|
|
1.IntroductionOne of the major challenges in characterizing cardiac tissue at the cellular level is accounting for the inherent three-dimensional architecture of the heart. To assess hypertrophy, for example, most studies use cross-sectional areas from serial sections acquired via conventional histological processing. Alternatively, cells are dissociated from the intact heart, eliminating the natural environment of the cells and discarding multicellular structural information. The precise nature of hypertrophy is thus difficult to characterize; for example, most studies of hypertrophic cardiomyopathy based on genetic mutations use terms such as “disarray” to describe changes in cardiac architecture. However, there is currently no clear description of the three-dimensional cardiac architecture at the microscopic level in either healthy or diseased hearts.1, 2, 3, 4, 5 One of the reasons for the absence of three-dimensional microscopic data is that cardiac myocytes are optically dense, arranged in varying three-dimensional orientations within the heart, and are longer than the thickness of standard histological sections. Some deep-tissue scanning techniques are available (e.g., ultrasound, magnetic resonance imaging) but lack the resolution to accurately characterize single cardiac cells or fibers, although improvements are continuing.6, 7 Wide-field stitching of serial sections is possible but can be subject to distortions from multiple cutting and has a low result-to-labor ratio, given that each section must be mounted and imaged successively.8 Two-photon microscopy is capable of imaging deeper into cardiac tissue compared to wide-field florescence techniques, but generally, the depth is not quite sufficient to obtain adequate architectural data (typical depths of of clear data). Additionally, for measurements of specific structural components, autofluorescence is generally not as useful as targeted labeling (such as with Sirius Red for histological slide processing), but fluorescence labeling of individual cells within intact whole organs is not well established. To overcome these difficulties, we have recently developed a protocol for intravital labeling of tissues in situ, after which the tissues are extracted, fixed, and embedded in paraffin for histological processing.9 This technique allows for uniform staining of the extracellular matrix surrounding cells using a fluorescent conjugated maleimide delivered by tail-vein injection, thereby labeling the entire intact heart. To complement this labeling, we have also recently developed an innovation for two-photon microscopy that combines a fast-scanning two-photon microscope using a polygonal mirror coupled to a mill-based microtome.10 This two-photon microtome is capable of imaging large volumes (up to several cubic millimeters) using a motorized stage, and repeated removal of already-imaged sections from the top of the sample. Because the entire heart is labeled, the newly exposed section can be imaged immediately. In the preliminary study by Ragan, the proof of concept of the overall technique was presented, but only general results were extracted from the data.10 A similar study was recently reported using confocal instead of two-photon microscopy, which examined the distribution of collagen in intact heart volumes;11 that study did not examine cell size or orientation, but focused on the different forms of collagen in different parts of the heart. In another study, second harmonic multiphoton imaging was used to elucidate extracellular matrix structure, with the advantage of bypassing tissue fixation and embedding.12 In this paper, we present results demonstrating that the combination of intravital staining and two-photon microtomy allow imaging of three-dimensional volumes of cardiac tissue while maintaining quantifiable, individual cellular information. By using custom postprocessing registration and segmentation algorithms, we obtain structural information based on assembly of acquired imaging sections. We then examine whether cardiac myocytes from mice expressing a mutant variant of -crystallin that causes cardiac hypertrophy13 exhibit different orientations compared to hearts from wild-type (WT) animals. Specifically, we test the hypotheses that (i) cross-sectional areas of the mutant myocytes are greater, when the orientation of the cells are accounted for, and (ii) the orientations of the mutant myocytes are more broadly distributed (i.e., have greater variances) compared to cells from WT hearts. 2.Materials and Methods2.1.Sample PreparationWT, WT transgenic (WT-CRY-AB), and -crystallin mutant transgenic (R120G) mice were kindly donated by J. Robbins (Childrens Hospital Medical Center, Division of Molecular Cardiovascular Biology, Cincinnati, Ohio).13 Transgenic mice were made and kept in the Friend Virus B background. Genotyping was performed by extracting deoxyribonucleic acid (DNA) from buccal cells and amplifying CryAB DNA using a polymerase chain reaction with the following primers: CryAB-forward: ctggcgttcttcgtgcttgccgtg- ; CryAB-reverse: gagtctgacctcttctcaacagcc- . At , hearts were labeled using tail-vein injection as previously described.9 Briefly, the hearts were labeled with Texas-Red Maleimide ( in dimethyl sulfoxide) and Hoechst ( in water) (Invitrogen, Carlsbad, California), harvested and fixed in 4% paraformaldehyde and embedded in paraffin. This time point was chosen to assess whether significant hypertrophy, typically seen at three-plus months, may be preceded by disorganization of the myocytes. Hearts were oriented similarly for all specimens, with the apex exposed. Thus, scanning was performed from apex to base. Sample sections were removed from the paraffin blocks to assess staining quality using an Olympus IX70 fluorescence microscope (Olympus, Center Valley, Pennsylvania), prior to imaging with two-photon microscopy. The left ventricles of three hearts from each group (Wt, WT-CRY-AB, and R120G, total nine mice) were imaged. The work was carried out in accordance with the Standing Committee at Harvard. 2.2.Two-Photon MicrotomyThe two-photon microtome was previously described.10 Two-photon microscopy images were acquired at at from a mode-locked titanium-sapphire laser, with the emission signal passed through a short-pass filter. Briefly, the paraffin sample was trimmed to fit under the oil immersion objective on a custom-built adapter to the motorized stage (Fig. 1 ). After scanning a desired area to deep (to allow for registration) at intervals using a piezoelectric objective positioner (PolyTec PI, Auburn Massachusetts), the sample was shuttled across to a mill, which removed the top of the sample. An air blower removed debris from the cut, and an automated syringe pump deployed more immersion oil onto the sample prior to shuttling the sample back under the objective. Synchronization of all data acquisition, hardware, and imaging controls was coordinated by LabView (National Instruments, Austin, Texas). Horizontal imaging based on the motorized stage was done with overlaps in both the and directions in order to facilitate image registration for postprocessing. Fig. 1The two-photon microtome setup has a rapid-spin polygonal mirror (PM) that passes the Ti-Saph laser beam into the left side of the microscope. The specimen is mounted on a special holder under the objective on a motorized stage. After scanning a desired volume, the specimen is shuttled across to the mill, which removes part of the scanned region, and the specimen is shuttled back to continue scanning.  After data acquisition, volumes were assembled using custom scripts in MATLAB (The MathWorks, Natick, Massachusetts). The data were converted from raw binary files to tagged image-file format, normalized by the average intensity, filtered with the wiener2 filter from MATLAB, and registered as individual stacks to eliminate major shifts resulting from polygonal mirror variations. Subsections of the imaged volume were selected to exclude blank spaces or unusable regions. The remaining sections were then stitched together using a fixed overlap to accommodate the overlaps from the horizontal imaging. Sections exhibiting strong cross-sectional signals were then used to register in (i.e., to couple the different stacks together from each microtome cut). To do this, images near the bottom of the top stack were registered with all of the images in the bottom stack. Each image resulted in a cross-correlation coefficient; the mode overlap of the peak cross-correlation for each image was considered the overlap, and the stacks were merged. 2.3.Data ExtractionAfter registration and assembly of the image stack from each imaged heart, relevant sections were isolated for review. Movies were generated of the image stacks where the fluorescence signal was high enough to identify features, which ranged from deep. Although the transverse outline of the cardiac myocytes were generally clear, the ends of the myocytes were generally not clearly visible. As a result, quantitative analysis was carried out on cardiac strands; that is, myocytes that are end-to-end adjacent, which have apparently continuous cross sections. Cardiac strands that presented clear cross sections were traced using a Wacom pen tablet (Wacom, Vancouver, Washington), and corresponding cardiac strands (identified visually) were traced above and below the initial section. The areas and area centroids of the cells were extracted from the tracing. Quantitative information from strands that underwent significant changes (e.g., disappearing, splitting, etc.) or could not otherwise be correlated to the original cell was excluded from cross-sectional area analysis. To adjust the cross-sectional area based on the angle of the cell, the in-plane translation distance was calculated using the area centroids for each section of the same cell, between the top and center slices, and the center and bottom slices (Fig. 2 ). The cell angle was then approximated by taking the average angle between the top and bottom angles. The cell’s cross-sectional area was adjusted by multiplying the raw area of the center section by the cosine of the average angle. Fig. 2Calculating the adjusted area. Cells are shown as line segments (profiles of the areas). The centroids are calculated, and then the angle to vertical is computed using the distance between centroids and the known spacing between the sections. The average is used to adjust the area of the center cell section. 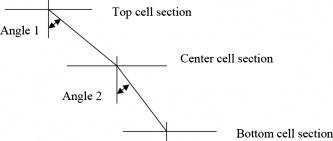 For further analysis of cardiac strand organization, sections of imaged volumes were divided into deep image blocks for a representative section of each type of heart (WT, WT-CRY-AB, and R120G). A three-dimensional fast-Fourier transform (FFT) was performed on each block in the section. The blocks were not perfectly cubical due to computer memory limitations. The resulting three-dimensional FFTs are projected onto the top and front views, with the top view composed of the imaging plane. Where possible, the long axes of the FFT projections were extracted and the angle to a fixed axis was measured. These long axes angles were used as a more general measure of cardiac strand orientation (as opposed to measuring single strands) in each block. 2.4.StatisticsIndividual cell areas, adjusted areas, and angles were compared using one-way ANOVA and Tukey posttest, with significance defined as . Standard deviations were compared using the Bartlett test, with indicating significantly different standard deviations. The authors had full access to the data and take responsibility for its integrity. All authors have read and agree to the paper as written. 3.Results3.1.Two-Photon Microtomy Can Acquire Three-Dimensional Cardiac Strand InformationTwo-photon microtomy is a powerful technique to obtain three-dimensional architecture of tissues. For this study, we imaged nine heart volumes using intravital staining and two-photon microtomy. The scan volumes were typically in cross-sectional area and ranged from deep (mostly over ). Representative images of WT, WT transgenic (WT-CRY-AB), and -crystallin mutant transgenic (R120G) hearts are shown in Fig. 3 . Gross morphology suggests no obvious differences in architecture at , based on a single slice. Visible in these images are the maleimide stain, outlining the cardiac strands, with some major vasculature. This technique of intravital staining coupled with two-photon microtomy is clearly capable of acquiring detailed structural information. Because still images do not convey the three-dimensional structural information clearly, representative acquired data volumes are posted as movie files ( Video 1, Video 2 Video 3 ), which will be discussed further in Sec. 3.2. Fig. 3Representative two-photon microtome cross-sectional images of (a) WT, (b) WT-CRY-AB, and (c) R120G left ventricles scanned with two-photon microtomy after intravital labeling with maleimide. The cross sections of the cardiac myocytes are clearly visible in several parts of the images. The R120G mutant exhibited waviness in its fibers in (c) but this was not apparent in all R120G hearts scanned. Scale bar at the lower left of each . 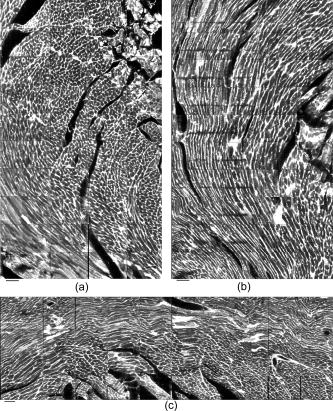 Video 1A movie of data stacks scanning through a WT left ventricular section. This is the movie of the heart section depicted in Fig. 3a. Scan direction is from apex to base, slices are separated by . Black borders are postprocessed artifacts to allow for image shifting due to imperfect realignment during stage motion (QuickTime, 5MG). .  Video 2A movie of data stacks scanning through a WT-CRY-AB left ventricular section. This is the movie of the heart section depicted in Fig. 3b. Scan direction is from apex to base, slices are separated by . Similar to Video 1, black borders are postprocessed artifacts to allow for image shifting due to imperfect realignment during stage motion (QuickTime, 5MG). .  Video 3A movie of data stacks scanning through a R120G mutant left ventricular section. This is the movie of the heart section depicted in Fig. 3c. Scan direction is from apex to base, slices are separated by . Similar to Video 1, black borders are postprocessed artifacts to allow for image shifting due to imperfect realignment during stage motion. Cardiac strand “waviness” can be seen in Video 3, as well as what may be characterized as disarray, with cells appearing to converge or diverge from certain shifting locations. The exact cause of this disarray is not known as the other two mutant hearts do not exhibit this behavior (QuickTime, 5MG). .  Once the image stacks are assembled as a three-dimensional image block, it is possible to display data from custom perspectives. Figure 4a shows an imaged WT heart section with the transverse cells on the top surface (as oriented) representing the plane of a single acquired image (in-plane image). The longitudinal aspects of the cells are clearly visible on the side surfaces, which are derived from reconstruction and thus have slightly lower resolution compared to the in-plane image. This reconstruction demonstrates that, where the heart exhibits transverse cross sections in the in-plane images, the longitudinal sections can be visualized from the reconstruction. Next, we examined data volume where the in-plane image was longitudinal, making it possible to characterize the orientation of cells deep within the volume [Fig. 4b]. By selecting some of the more clearly delineated cells within the reconstructed volume, the orientation of those cells can be seen to be horizontal [Fig. 4c]. One of the R120G mutant hearts displayed less uniform behavior. Figure 4d shows the transverse section similar to Fig. 4a for this R120G mutant heart, with similar longitudinal cutaways on the side view. Selecting a longitudinal section, however, revealed that the cells had more varying orientation within the block compared to the wt section [Fig. 4b and 4f]. It is unclear why only horizontally oriented, and not vertically oriented cells, exhibited this variance in this particular heart. Fig. 4Postprocessed data blocks (false color) of various cardiac sections. In the WT heart, (a) transverse sections reveals the cardiac strands are well aligned, as can be seen in the lateral cut-away fields. (b) The sides of the longitudinal sections display fewer clear structures but the cardiac strands that are visible tend to exhibit similar orientations as outlined in (c). In one of the R120G mutant hearts, (d) transverse sections reveal similar alignments compared to WT hearts. However, longitudinal sections (e) exhibit slightly more heterogeneity in cell angles, as can be seen in the cells outlined in (f) whereby the angles switch back and forth. This phenomenon is clearly visible in the movies of the heart sections ( Videos 1 – 3). (Color online only.) 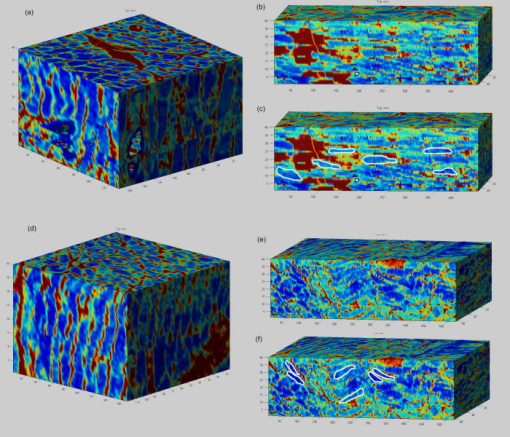 WT myocardial tissue displayed many morphological features that demonstrate the complexity of myocyte architecture. First, cardiac strands are only roughly circular in cross section, and the cross-sectional geometry changes as a function of depth [shape, not just size, Fig. 5a ]. Second, cardiac strands are never composed solely of continuous cardiac myocyte loops, with new strands appearing spontaneously [Fig. 5b]. Third, multiple strands appear to merge into single strands [Fig. 5c, as well as single strands separating into multiple strands, Fig. 5a], revealing that the organization of the tissue has local variance. Finally, the vasculature does not necessarily follow the direction of the cardiac strands in the heart [Fig. 5d]. To see these features more clearly, these extracted sections can be viewed as movies posted as Video 4, Video 5, Video 6, Video 7 . Fig. 5Two-photon microtomy reveals three-dimensional structural information about cardiac tissue. (a) Cells do not have circular cross-sections, and the cross-sectional geometry changes with depth (spacing between images, reading left to right, top to bottom). The arrow points to a cardiac strand that appears to split into two, each of which develops its own morphology. (b) A new cell inserts itself between two cells (location of insertion designated by the arrow). Thus, not all strands are linked end to end; there are several instances of new strand insertion and existing strand disappearance in the imaged sections. Spacing is between slices from left to right. (c) Example of two or three strands initially separated that merge into a single strand, in the cell slightly right of center. Spacing is from left to right. (d) Image sequence of a blood vessel moving up through the tissue, while the cells are generally moving down (while some cell tracking is possible through the sequence of images, see Videos 7). Spacing is from left to right. (e) False-colored merged image of the maleimide stain (in green) and the nuclei (in red; appears orange or yellow in overlap regions). The images were brightness and contrast adjusted prior to merging. The nuclear location is predominantly at the junctions between the cells. In all images, the first image in the sequence (upper left image) has a scale . (Color online only.) 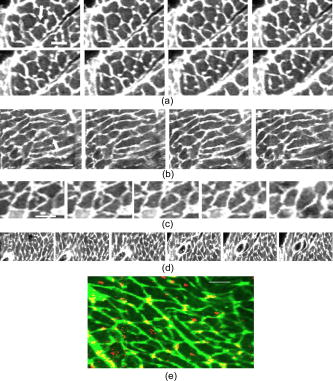 Video 4Corresponding to Fig. 5a, a cardiac strand can be seen developing a partition that divides the strand into two (QuickTime, 188KB). . 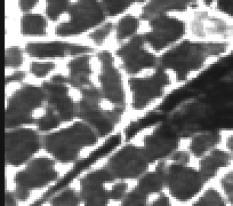 Video 5Corresponding to Fig. 5b, a new cardiac strand can be seen forming in the right-of-center region. Although this can be partially due to the “fountain” of cells appearing, the small size at onset, coupled with the subsequent increase in cross-sectional area appears to support the observation that a new strand is forming (QuickTime, 318KB). . 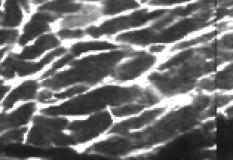 Video 6Corresponding to Fig. 5c, three cardiac strands can be seen merging into the same strand (QuickTime, 172KB). .  Video 7Corresponding to Fig. 5d a blood vessel can be observed that is oriented differently from the local cardiac strands. Specifically, the vessel appears to “move” upward in the images, while the cells are all “moving” downward. This corresponds to a vessel that is oriented at a significant angle to, and not parallel to, the majority of the local cardiac strands (QuickTime, 1MG). .  In some specimens, Hoechst stain was used and revealed location of nuclei in the tissue [Fig. 5e]. However, it is not possible with this technique to distinguish between nuclei from cardiomyocytes versus from other cells (such as the endothelial cells in the microvasculature). The majority of the visible nuclei are located near the junctions between adjacent cells. 3.2.Two-Photon Microtome StacksBy merging scan images into movies, it is possible to follow the overall cardiac architecture more easily. These movies (see Video 1, Video 2, Video 3, Video 4, Video 5, Video 6, Video 7) show changes in the cardiac architecture. Sometimes, the cells are oriented nearly vertically in the plane of the slices, whereas other times the cells exhibit more horizontal orientation, appearing to “shift” laterally as the movie is played. One of the R120G hearts exhibited particularly anomalous geometry ( Videos 3, compared to Videos 1 and 2). In particular, the myocytes appeared to be in three-dimensional disarray. First, the images exhibited waviness in the more longitudinal regions (Fig. 3). Second, various regions exhibited cells appearing to converge or diverge [that is, when scanning through the stack, cells appear to converge or diverge from fixed points (see Videos 3)]. This behavior is consistent with the zigzag pattern exhibited by the cells in the longitudinal blocks [Figs. 4e and 4f]. It should be noted that of the three R120G hearts examined, this one was the only one that manifested such unusual geometry. As a result, it cannot be concluded that this morphology is pathognomonic of the -crystallin mutation. However, these stacks clearly show a great deal of structural information and allow us to assess the native three-dimensional arrangement of the cells, so that we can begin identifying and characterizing potential disarray, as in Videos 3. 3.3.Quantitative AnalysisOne of the key characteristics of the R120G mutation is the development of cardiac hypertrophy. Typically, this is determined by measuring cell cross-sectional area or cell volume in dissociated cardiomyocytes. Using two-photon microtomy, we can obtain a more accurate representation of the true (in situ) cross-sectional areas by accounting for the angle of the cell. We initially used a t-test to compare the WT and R120G mutant hearts to verify that the R120G cells had larger cross sections ( , ) in a single plane, which is consistent with previous findings.13 For quantitative analysis of individual cell properties, we analyzed each cell strand as an independent unit, combining cells from hearts of the same genotype. When the cross-sectional areas were measured in a single transverse section, both the WT-CRY-AB and R120 hearts appeared to exhibit hypertrophy via increased cross-sectional area [Fig. 6a, WT-CRY-Ab versus WT, R120G versus WT, WT-CRY-AB versus R120G]. However, once the cell orientation was taken into account, only the R120G hearts were statistically significantly different from either the WT or WT-CRY-AB cellular cross-sectional areas [Fig. 6b, for R120G versus either WT or WT-CRY-AB, for WT versus WT-CRY-AB]. Furthermore, the average angles the cells make with the imaging plane was significantly lower for the R120G versus either WT or WT-CRY-AB [Fig. 6c, for each], suggesting the R120G cells in the imaged sections were arranged more transversely compared to the WT and WT-CRY-AB hearts. Changes in angles to the vertical axis are not likely due to artifacts of heart placement because the embedding process was done blinded to the heart genotype. However, the ability to account for angle reduces or eliminates errors that may be associated with differing protocols and variances that exist during embedding. Fig. 6(a) Extraction of cross-sectional areas of the myocytes revealed initially that both WT-CRY-AB and R120G had statistically significantly increased areas compared to WT samples. (b) However, on adjustment of angles using three-dimensional data, only the R120G mutant hearts exhibited significantly increased cross-sectional area compared to either WT or WT-CRY-AB hearts. (c) Analysis of the angles revealed that the R120G cardiac strands were at significantly shallower angles compared to either WT or WT-CRY-AB strands, suggesting that three-dimensional information is vital for proper characterization of heart structure. 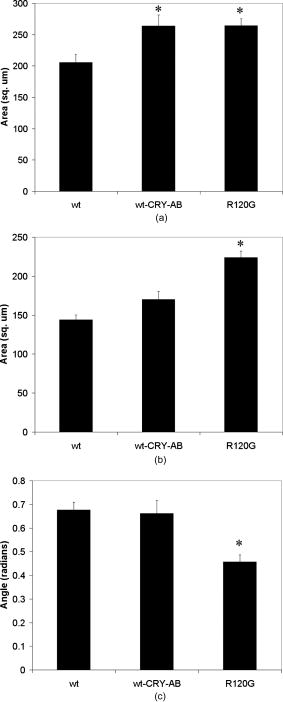 The standard deviations of the cellular corrected cross-sectional areas were also compared and found to be significantly greater for the WT-CRY-AB and R120G cells compared to the WT cells . However, the amplification of the standard deviation (1.52 times) was comparable to the increase in adjusted cross-sectional area (1.55 times) for R120G versus WT. Thus, we conclude that the adjusted cross-sectional areas are being amplified proportionally rather than arithmetically (i.e., hypertrophy of R120G mutations is equivalent to multiplying the areas by a constant rather than adding a constant). To examine the orientation of the fibers over larger volumes, we used three-dimensional FFT on subvolumes of the imaged heart sections, using volumes of cubed. The FFT was then projected to a top view and a side view (the scanning direction is considered the top view). Where the FFT projections exhibited a long axis, the angle of the long axis was measured [Figs. 7a and 7b ]. These angles were then arrayed over a slice of data for each of the imaged hearts [WT, Fig. 7c; mutant, Fig. 7d]. In most of the hearts, the angle arrays were even and showed smooth changes, similar to Fig. 7c. In a few cases, the arrays had two different regions of similar angles, suggesting that the regions were aligned differently. In the R120G mutant heart shown in Fig. 7d [same heart as in Fig. 3c], the fibers’ are clearly arranged in an oscillatory pattern, although this was not observed in the other mutant hearts. Fig. 7(a) A typical cross section of two-photon data, size , and (b) its FFT projected in the same plane. Note that the long axis of the FFT is perpendicular to the dominant fiber direction. (c) Top-view FFT long axes projection display shows a smooth change in fiber orientation in a WT ventricular section. In contrast (d) top-view FFT long axes projection displays some oscillations in fiber orientation in a R120G mutant ventricular section. Not all of the mutant ventricles displayed oscillations. 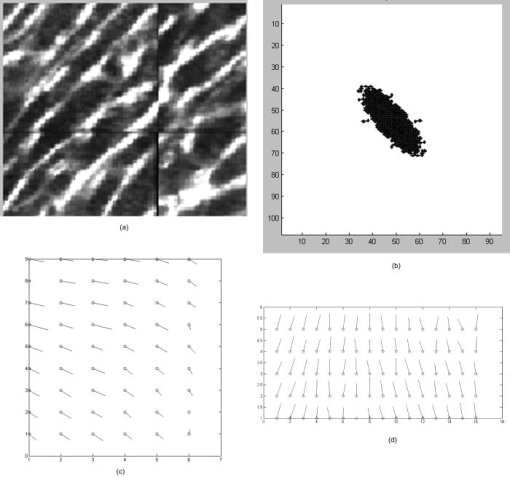 The standard deviations of the angles were compared and showed no significant difference among the three different hearts examined (standard deviations 22.6 for WT, 19.6 for WT-CRY-AB, and 19.5 for R120G mutant, projected to the top, , side projections not compared because their distribution was not normal), suggesting that the fiber orientations have similar variances in these hearts. The actual angles were not compared statistically because the orientation of the heart volume being scanned is random, resulting in differences in cardiac strand orientation. Note that this is in contrast to the angles being compared previously to correct for cross-sectional area. The hearts are embedded in paraffin so that the apex-base angles are all similar. However, the rotation about the apex-base axis is not predetermined. These results demonstrate that cardiac strand orientation and distribution can be extracted from the three-dimensional data and provides a way of examining potential disarray in a more quantitative fashion. 4.DiscussionThe use of two-photon microscopy for deep-tissue imaging, coupled with an automated mill/air/oil system has previously been shown to be capable of imaging an entire heart, preserving its three-dimensional structure. 10 Here, we demonstrate that the system is capable of obtaining detailed, three-dimensional structural information about the cardiomyocytes in mutant and normal hearts. We show that this information can be used for qualitative analysis of geometry, in terms of cell organization, changes in the cross sections, and the arrangement of adjacent cells, as well as for quantitative information, in terms of determining a more accurate cross-sectional area and the orientation of cardiac strands. By correcting the cell areas using angles derived from the three-dimensional data, this method allows for a broader selection of cells that may be at shallower angles to the transverse plane and corrects for variances in embedding. Additionally, this method also allows for the visualization of the tissue geometry in the native environment, postfixation. By analyzing the results of the imaged hearts, we can make some preliminary conclusions. First, at ten weeks, some form of cardiac alteration is already occurring; at least, the cells’ cross-sectional areas are increasing in only the R120G mutant hearts, indicating cardiac hypertrophy. We cannot definitely conclude that the cells are increasing in volume, because the current method does not permit length measurements. Second, while one R120G mutant heart exhibits disarray [Fig. 7d], not all do. Thus, we suspect that cell enlargement may precede the development of gross fiber disarray, as evaluated by FFTs [Fig. 7d]. The standard deviation of the cross-sectional areas was significantly lower for WT compared to the R120G cells, by the same scale as the areas. This suggests that observed disarray might not result from more diverse cardiac strand orientations, but rather cell size heterogeneity. We believe this is the first study that has examined and attempted to characterize the three-dimensional nature of cardiac hypertrophy in genetic mutants. Although the sample size of the current study is relatively small ( for each genotype), we believe we have successfully demonstrated a proof of concept that can be used more broadly for examining hypertrophy in general. In the previous study on these hearts,13 Wang did not find any increase in cross-sectional area in the WT-CRY-AB cardiac cells. Their method for determining cross-sectional area was to find the cell volumes in dissociated cells and then divide by the cell length. Because they depended on dissociation and averaged the cross section across the entire cell, their technique would not be affected by angular discrepancies. Our finding that, without correction for angles, the WT-CRY-AB cells exhibit increased cross-sectional areas, should be taken as a cautionary remark on other studies that use conventional histological sectioning techniques for measuring cell cross-sectional area. That finding also suggests that there may be other alterations in cardiac architecture associated with the WT-CRY-AB models, although a specific examination of that is beyond the scope of the current study. Despite the power of this technique, there are still several limitations that prevented us from taking full advantage of this method. From a hardware perspective, the polygonal mirror is imperfect and, as such, there are shifts in the data set that could not be removed. These shifts likely cause an error of a few in the horizontal plane, as estimated by observing the shifts in individual data stacks. This may be improved by switching to a fixed imaging system, such as with a multifocal lenslet, which requires a high-powered laser. Scanning time can be an impediment to high-volume use, especially of larger volumes (this was one of the reasons the sample size was small), despite the use of the high-speed polygonal mirror configuration. The heart, being round with empty spaces at the ventricles, has a particularly difficult geometry to image using an automated scanning algorithm in a rectangular array. The use of analog controls (such as the polygonal mirror control and the motorized stage controls) resulted in some undesirable shifting in the data (see Video 1, Video 2, Video 3, Video 4, Video 5, Video 6, Video 7). Furthermore, not all hearts that were imaged resulted in usable data, due to fluorescence label degradation or poor sample positioning; thus, while more than three hearts in each group were imaged, only three in each group ended up being usable. The typical resolution of the objective used in this study is . Given the imaging errors from hardware and software, we estimate the errors on measurements to be approximately in the horizontal direction. Errors in the vertical direction are likely much smaller due to the piezoelectric objective positioner; however, during vertical registration, image overlap could produce errors of , resulting from poor registration or distortions from the mill-microtome unit. Overall, however, the ability to acquire geometric features is likely superior to that of MRI and ultrasound in their current deployment. 5.ConclusionWe have shown that using a two-photon microtome is an extremely valuable tool for characterizing cardiac architecture in three dimensions. For the first time, morphological features that follow strands could be identified. Quantitative extraction can be derived from an imaging volume rather than an arbitrarily chosen cross-sectional area. The contribution of genetic alterations to hypertrophic disarray is more clearly presented in a stack of images rather than relying on individually processed serial sections, and the types of disarray can potentially be categorized using this method, based on observed three-dimensional structure, hopefully leading to a more specific understanding of how different mutations can lead to cardiac hypertrophy. AcknowledgmentsThis work was supported by NIH Grant No. R21 EB004646 to H.H. and NIH Grant No. R01HL081404 to R.T.L. ReferencesE. A. Aiello, M. C. Villa-Abrille, E. M. Escudero, E. L. Portiansky, N. G. Perez, M. C. de Hurtado, and H. E. Cingolani,
“Myocardial hypertrophy of normotensive Wistar-Kyoto rats,”
Am. J. Physiol. Heart Circ. Physiol., 286
(4), H1229
–1235
(2004). https://doi.org/10.1152/ajpheart.00779.2003 0363-6135 Google Scholar
G. W. Dorn 2nd, J. Robbins, and P. H. Sugden,
“Phenotyping hypertrophy: eschew obfuscation,”
Circ. Res., 92
(11), 1171
–1175
(2003). https://doi.org/10.1161/01.RES.0000077012.11088.BC 0009-7330 Google Scholar
D. Fatkin, B. K. McConnell, J. O. Mudd, C. Semsarian, I. G. Moskowitz, F. J. Schoen, M. Giewat, C. E. Seidman, and J. G. Seidman,
“An abnormal response in mutant sarcomere protein-mediated familial hypertrophic cardiomyopathy,”
J. Clin. Invest., 106
(11), 1351
–1359
(2000). https://doi.org/10.1172/JCI11093 0021-9738 Google Scholar
J. P. Schmitt, C. Semsarian, M. Arad, J. Gannon, F. Ahmad, C. Duffy, R. T. Lee, C. E. Seidman, and J. G. Seidman,
“Consequences of pressure overload on sarcomere protein mutation-induced hypertrophic cardiomyopathy,”
Circulation, 108
(9), 1133
–1138
(2003). https://doi.org/10.1161/01.CIR.0000086469.85750.48 0009-7322 Google Scholar
X. Wang, H. Osinska, G. W. Dorn 2nd, M. Nieman, J. N. Lorenz, A. M. Gerdes, S. Witt, T. Kimball, J. Gulick, and J. Robbins,
“Mouse model of desmin-related cardiomyopathy,”
Circulation, 103
(19), 2402
–2407
(2001). 0009-7322 Google Scholar
W. G. O’Dell and A. D. McCulloch,
“Imaging three-dimensional cardiac function,”
Annu. Rev. Biomed. Eng., 2 431
–456
(2000). https://doi.org/10.1146/annurev.bioeng.2.1.431 1523-9829 Google Scholar
W. Y. Tseng, J. Dou, T. G. Reese, and V. J. Wedeen,
“Imaging myocardial fiber disarray and intramural strain hypokinesis in hypertrophic cardiomyopathy with MRI,”
J. Magn. Reson Imaging, 23
(1), 1
–8
(2006). https://doi.org/10.1002/jmri.20473 1053-1807 Google Scholar
J. Yoshioka, R. N. Prince, H. Huang, S. B. Perkins, F. U. Cruz, C. MacGillivray, D. A. Lauffenburger, and R. T. Lee,
“Cardiomyocyte hypertrophy and degradation of connexin43 through spatially restricted autocrine/paracrine heparin-binding EGF,”
Proc. Natl. Acad. Sci. U.S.A., 102
(30), 10622
–10627
(2005). https://doi.org/10.1073/pnas.0501198102 0027-8424 Google Scholar
C. MacGillivray, J. Sylvan, R. T. Lee, and H. Huang,
“Time-saving benefits of intravital staining,”
J. Histotechnol., 31
(3), 129
–134
(2008). Google Scholar
T. Ragan, J. D. Sylvan, K. H. Kim, H. Huang, K. Bahlmann, R. T. Lee, and P. T. So,
“High-resolution whole organ imaging using two-photon tissue cytometry,”
J. Biomed. Opt., 12
(1), 014015
(2007). https://doi.org/10.1117/1.2435626 1083-3668 Google Scholar
A. J. Pope, G. B. Sands, B. H. Smaill, and I. J. LeGrice,
“Three-dimensional transmural organization of perimysial collagen in the heart,”
Am. J. Physiol. Heart Circ. Physiol., 295
(3), H1243
–H1252
(2008). https://doi.org/10.1152/ajpheart.00484.2008 0363-6135 Google Scholar
K. Schenke-Layland, I. Riemann, U. A. Stock, and K. Konig,
“Imaging of cardiovascular structures using near-infrared femtosecond multiphoton laser scanning microscopy,”
J. Biomed. Opt., 10
(2), 024017
(2005). https://doi.org/10.1117/1.1896966 1083-3668 Google Scholar
X. Wang, H. Osinska, R. Klevitsky, A. M. Gerdes, M. Nieman, J. Lorenz, T. Hewett, and J. Robbins,
“Expression of R120G-alphaB-crystallin causes aberrant desmin and alphaB-crystallin aggregation and cardiomyopathy in mice,”
Circ. Res., 89
(1), 84
–91
(2001). https://doi.org/10.1161/hh1301.092688 0009-7330 Google Scholar
|

