the globlal class separability measurement, , requires one to calculate the difference between every two subtypeswhere is each subtype probability and is the distance between subtypes and , as stated in Eq. 4.2.3.Quantitative Evaluation of Tissue Discrimination CapabilityIn order to accurately estimate the performance of the discrimination methodology a threefold cross-validation technique or procedure24, 25 was applied both in the tumor versus normal and tumor region discrimination. A total of within the regions of interest of three normal samples and within the regions of interest of five tumor samples were considered. This data set was divided into three nonoverlapping sets containing 6653 data points each (5533 of normal tissue and 1120 of tumor). Two of these sets were employed as the training set, and the other one, the so-called validation set, was used to calculate the error of the classifier. The latter is mathematically given by where misclassification means that the tissue or tumor type assigned in an automated manner to that pixel localization does not match the pathologist criterion. This procedure is repeated three times, each time with different training and validation sets. Finally, the estimated performance of the classifier was calculated by averaging the three resulting errors.Pixels corresponding to locations where the scatter data could not be reliably measured were tagged as masked pixels. These pixels were neither included in the training nor the validation sets of the cross-validation procedure. 3.Results3.1.Tumor versus Normal Tissue DiscriminationFigure 5 shows the actual three-dimensional map representing the pixel locations in the space [Fig. 5a] and its three corresponding two-dimensional maps: versus [Fig. 5b] versus [Fig. 5c] and versus [Fig. 5d]. Normal pixels appear well grouped, whereas a remarkable spreading is observed in the scattering parameters of tumor pixels. Fig. 5Grouped scatter plots for normal and tumor pixel localizations in the three input data parameters, , (a) space and (b–d) its corresponding two-dimensional spaces. 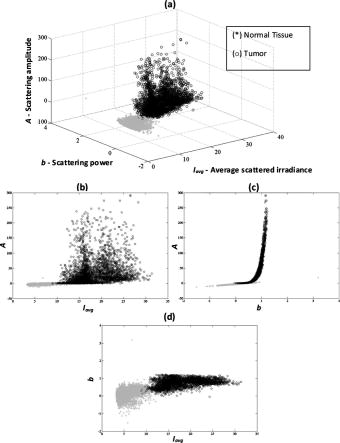 Figure 6 depicts the behavior of the KNN algorithm in the discrimination of normal and tumor tissue pixel locations as a function of the number of considered neighbors. Classification accuracy should be expected to increase with the number of neighbors because this reduces the influence of outliers (i.e., training data points assigned to a wrong class). There were no outliers here because the distinct sample regions were segmented as indicated by the pathologist. This absence of outliers, together with the spreading of tumor scattering parameters, caused an increase in the classification error percentage with the number of neighbors. This influence of the scattering parameter spreading on classification has been proved, employing as training set those pixels whose scattering parameters are close to the mean of each parameter and an independency of error probability on the number of neighbors has been obtained. Finally, discrimination results for all pixel locations in both normal and tumor samples, while considering only one neighbor, are presented in Figs. 7 and 8 . An absolute correlation was achieved between automatic and expert-based classification within the predefined regions of interest. In fact, the three normal samples in Fig. 7 were entirely correctly classified as well as the four tumor samples in Fig. 8. Only some errors are encountered within the regions of interest in the fifth tumor sample. A quantitative measurement of tissue-type discrimination, including percentages of pixels lost as masked pixels (inside the samples) and number of correctly identified localizations per sample is presented in Table 1 . Tissue discrimination capability of the approach is, however, measured in terms of the quantities presented in Fig. 6 because they are obtained through the cross-validation procedure described before, which removes the dependency of classification results on the training or test sets. Fig. 6Behavior of the methodology in normal versus tumor discrimination when the number of considered neighbors increases. 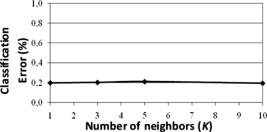 Fig. 7Normal and tumor pixel discrimination in normal samples using the three scatter measured parameters. 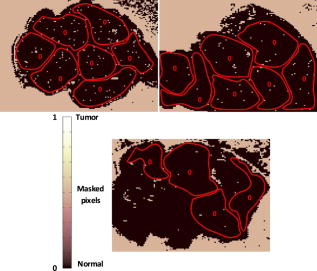 Fig. 8Normal and tumor pixel discrimination in tumor samples using the three scatter measured parameters.  Table 1Quantitative measurement of tissue-type discrimination.
3.2.Segmentation of Pathologically Distinct Tumor RegionsFigure 9 depicts some pixel localizations of the tumor samples in the space [Fig. 9a] and its three corresponding two-dimensional maps: versus [Fig. 9b], versus [Fig. 9c] and versus [Fig. 9d]. For visualization purposes only, those tumor pixels whose scattering parameters were within the interval , where is each of the three scattering parameters and and are respectively its mean and variance, are represented in the map. As expected, because of the subtle changes in the scattering parameters, pixel localizations do not appear well grouped according to their tissue subtype. The behavior of the KNN methodology in the discrimination of these pathologically distinct tumor regions as a function of the number of neighbors is shown in Fig. 10, where an increase in the classification error percentage occurs as the number of neighbors grows. Figure 11 displays the segmentation of the different regions in tumor samples by means of the KNN with , while Table 2 summarizes the same information as a confusion matrix, where each row represent the total number of pixel localizations in each tumor subtype (as defined by the pathologist inside the regions of interest). It seems that the automated classification accurately imitates the regions-of-interest identification process performed by the pathologist. However, once generality on the training and test sets is attained by means of the cross-validation procedure, an classification error occurs within these regions of interest, as shown in Fig. 10. Statistical data, as described in Sec. 2.2, was added to improve the performance of the methodology. Fig. 9Grouped scatter plots for all tumor sub-types in the (a) space and (b–d) its corresponding two-dimensional spaces. 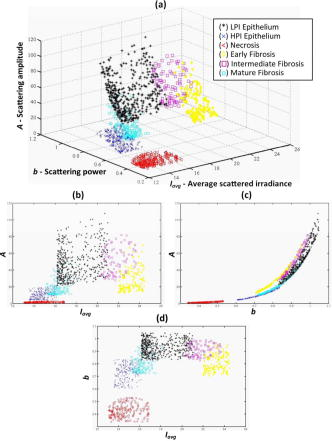 Fig. 10Behavior of the KNN methodology in tumor subtype discrimination as a function of the number of neighbors. 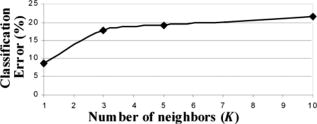 Table 2Overall confusion matrix in tumor subtype discrimination in the three input data parameters, A-b-Iavg space.
Classification error percentages in both 12-dimensional and 15-dimensional spaces are depicted in Fig. 12, which shows how accuracy decreases for window sizes smaller than 4, because statistics do not provide discriminant information and, then, the error decreases as the window size increases, up to a size of 12. In Fig. 12, a null window size means that no statistics calculation is performed (i.e., only the scattering parameters are employed in the discrimination). Classification accuracy is slightly better when statistics are concatenated with the scattering parameters. Therefore, this is the considered case in the study on the number of neighbors. Figure 13 shows again that the performance of the methodology decreases with the number of neighbors. Fig. 12Behavior of the KNN methodology in the higher dimensional spaces as a function of the window size employed in statistics calculation . 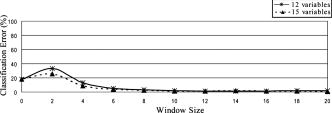 Fig. 13Classification error dependence on the number of neighbors, when both scattering parameters and their statistics were employed for discrimination.  3.2.1.Classification with spatial statistical data includedFigure 14 depicts the segmentation of one of the tumor samples into its distinct tumor regions in three different cases: employing only the scattering parameters (three-dimensional space), only their statistics (12-dimensional space) and both of them concatenated (15-dimensional space), while the corresponding confusion matrices are summarized in Table 3 . The improvement in the correlation between automated and expert-based sample segmentation within the regions of interest is barely perceptible. Sample boundary detection is significantly improved when statistics calculation is employed. Because when only the scattering parameters were considered, boundaries were improperly classified as mature fibrosis. The segmentation of the other four tumor samples in their distinct tumor regions is presented in Fig. 15 . These results are obtained employing a window size, concatenating the new calculated statistical parameters and the fitted scattering parameters, and their overall confusion matrix is presented in Table 4 . In this process, unclassified pixels were assigned to the same class as the closest one in the training set. Window sizes had to be as large as possible in such a way that the four statistical moments became relevant, but also small enough to assure that the moments are mostly computed for pixel localizations inside the same tissue subtype. A window size of assured the latter in most situations because it implies a window side of and, apart from that, classification error remains nearly constant for greater window sizes.26 Fig. 14Qualitative comparison among segmentations of one tumor sample in the distinct feature spaces are shown (scattering parameter feature space, statistical data, and both scattering parameters and their statistical values). 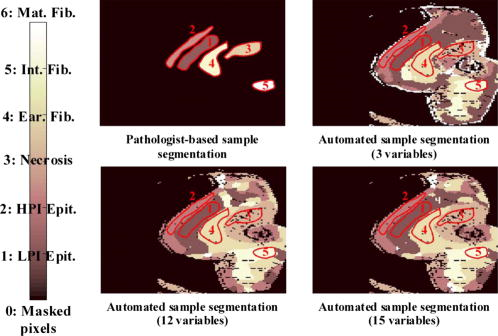 Fig. 15Segmentation performance of some tumor samples are illustrated, employing a window size in statistical calculation, with a 15-dimensional feature space . 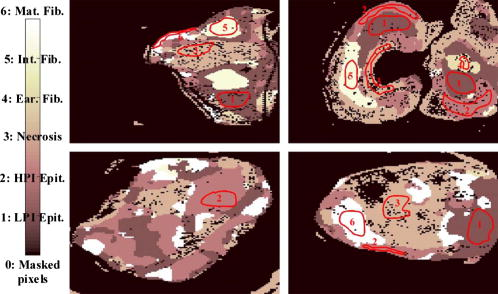 Table 3Tumor sample number 2 confusion matrices in tumor subtype discrimination in the distinct feature spaces: (a), scattering parameter feature space (b) statistical data, and both scattering parameters and their statistical values.
Table 4Overall confusion matrix in tumor subtype discrimination in the 15-dimensional feature space (K=1) .
The subtype separability measurements through the SFFS algorithm, as described in Sec. 2, were computed for the 15 features, the scattering parameters, and their statistics. The first five features selected according to this criterion and as a function of the window size are summarized in Table 5, where and are, as indicated before, the mean and variance of the scattering parameter , and stands for its kurtosis moment. For smaller window sizes, which means that mostly vicinity regions will be within the same tissue subtype, the mean scattering power is always selected as the most discriminant feature. Table 5Sorting of the scattering parameters and their statistics as a function of their tissue subtype discrimination capability.
4.DiscussionA strong correlation exists between the automated and expert-based normal versus tumor tissue segmentation of the samples as is seen in Figs 7 and 8. It is then justified to conclude that a consistent trend exists in the scattering parameters across these types. In the previous study without automated classification of the data;6 there was no obviously consistent trend in the scatter power images across the different tumor samples, while scatter amplitude exhibited a high corrupting influence because of coupling artifacts leading to the integrated scatter intensity variations. As a consequence, prior to the current work, this data set has been insufficient to achieve a proper identification of the different tumor regions. An classification error was then encountered in the interpretation of scatter changes according to their tissue subtypes without any statistical preprocessing. The concatenation of the scattering parameters of a pixel localization with their first four statistical moments or, even the employment of the statistical values on their own, allowed the achievement of a classification error of . This presumably indicates that it is necessary to smooth pixel-to-pixel variations to achieve reliable tumor region segmentations. This is a critically important finding because it means that in a raster-scanning approach the pixel-to-pixel variations could be high, but may actually not be useful information if the sampling volume is too small to provide a consistently smooth signal through a given tissue subtype. A great increase in the classification error was expected for large window sizes because it was assumed that distinct tumor regions were combined inside the same spatial vicinity region. However, classification error remains nearly constant unless the entire image was considered as one window and, then, as expected, the error matched the one achieved only when the three initial scattering parameters were considered. This is perhaps due to the fact that the distributions of moments are dominated by pixel localizations within the window that belong to the same tissue type. The proposed solution was to employ meaningful window sizes from the tissue morphology point of view. To assure this, the histograms of the sizes, width, and length, of the regions of interest identified by the pathologist were obtained. Most of them were shorter than , and therefore, larger window sizes would not be advisable to attain relevant statistical information. As shown in Table 5, the mean region values of scattering parameters turned out to be the strongest data set for classification of the distinct tumor pathologies. This agrees with the improvement achieved in the classification error by the introduction of statistical calculations and confirms that pixel-to-pixel variations in the remitted spectra need to be minimized for reliable classification approaches. This has to be taken into consideration for future system improvements. In addition, when small window sizes are employed in the statistics calculation, the mean scattering power turned out to be the strongest data feature for discrimination. It is believed from the previous study6 that the scatter power should be more reliable than the remitted intensity because it is independent of coupling errors in the imaging system. All acquired scatter images were reflectance corrected to minimize referencing artifacts, but average scattered irradiance and scatter amplitude images are not entirely free from such artifacts due to small sample positioning and other instrumental issues. The scatter power is related to the wavelength-dependent scatter function and hence is relatively free from referencing artifacts. A mathematical proof of this intuitive point of view has been provided by the SFFS study on the discrimination capability, and as a consequence, future system designs have to pursue more accurate fitting of this particular parameter. 5.ConclusionsThis study reports the development of an automated interpretation methodology of scatter changes in tissue. The capability of the methodology to mimic the identification of regions of interest performed by a veterinary pathologist has been shown in two different situations: discrimination between normal and tumor tissue and segmentation of pathologically distinct tumor regions. In both of them, a correlation between automated and expert-based segmentation of correlation error has been achieved across all regions identified and all samples used in the study. Calculations of the statistical moments of these scattering parameters in a vicinity region of every pixel location were required to achieve a reliable discrimination of the distinct tumor regions. That means that future system designs should somehow try to minimize pixel-to-pixel variations. In order to justify this requirement, the SFFS algorithm has been used to determine which parameters exhibited more consistent trends across the different tumor subtypes. For physically reasonable window sizes, mean scattering parameters are more significant than the parameters themselves, as shown in Table 5, which explains the increase in tumor region determination accuracy. Statistical calculations could be avoided by including polarization measurements because this would give us more degrees of freedom. However, the opposite problem could also be encountered. As mentioned above, three scatter measurements were initially found to be insufficient to achieve reliable classifications and this situation was solved including a feature extraction procedure based on the spatial statistical values calculated from preprocessing of the data. If polarization is included in the measurements, the opposite situation could be encountered. Some kind of dimensionality reduction process would be demanded in order to retain usability in real time. The feasibility of the SFFS algorithm to extract the physically relevant parameters has been demonstrated in the discrimination capability study, and therefore, it would be a suitable candidate to perform this dimensionality reduction. Finally, it is worth it to reiterate that the performance of this discrimination capability study concluded that the mean scattering power was the strongest data set for discrimination. Hence, future system improvements should focus on an approach that provides the most accurate fitting of this important parameter. Future work studying analysis of larger samples and a wider variety of tumors is ongoing. AcknowledgmentsP. Beatriz Garcia-Allende is grateful for financial support from the Spanish Government’s Ministry of Science and Technology through the Projects No. TEC’2005-08218-C02-02 and No. TEC’2007-67987-C02-01, allowing her to spend three months in the Thayer School of Engineering at Dartmouth College. Funding at Dartmouth for this work came from NIH Research Grants No. PO1CA84203 and No. PO1CA80139. ReferencesT. Kuroiwa, Y. Kajimoto, and T. Ohta,
“Development of a fluorescein operative microscope for use during malignant glioma surgery A technical note and preliminary report,”
Surg. Neurol., 50
(1), 41
–49
(1998). https://doi.org/10.1016/S0090-3019(98)00055-X 0090-3019 Google Scholar
A. Bogaards, A. Varma, S. P. Collens, A. Lin, A. Giles, V. X. D. Yang, J. M. Bilbao, L. D. Lilge, P. J. Muller, and B. C. Wilson,
“Increased brain tumor resection using fluorescence image guidance in a preclinical model,”
Lasers Surg. Med., 35
(3), 181
–90
(2004). https://doi.org/10.1002/lsm.20088 0196-8092 Google Scholar
J. C. Ramella-Roman, K. Lee, S. A. Prahl, and S. L. Jacques,
“Design, testing, and clinical studies of a handheld polarized light camera,”
J. Biomed. Opt., 9 1305
–1310
(2004). https://doi.org/10.1117/1.1781667 1083-3668 Google Scholar
S. G. Demos, H. B. Radousky, and R. R. Alfano,
“Deep subsurface imaging in tissues using spectral and polarization filtering,”
Opt. Express, 7
(1), 23
–28
(2000). 1094-4087 Google Scholar
V. X. D. Yang, P. J. Muller, P. Herman, and B. C. Wilson,
“A multispectral fluorescence imaging system: design and initial clinical tests in intra-operative Photofrin-photodynamic therapy of brain tumors,”
Lasers Surg. Med., 32
(3), 224
–232
(2003). https://doi.org/10.1002/lsm.10131 0196-8092 Google Scholar
V. Krishnaswamy, P. J. Hoopes, K. Samkoe, J. A. O’Hara, T. Hasan, and B. W. Pogue,
“Quantitative imaging of scattering changes associated with epithelial proliferation, necrosis and fibrosis in tumors using microsampling reflectance spectroscopy,”
J. Biomed. Opt., 14
(1), 014004
(2009). https://doi.org/10.1117/1.3065540 1083-3668 Google Scholar
J. R. Mourant, T. J. Bocklage, T. M. Powers, H. M. Greene, K. L. Bullock, L. R. Marr-Lyon, M. H. Dorin, A. G. Waxman, M. M. Zsemlye, and H. O. Smith,
“In vivo light scattering measurements for detection of precancerous conditions of the cervix,”
Gynecol. Oncol., 105
(2), 439
–445
(2007). https://doi.org/10.1016/j.ygyno.2007.01.001 0090-8258 Google Scholar
A. Wax,
“Low-coherence light scattering calculations for polydisperse size distributions,”
J. Opt. Soc. Am. A, 22
(2), 256
–261
(2005). https://doi.org/10.1364/JOSAA.22.000256 0740-3232 Google Scholar
A. Wax, C. H. Yang, M. G. Muller, R. Nines, C. W. Boone, V. E. Steele, G. D. Stoner, R. R. Dasari, and M. S. Feld,
“In situ detection of neoplastic transformation and chemopreventive effects in rat esophagus epithelium using angle-resolved low-coherence interferometry,”
Cancer Res., 63 3556
–3559
(2003). 0008-5472 Google Scholar
Y. Liu, R. E. Brand, V. Turzhitsky, Y. L. Kim, H. K. Roy, N. Hasabou, C. Sturgis, D. Shah, C. Hall, and V. Backman,
“Optical markers in duodenal mucosa predict the presence of pancreatic cancer,”
Cancer Res., 13 4392
–4399
(2007). https://doi.org/10.1158/1078-0432.CCR-06-1648 0008-5472 Google Scholar
H. K. Roy, Y. L. Kim, Y. Liu, R. K. Wali, M. J. Goldberg, V. Turzhitsky, J. Horwitz, and V. Backman,
“Risk stratification of colon carcinogeneses through enhanced backscattering spectroscopy analysis of the uninvolved colonic mucosa,”
Clin. Cancer Res., 12 961
–968
(2006). https://doi.org/10.1158/1078-0432.CCR-05-1605 1078-0432 Google Scholar
Y. L. Kim, V. M. Turzhitsky, Y. Liu, H. K. Roy, R. K. Wali, H. Subramanian, P. Pradhan, and V. Backman,
“Low-coherence enhanced backscattering: review of principles and applications for colon cancer screening,”
J. Biomed. Opt., 11
(4), 041125
(2006). https://doi.org/10.1117/1.2236292 1083-3668 Google Scholar
B. W. Pogue and G. C. Burke,
“Fiber-optic bundle design for quantitative fluorescence measurement from tissue,”
Appl. Opt., 37 7429
–7436
(1998). https://doi.org/10.1364/AO.37.007429 0003-6935 Google Scholar
A. Amelink and H. J. Sterenborg,
“Measurement of the local optical properties of turbid media by differential path-length spectroscopy,”
Appl. Opt., 43 3048
–3054
(2004). https://doi.org/10.1364/AO.43.003048 0003-6935 Google Scholar
A. Amelink, H. J. Sterenborg, M. P. Bard, and S. A. Burgers,
“In vivo measurement of the local optical properties of tissue by use of differential path-length spectroscopy,”
Opt. Lett., 29 1087
–1089
(2004). https://doi.org/10.1364/OL.29.001087 0146-9592 Google Scholar
S. A. Prahl,
“Oregon Medical Laser Center website,”
(2007) http://omlc.ogi.edu/spectra/hemoglobin/ Google Scholar
K. Fukunaga, Introduction to Statistical Pattern Recognition, 2nd ed.Academic Press, New York
(1990). Google Scholar
V. Vapnik, The nature of Statistical Learning Theory, Springer, New York
(1995). Google Scholar
A. Oliver, J. Freixenet, R. Martí, J. Pont, E. Pérez, E. R. E. Denton, and R. Zwiggelaar,
“A novel breast tissue density classification methodology,”
IEEE Trans. Inf. Technol. Biomed., 12
(1), 55
–65
(2008). https://doi.org/10.1109/TITB.2007.903514 1089-7771 Google Scholar
G. F. Hughes,
“On the mean accuracy of statistical pattern recognizers,”
IEEE Trans. Inf. Theory, 14
(1), 55
–63
(1968). https://doi.org/10.1109/TIT.1968.1054102 0018-9448 Google Scholar
J. Kittler,
“Feature selection and extraction,”
Handbook of Pattern Recognition and Image Processing, 59
–83 Academic, New York
(1986). Google Scholar
L. Gomez-Chova, J. Calpe, G. Camps-Valls, J. D. Martin, E. Soria, J. Vila, L. Alonso-Chorda, and J. Moreno,
“Feature selection of hyperspectral data through local correlation and SFFS for crop classification,”
555
–557
(2003). Google Scholar
B. Deronde, P. Kempeneers, and R. M. Forster,
“Imaging spectroscopy as a tool to study sediment characteristics on a tidal sandbank in the Westerschelde,”
Estuarine Coastal Shelf Sci., 69 580
–90
(2006). https://doi.org/10.1016/j.ecss.2006.05.048 0272-7714 Google Scholar
C. Goutte,
“Note on free lunches and cross validation,”
Neural Comput., 9 1211
–1215
(1997). https://doi.org/10.1162/neco.1997.9.6.1245 0899-7667 Google Scholar
H. Zhu and R. Rohwer,
“No free lunch for cross-validation,”
Neural Comput., 8 1421
–1426
(1996). https://doi.org/10.1162/neco.1996.8.7.1421 0899-7667 Google Scholar
P. B. Garcia-Allende, V. Krishnaswamy, K. S. Samkoe, B. W. Pogue, O. M. Conde, and J. M. Lopez-Higuera,
“Automated segmentation based upon remitted scatter spectra from pathologically distinct tumor regions,”
Proc. SPIE, 7187 718717
(2009). https://doi.org/10.1117/12.808322 0277-786X Google Scholar
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||