|
|
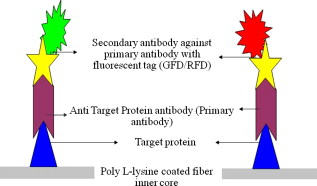
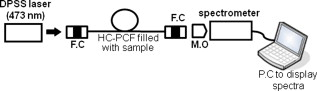
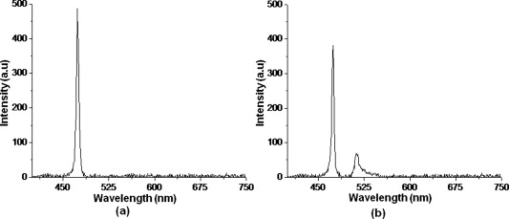
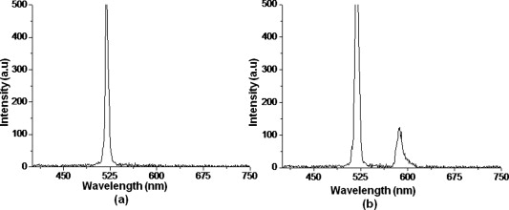
1.IntroductionBiomedical research is mainly focused on detection, diagnosis, treatment, and prevention of diseases that ultimately leads to better health. Ultrasensitive detection and imaging methods are enabling technologies for the improvement of the same to take the arena of diagnostics methodologies to the next level. The steady advancement of biology and medicine toward diagnostics based on molecular markers leads to the exploration of high-throughput methods for the detection of biomolecules and their interactions in a biological system. Protein microarrays are an imperative tool for proteomics research and are also used in biomedical applications to determine the presence and/or amount of proteins in a biological sample.1 In recent years, fluorescence assay technologies have played a pivotal role in the high-throughput analysis of proteins and protein interactions. The precise measurement of a fluorescence signal is a critical parameter in analyzing the functional response of the biological sample.2 Fluorescence-based bioassays are novel diagnostic tools to the clinicians for deciding on further treatment and to the researchers for monitoring biological functions that lead to novel investigations. Detection at the molecular level, such as reporter assays and gene expression studies, are achieved by improved techniques using fluorescent proteins as biomarkers.3 A nanotechnology-based platform offers promise for multiplexed detection of proteins and nucleic acids to bring substantial advances in molecular medicine. A broad spectrum of highly innovative approaches of nanodevices was critically reviewed.4 These include surface nanotexturing for mass spectrophotometry and reverse phase protein microarrays, the biobar code assay, biologically gated nanowire sensors, and silicon-based nanostructures for the transduction of molecular binding in to an electrical and a mechanical signal, respectively. Recent progress of optical fiber-based biosensors and their sensing performances have found good potential and were elaborately reviewed in the literature.5 Photonic crystal fibers (PCFs) are a special class of silica-based microstructured optical fibers (MOFs) with tiny air holes running along the length of a silica fiber.6 Among them, hollow-core PCF (HC-PCF) can achieve air guiding and can be used for efficient biosensing application due to the increased light-matter interaction in the HC.7 These fibers has been used for evanescent wave sensing or highly efficient sensing of biomolecules, such as DNA, enzymes, antigen, antibodies, and proteins.8, 9, 10, 11, 12 Hence, these HC-PCFs can be used as “immobilization nanoholes” and it has been proven that photonic bandgap can be preserved by filling the center core with the solution and, thereby, making it an ideal probe for sensing applications.13 Breast cancer is the fifth most common cancer, which leads to 502,000 deaths worldwide per year, and it has already been linked with the steroid hormone estrogen.14 It has long been known that many human breast cancers are initially hormone dependent, which has led to the utilization of antiestrogens in the treatment of breast cancer.15, 16 Discovery of the estrogen receptor (ER) has given insight, not only as a powerful diagnostic and predictive marker, but also as a proficient target for the treatment of hormone-dependent breast cancer.17, 18 Several techniques are available for the specific detection of the said protein. The immunosensing technique is a powerful and flexible tool, used to identify targeted an antigen with desired specific antibodies.19, 20, 21 Even though enzyme-linked immunosorbent assay (ELISA) is an old method, it still in use to detect proteins,22, 23 but it has the disadvantage of showing nonspecific interaction leading to false positives. The gel shift assay is another approach for detecting the ER protein; its intracellular localization and expression levels are reported elsewhere.24 This assay needs a precise radioactive tracer and a specific protected area with federal certification approval and supervision. Immunohistochemistry (IHC) is another technique for assessing the ER status of breast cancers.25, 26 It has a repeated disadvantage in deciding the level of expression, the Allred’s IHC score between 2 and 3. In this context, we have made an approach to develop a technique to detect the protein. This method is mainly proposed and illustrated using a HC photonic crystal fiber based on immunobinding, which can be an efficient biosensor in clinical diagnosis. 2.Materials and MethodsThe ER-positive MCF-7 Breast carcinoma cells and ER-negative MDA-MB 231 cells were chosen for the study. 2.1.Cell Culture and Sample PreparationBoth MCF-7 and MDA-MB 231 cells were grown to confluence in Dulbecco’s modified-eagle-medium–high glucose supplemented with 10% fetal bovine serum and 1% penicillin and streptomycin. The media were removed from the 12 well culture plates in which the cells were grown, and the cells were washed twice with phosphate buffered saline (PBS). These cells were lysed in lysis buffer (1X), kept at rotator for . After incubation, of phenylmethylsulfonyl fluoride was added and centrifuged at for . The supernatant-containing protein was extracted and stored at until use. 2.2.Western Blot Analysis Using Anti-ER AntibodyThe concentration of proteins in the cell lysates were quantified using the Bradford method.27 The endogenous estrogen receptor levels in the positive cells were detected using the antibody raised against . For this purpose, protein was resolved by 4–12% gradient sodium dodecyl sulfate-polyacrylamide gel electrophoresis and was transferred to nitrocellulose membrane. The membrane was blocked in 1X Tris buffered saline with tween-20 (TBST) containing 5% milk powder for . The membrane was further incubated overnight in the same buffer containing anti- antibody (Acris Antibodies, GmbH, Germany) at . The membrane was washed and incubated with the secondary anti-Rabbit antibody conjugated with horseradish peroxidase (HRP) enzyme for . The washed membrane was incubated with chemiluminescent substrate for HRP and exposed to X-ray film and developed. The stripped membrane was detected for protein -actin and used as internal loading control. 2.3.ELISAThe same crude extracts protein and antibody (Acris Antibodies GmbH, Germany) were used to perform ELISA. The protein was diluted to , , , , and in coating buffer ( , pH 8.6). Twenty-five microliters of each diluted protein was added to each well of a pretreated ELISA plate, and it was then covered with plastic film and incubated at overnight. The wells were blocked by adding of 3% bovine serum albumin (BSA)/PBS and incubated for at in a moist, sealed container. Then of 1% BSA/PBS mixed with primary antibody was added to each well. The plates were shacked well with the antibody solution and washed with 1X PBS/Tween 20 (0.05%). Antibody enzyme conjugates (antimouse alkaline phosphatase at 1:1000 in 1% BSA/PBS) was added to each well and again incubated at for . Again, each well was washed with 1X PBS/Tween 20 (0.05%). Finally, of substrate solution (one tablet of Sigma 104® phosphatase in alkaline phosphatase developing buffer) was added to each well. The developed color was read at 15, 30, and , respectively. 3.Implementation and Experimental Analysis3.1.Photonic Crystal FiberHC photonic crystal fiber was chosen for this sensor development (Crystal Fiber A/S–AIR-6-800). The fiber was cut into segments of length, and one end of the fiber was cleaved approximately into length [Fig. 1 ]. This fiber has a center core size of diameter and cladding diameter of [Fig. 1]. 3.2.Protein ImmobilizationThe different steps involved in the process were schematically represented in Fig. 2 . The tip of the fiber was dipped in the 0.01% Poly L-lysine (Sigma Aldrich, USA) solution for , and the solution is allowed to get into the fiber by simple capillary forces. The fiber was permitted to dry out at room temperature for . After drying, it was washed twice with PBS for ; thus, the inner core was activated for further process. Then, the cell lysate solutions from MCF-7 and MDA-MB-231 were immobilized separately into different fibers for protein binding . After that, the fibers were incubated at for then washed thrice briefly in TBST buffer. In the next step, the primary antibody binding was done using antiestrogen [primary antibody (Acris antibodies, GmbH, Germany)] solution diluted to a final concentration of 1:1000 in TBS for and incubated for at . Concurrently, sodium iodide symporter antibody (primary antibody; Whatman®) was also used to check the nonspecific binding of the protein. Then the fibers were washed with TBST buffer (thrice) for and dried at for The experiment was completed by adding a secondary antibody solution made in TBS containing Alexa™ Fluor 488 (green fluorescent dye) or 555 (red fluorescent dye) labeled Goat anti-rabbit IgG Invitrogen™ diluted to 1:100, the same way as that of the earlier step. Appropriate controls were performed simultaneously for both green/red dye detection. Now the immuno binding of the protein was completed and the fibers were ready for imaging sensing. The fibers were stored in dark ambience at . 3.3.Imaging and Sensing AnalysisThe cleaved end of the fiber carrying immobilized protein was focused under the microscope (Olympus fluorescence microscope CKX41) and checked for the fluorescence signal. The schematic setup used for the spectral analysis of fluorescent proteins was shown in Fig. 3 . For fluorescence analysis, diode pumped solid state (DPSS) laser (output power ) was coupled to the proximal end of the HC photonic crystal fiber. The beam emerging from the distal end of the fiber was focused to a high-quantum-efficiency spectrophotometer (Ocean Optics, QE65000) using a microscope objective ( , ). In this study, lasers with wavelength of 473 and were used for the analysis of green fluorescent dye and red fluorescent dye, respectively. 4.Results4.1.Protein Concentration and Western Blot AnalysisThe estimated amount of protein in MCF-7 and MDA-MB-231 cell lysates by Bradford’s method is found to be . The amounts of protein immobilized in the fibers were calculated according to the protein quantity in the cell lysate. To confirm the ER protein signal, the cells are analyzed for Western blot using anti- antibody for cellular ER, and anti- -actin antibody for -actin. The result obtained is positive in the MCF-7 cells and negative in the MDA-MB-231 cells. This is the qualitative confirmation for the presence of ER protein in two different cells (Fig. 4 ). Because the -actin is the housekeeping gene, it gives a signal in both cell lines, whereas the negative cell line MDA-MB-231 has failed to produce a signal due to lack of a receptor. 4.2.ELISA AssayAmong the different concentrations of samples used, a positive signal is observed in of protein by the ELISA technique, whereas the detection failed in 1, 5, 20, and of protein. This shows the significance of the results obtained with PCF, where only protein is needed for obtaining the positive signal, whereas several fold more protein is needed for the detection through ELISA. 4.3.Fluorescence AnalysisThe green and the red signals from the fiber imaged under the microscope are shown in Figs. 5, 5, 5, 5, 6, 6, 6, 6 , respectively. The fluorescent signal is found to increase linearly with the concentration of cell lysate as a result of a greater number of binding surfaces inside. The protein binding inside the fiber is further confirmed by spectroscopic analysis. Both the control fibers MDA MB 231 and the sample fiber MCF-7 are analyzed for the emission signal. The control fibers (MDA-MB-231) showed no emission peak, whereas a green-dye emission is observed in MCF-7 fibers at , as given in Figs. 7 and 7 and a red dye emission around as given in Figs. 8 and 8 . These spectral signatures authenticate the strong binding of protein inside the photonic crystal fiber. Fig. 5Fluorescent microscopic images showing protein binding inside the fiber: Alexa Fluor 488 images: (a) negative control and (b–d). positive binding of ER protein ( , and , respectively). 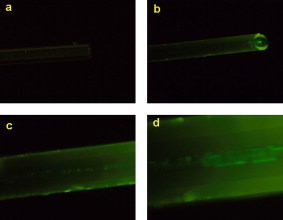 Fig. 6Fluorescent microscopic images showing protein binding inside the fiber: Alexa Fluor 555 images: (a) negative control and (b–d). positive binding of ER protein ( , and , respectively). 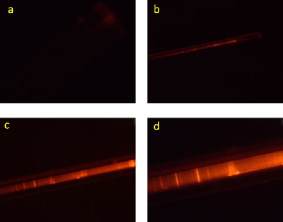 5.DiscussionThe most important aspect of this work is to design a sensor with competent immobilization performance to achieve the highest sensitivity of protein detection. The proposed methodology is focused on an array format immuno recognition of protein using a HC photonic crystal fiber. The primary step of the experiment is to activate the silica inner core of the fiber to facilitate the detection of ER protein. To attach proteins on a solid surface, the surface of the substrate has to be modified to achieve a maximum binding capacity.28, 29 Poly (ethylene glycol) derived materials are generally considered to be effective candidates for fabrication of protein-resistant materials. In particular, polycations are found to form stable adsorbed layers on negatively charged oxides, such as silicon dioxide and titanium dioxide.30 Kenausis described the surface functionalization method and its importance in the protein resistance.31 In their immobilization study, they observed that poly (L-lysine)-g-poly (ethylene glycol) (PLL-g-PEG) complex makes polymer combs backbone with the negatively charged metal surface, producing a protein-resistant surface. PLL-g-PEG surface is considered to be a promising sensor platform for binding His-tagged proteins and high specificity and quantitative reversibility of the protein was observed.32 A biotin-labeled DNA and streptavidin is captured efficiently in PLL and gluteraldehyde coated HC fibers.12 PLL showed no interference with the target protein signal because it does not contain intrinsically fluorescent amino acids.33 The binding of protein mainly depends on the efficient surface attachment procedures, which is essential for biosensor application. In the work reported in this paper, PLL is used to precoat the inner wall of the fiber to create an activated surface that can bind the targeted protein effectively. The second step of the sensor construction is very crucial which involves the protein antigen on silica glass fiber in a way that it conserves its characteristics and confirmations. Initially, the positive (MCF-7) and negative cell (MDA-MB-231) lysates are allowed to immobilize inside the core. Second, the primary antibody raised against the protein (antirabbit) is subsequently used to recognize the protein even in fragments, which is available on the inner core surface. Finally, we used the Alexa Fluor 488 goat antirabbit IgG (green fluorescent dye) as a secondary antibody compatible to the anti- protein. We observed under a microscope that the Alexa-488 IgG quenches fluorescence of the fiber core, which confirms the presence of the specific protein, . Concurrently, we have used another secondary antibody Alexa Fluor 555 goat antirabbit IgG tagged with red fluorescent dye to validate the protein, and a similar profile was noted, evidencing the specific protein binding. Recently, few studies have been reported regarding the immobilization and detection of a biomolecule using microstructured optical fibers. Hoiby performed DNA hybridization and specific recognition of antistrepdavidin with streptavidin was observed.34 Similarly, fluorescently labeled streptavidin cy3 and CRP-cy3 were achieved by immobilizing inside the polymer optical fibers by using sample of .35 Ruan achieved a good immobilization and binding of Q dot 800 goat F (ab’)2 antimouse IgG conjugate using a sample in soft glass optical fiber.11 Multiple toxic agents, such as ricin , F1 antigen from Yersinia pestis , and staphylococcal enterotoxin B were detected in a single sample using planar array immunosensor.36 Fiber optic evanescent wave immunosensors have been developed for the detection of toxins, hazardous materials, explosives, and clinical analytes using monoclonal and poly clonal antibodies.37, 38, 39, 40 Human chorionic gonadotropin was detected through planar waveguide fluorescent immunosensors by sandwich assay using monoclonal antibody.41, 42 Comparatively, the newly developed assay significantly detect the protein by direct immuno binding. In addition, the architecture of this sensor design was strengthened by implementation of strong light sources, precise lenses, and a spectrophotometer to analyze the spectral signatures of the fluorescence images. The emission peaks of green and red dyes were measured at 515 and , respectively. This spatial information strongly confirms the specific binding of protein. It is well understood that the great advantage of the sensor is the capability of analyzing both biological and physical properties of targeted molecules. Though conventional optical fibers are used for immunoassays, photonic crystal fibers are more biocompatible and chemically inert because it is made up of pure silica without any doping. Here, we describe the potentiality of silicon made, evanescent-based HC photonic crystal fiber as a biosensor for the detection of protein using specific fluorescent-labeled antibody. The proposed HC-PCF–based biosensors is advantageous over waveguide-based array sensors as a result of the easy light coupling to the sample region. Also, it is possible to change the excitation laser light source more easily by making use of an optical connector. Although we have used an antigen antibody reaction to recognize the protein, in general, this sensor can be applied to other proteins of interest. Our experimental data of ELISA method shows that it needs more quantity of samples for the detection, whereas our microstructured fiber optical detection needs very low sample quantity. In comparison to the available techniques, our new method is simple, safer, less expensive, sensitive, and highly specific in assessing the ER status in breast cancer research. This method can be applied for high-throughput biosensor application in biological research and also be used as a diagnostic tool in future translational medicine. 6.ConclusionsA biosensing system using a HC photonic crystal fiber in a total internal reflection configuration for monitoring the fluorescence response of the specific protein is established. We demonstrate an antigen-antibody binding method to detect protein in an ultrasmall quantity of sample. The recognition of the protein by secondary antibody is visualized by the fluorescent signal (green and red) and measured emission spectrum confirms the presence of protein. This method involves highly sensitive and simple procedures that can detect of protein in sample volume. Consequently, the photonic crystal fibers are emerging as potential platforms for biosensing applications in biology and medicine. AcknowledgmentsThe authors acknowledge the financial support received through ASTAR-SERC. One of the authors, V. K. Shinoj, thanks the NTU for a research scholarship award. Dr. Parasuraman Padmanabhan acknowledges Sir George Radda, A*STAR-BMRC for extending his support toward this collaborative project. ReferencesE. Phizicky,
P. I. H. Bastiaens,
H. Zhu,
M. Snyder, and
S. Fields,
“Protein analysis on a proteomic scale,”
Nature, 422
(6928), 208
–215
(2003). https://doi.org/10.1038/nature01512 0028-0836 Google Scholar
W. Chen,
“Nanoparticle fluorescence based technology for biological applications,”
J. Nanosci. Nanotechnol., 8
(3), 1019
–1051
(2008). https://doi.org/10.1166/jnn.2008.301 1533-4880 Google Scholar
R. Y. Tsien,
“The green fluorescent protein,”
Annu. Rev. Biochem., 67
(1), 509
–544
(1998). https://doi.org/10.1146/annurev.biochem.67.1.509 0066-4154 Google Scholar
M. M. C. Cheng,
G. Cuda,
Y. L. Bunimovich,
M. Gaspari,
J. R. Heath,
H. D. Hill,
C. A. Mirkin,
A. J. Nijdam,
R. Terracciano,
T. Thundat, and
M. Ferrari,
“Nanotechnologies for biomolecular detection and medical diagnostics,”
Curr. Opin. Chem. Biol., 10
(1), 11
–19
(2006). https://doi.org/10.1016/j.cbpa.2006.01.006 1367-5931 Google Scholar
X. Fan,
I. M. White,
S. I. Shopova,
H. Zhu,
J. D. Suter, and
Y. Sun,
“Sensitive optical biosensors for unlabeled targets: a review,”
Anal. Chim. Acta, 620
(1–2), 8
–26
(2008). https://doi.org/10.1016/j.aca.2008.05.022 0003-2670 Google Scholar
P. St. J. Russell,
“Photonic crystal fibers,”
Science, 299 358
–362
(2003). https://doi.org/10.1126/science.1079280 0036-8075 Google Scholar
R. F. Cregan,
B. J. Mangan,
J. C. Knight,
T. A. Birks,
P. St. J. Russel,
P. J. Roberts, and
D. C. Allan,
“Single-mode photonic band gap guidance of light in air,”
Science, 285 1537
–1539
(1999). https://doi.org/10.1126/science.285.5433.1537 0036-8075 Google Scholar
J. A. Ferguson,
“A fiber-optic DNA biosensor microarray for the analysis of gene expression,”
Bio/Technology, 14
(13), 1681
–1684
(1996). https://doi.org/10.1038/nbt1296-1681 0733-222X Google Scholar
J. B. Jensen,
L. H. Pedersen,
P. E. Hoiby,
L. B. Nielsen,
T. P. Hansen,
J. R. Folkenberg,
J. Riishede,
D. Noordegraaf,
K. Nielsen,
A. Carlsen, and
A. Bjarklev,
“Photonic crystal fiber based evanescent-wave sensor for detection of biomolecules in aqueous solutions,”
Opt. Lett., 29
(17), 1974
–1976
(2004). https://doi.org/10.1364/OL.29.001974 0146-9592 Google Scholar
M. M. Orosco,
“Protein-coated porous-silicon photonic crystals for amplified optical detection of protease activity,”
Adv. Mater., 18
(11), 1393
–1396
(2006). https://doi.org/10.1002/adma.200502420 0935-9648 Google Scholar
Y. Ruan,
E. P. Schartner,
H. E. Heidepriem,
P. Hoggmann, and
T. M. Monro,
“Detection of quantum-dot labeled proteins using soft glass microstructured optical fibers,”
Opt. Express, 15
(26), 17819
–17826
(2007). https://doi.org/10.1364/OE.15.017819 1094-4087 Google Scholar
L. Rindorf,
“Towards biochips using microstructured optical fiber sensors,”
Anal. Bioanal. Chem., 385
(8), 1370
–1375
(2006). https://doi.org/10.1007/s00216-006-0480-8 1618-2642 Google Scholar
N. Skivesen,
A. Tetu,
M. Kristensen,
J. Kjems,
L. H. Frandsen, and
P. I. Borel,
“Photonic-crystal waveguide biosensor,”
Opt. Express, 15
(6), 3169
–3176
(2007). https://doi.org/10.1364/OE.15.003169 1094-4087 Google Scholar
, “Fact Sheet No. 297: Cancer 2006,”
(2006) http://www.cancer-sos.com/content.php Google Scholar
P. Picard,
G. Bunone,
J. W. Liu, and
O. Donze,
“Steroid-independent activation of steroid receptors in mammalian and yeast cells in breast cancer,”
Biochem. Soc. Trans., 25 597
–602
(1997). 0300-5127 Google Scholar
V. J. Elwood,
“Steroid hormones, receptors, and antagonists,”
Ann. N.Y. Acad. Sci., 784 1
–17
(1996). https://doi.org/10.1111/j.1749-6632.1996.tb16223.x 0077-8923 Google Scholar
D. R. J. Snead,
“Methodology of immunohistological detection of oestrogen receptor in human breast carcinoma in formalin-fixed, paraffin-embedded tissue: A comparison with frozen section methodology,”
Histopathology, 23
(3), 233
–238
(1993). https://doi.org/10.1111/j.1365-2559.1993.tb01195.x 0309-0167 Google Scholar
S. M. Thorpe,
“Prognostic value of steroid hormone receptors: Multivariate analysis of systemically untreated patients with node negative primary breast cancer,”
Cancer Res., 47
(22), 6126
–6133
(1987). 0008-5472 Google Scholar
A. P. F. Turner,
“Biosensors—sense and sensitivity,”
Science, 290
(5495), 1315
–1317
(2000). https://doi.org/10.1126/science.290.5495.1315 0036-8075 Google Scholar
T. Vo-Dinh,
“Biosensors and biochips: advances in biological and medical diagnostics,”
J. Anal. Chem. USSR, 366
(6–7), 540
–551
(2000). 0021-8766 Google Scholar
R. I. Stefan,
“Immunosensors in clinical analysis,”
J. Anal. Chem. USSR, 366
(6–7), 659
–668
(2000). 0021-8766 Google Scholar
H. J. Geerligs,
“The influence of pH and ionic strength on the coating of peptides of herpes simplex virus type 1 in an enzyme-linked immunosorbent assay,”
J. Immunol. Methods, 106
(2), 239
–244
(1988). https://doi.org/10.1016/0022-1759(88)90203-7 0022-1759 Google Scholar
J. Yang,
H. Wang,
Y. Jiang,
Y. Sun,
K. Pan,
H. Lei,
Q. Wu,
Y. Shen,
Z. Xiao, and
Z. Xu,
“Development of an enzyme-linked immuno-sorbent assay (ELISA) method for carbofuran residues,”
Molecules, 13
(4), 871
–881
(2008). https://doi.org/10.3390/molecules13040871 1420-3049 Google Scholar
Y. Chien,
M. Ito,
Y. Park,
T. Tagami,
B. D. Gehm, and
J. L. Jamsen,
“A fusion protein of the estrogen receptor (ER) and nuclear receptor corepressor (NCoR) strongly inhibits estrogen-dependent responses in breast cancer cells,”
Mol. Endocrinol., 13
(12), 2122
–2136
(1999). https://doi.org/10.1210/me.13.12.2122 0888-8809 Google Scholar
J. M. Harvey,
G. M. Clark,
C. K. Osborne, and
D. C. Alfred,
“Estrogen receptor status by immunohistochemistry is superior to the ligand-binding assay for predicting response to adjuvant endocrine therapy in breast cancer,”
J. Clin. Oncol., 17
(5), 1474
–1481
(1999). 0732-183X Google Scholar
H. Gobbi,
“Predictive factors of breast cancer evaluated by immunohistochemistry,”
J. Brasil. Patologia Med. Lab., 44
(2), 131
–140
(2008). Google Scholar
M. M. Bradford,
“A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding,”
Anal. Biochem., 72
(1–2), 248
–254
(1976). https://doi.org/10.1016/0003-2697(76)90527-3 0003-2697 Google Scholar
H. Zhu and
M. Snyder,
“Protein arrays and microarrays,”
Curr. Opin. Chem. Biol., 5
(1), 40
–45
(2001). https://doi.org/10.1016/S1367-5931(00)00170-8 1367-5931 Google Scholar
H. Zhu and
M. Snyder,
“Protein chip technology,”
Curr. Opin. Chem. Biol., 7
(1), 55
–63
(2003). https://doi.org/10.1016/S1367-5931(02)00005-4 1367-5931 Google Scholar
S. Morgenthaler,
C. Zink,
B. Stadler,
J. Voros,
S. Lee,
N. D. Spencer, and
S. G. P. Tosatti,
“Poly(L-lysine)-grafted-poly(ethylene glycol)-based surface-chemical gradients: preparation, characterization, and first applications,”
BioInterphases, 1
(4), 156
–165
(2006). https://doi.org/10.1116/1.2431704 1559-4106 Google Scholar
G. L. Kenausis,
J. Vörös,
D. L. Elbert,
N. Huang,
R. Hofer,
L. R. Taylor,
M. Textor,
J. A. Hubbell, and
N. D. Spencer,
“Poly(l-lysine)-g-poly(ethylene glycol) layers on metal oxide surfaces: attachment mechanism and effects of polymer architecture on resistance to protein adsorption,”
J. Chem. Phys., 104
(14), 3298
–3309
(2000). Google Scholar
G. Zhen,
S. Zurcher,
D. Falconet,
F. Xu,
E. Kuennemann, and
M. Textor,
“NTA-functionalized poly(L-lysine)-g-poly(ethelyene glycol): A polymeric interface for binding and studying 6 His-tagged protiens,”
1036
–1038
(2005). Google Scholar
G. MacBeath and
S. L. Schreiber,
“Printing proteins as microarrays for high-throughput function determination,”
Science, 289
(5485), 1760
–1763
(2000). 0036-8075 Google Scholar
P. E. Hoiby,
L. B. Nielsen,
J. B. Jensen,
T. P. Hansen,
A. Bjarklev, and
L. H. Pedersen,
“Molecular immobilization and detection in a photonic crystal fiber,”
Proc. SPIE, 5317 220
–223
(2004). https://doi.org/10.1117/12.528891 0277-786X Google Scholar
J. B. Jensen,
P. E. Hoiby,
G. Emiliyanov,
O. Bang,
L. H. Pedersen, and
A. Bjarklev,
“Selective detection of antibodies in microstructure polymer optical fibers,”
Opt. Express, 13
(5), 5883
–5889
(2005). https://doi.org/10.1364/OPEX.13.005883 1094-4087 Google Scholar
R. M. Wadkins,
J. P. Golden,
L. M. Pritsiolas, and
F. S. Ligler,
“Detection of multiple toxic agents using a planar array immunosensor,”
Biosens. Bioelectron., 13
(3–4), 407
–415
(1998). https://doi.org/10.1016/S0956-5663(97)00113-9 0956-5663 Google Scholar
R. A. Ogert,
J. E. Brown,
B. R. Singh,
L. C. Shriver-Lake, and
F. S. Ligler,
“Detection of Clostridium botulinum toxin A using a fiber optic–based biosensor,”
Anal. Biochem., 205 306
–312
(1992). https://doi.org/10.1016/0003-2697(92)90440-I 0003-2697 Google Scholar
F. S. Ligler,
J. P. Golden,
L. C. Shriver-Lake,
D. Wijesuria,
R. A. Ogert,
J. E. Brown, and
G. P. Anderson,
“Fiber-optic biosensor for the detection of hazardous materials,”
Immunomethods, 3 122
–127
(1993). https://doi.org/10.1006/immu.1993.1046 1058-6687 Google Scholar
U. Narang,
P. R. Gauger,
A. W. Kusterbeck, and
F. S. Ligler,
“Multianalyte detection using a capillary-based flow immunosensor,”
Anal. Biochem., 255 13
–19
(1998). https://doi.org/10.1006/abio.1997.2411 0003-2697 Google Scholar
C. A. Rowe,
S. B. Scruggs,
M. J. Feldstein,
J. P. Golden, and
F. S. Ligler,
“An array immuno sensor for simultaneous detection of clinical analysis,”
Anal. Chem., 71 433
–439
(1999). https://doi.org/10.1021/ac980798t 0003-2700 Google Scholar
J. N. Herron,
K. D. Caldwell,
D. A. Christensen,
S. Dyer,
V. Hlady,
P. Huang,
V. Janatova,
H. K. Wang, and
A. P. Wei,
“Fluorescent immunosensors using planar waveguides,”
Proc. SPIE, 1885 28
–39
(1993). https://doi.org/10.1117/12.144735 0277-786X Google Scholar
D. A. Christensen,
S. Dyer,
D. Fowers, and
J. N. Herron,
“Analysis of excitation and collection geometries for planar waveguide immunosensors,”
Proc. SPIE, 1886 2
–8
(1993). https://doi.org/10.1117/12.144833 0277-786X Google Scholar
|