|
|
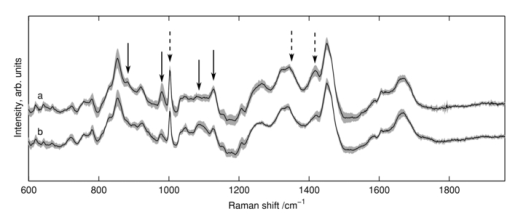
1.IntroductionRapid identification of microbial species is important to any field where bacterial contamination is a possibility. In medical diagnostics, it may allow for faster treatment of disease than would be possible with traditional microbiological techniques. For example, selective plating involves streaking a bacterial sample on a variety of agar plates containing different growth media and waiting up to 48 h to determine the species. Serial dilutions on these selective plates typically provide only an order-of-magnitude estimate of concentration, making it difficult to determine relative concentrations of two or more species. In addition, this method can identify only bacteria that are in a state capable of reproduction. Raman spectroscopy has the potential to identify and quantify bacterial species in a more timely manner, including those that have died/ceased replication, by probing the chemically specific differences between bacteria in a noninvasive manner. It has been used to successfully identify a variety of medically and clean-room-relevant bacteria.1, 2, 3 Raman spectroscopy is used here to distinguish between two species of streptococci S. sanguinis and S. mutans. These bacteria are of particular interest due to their relationship with oral health, as the two most common species found in human dental plaque.4 On one hand, S. mutans has been identified as the most cariogenic species, with elevated concentrations being linked to increased tooth decay.5, 6 On the other hand, S. sanguinis is the main constituent of healthy plaque.4 Examining the relative concentration balance between these two species may serve as an indicator of a patient's oral health and risk of tooth decay. In previous work, our group successfully discriminated between several species of oral bacteria, including S. sanguinis and S. mutans, and determined relative concentrations of multiple species in a mixed sample.7 For simplicity, the bacteria in these studies were centrifuged from planktonic suspension; this environment is not, however, a good model for the bacterial environment on the surface of teeth. The focus is now shifted to study S. sanguinis and S. mutans as they appear in biofilms as a simplified model for dental plaque. Cells growing as a biofilm exhibit a number of properties that are distinct from cells grown in suspension, including changes in protein production,6 making it important to study the bacteria in the form in which they are more commonly found. In this paper, the ability to distinguish between biofilm samples of S. sanguinis and S. mutans is demonstrated. A species-identification model was calibrated with spectra from pure biofilms and validated on a unique set of pure biofilms grown at a later date. Also, since a further goal of this work is to examine intact biofilms of a mixture of the two species, the spatial resolution at which correct classifications can be performed in a multispecies environment was examined. As a means of verifying the true species in mixed biofilms, “pseudo-mixed” biofilms were created, where one species is stained before stirring the two species together. 2.Methods2.1.Biofilm PreparationsStreptococcus mutans UA159 and S. sanguinis 10904 were the bacterial strains used in this study. Although the species have different preferred growth media, identical preparations were used to avoid any chance that classifications would be based on chemical information from the food source. Cells were streaked onto agar plates containing brain heart infusion medium (BD Difco, Franklin Lakes, New Jersey). Colonies were selected to inoculate liquid cultures containing Todd Hewitt (TH) broth (VWR International, West Chester, Pennsylvania) and 0.5% (w/v) sucrose. The introduction of sucrose enabled the bacteria to begin production of extracellular polysaccharides (EPS), which are necessary for the formation of biofilms. After approximately 24 h, 1 mL of the inoculant was added to ∼49 mL of fresh TH with 0.5% sucrose in a bottle containing a glass microscope slide standing upright. Twenty-four hours later, the slide was transferred to fresh TH with 0.5% glucose. The change of sugars was meant to enable the biofilms to continue growing in a manner that would make them rich in cellular content while limiting the amount of EPS generated. The media was changed daily to reach 4 days of biofilm growth. Since the biofilms were grown as models for dental plaque, samples of the biofilms were scraped from the microscope slides and transferred to CaF2 disks, much like a plaque scraping might be harvested from a tooth surface. The samples were allowed to dry at room temperature to prevent sample recession from the laser focus as a result of evaporation during longer scans. For the generation of pseudo-mixed biofilms, a staining protocol was created based on traditional Gram staining procedures. We centrifuged 2 mL of suspended planktonic cells from the liquid surrounding a biofilm slide at 10,000 rpm for 6 min. The supernatant was removed and 100 μL of crystal violet (CV) was added. After a few minutes, 1 mL of water was added to dilute the dye and the sample was recentrifuged. We added 100 μL of iodine to the pellet and allowed it to sit for a few minutes. We added 1 mL of water to dilute the iodine before recentrifugation. Since streptococci are Gram-positive, the staining procedure could be terminated at this point to avoid the cellular damage that might occur with the washing of cells with ethanol or acetone. The sample was, however, washed three times with water to remove loose stain/iodine. Inspection via microscope indicated uptake of the stain by the bacteria. 2.2.System DesignThe confocal Raman microscope used in this study was described previously8 and is shown schematically in Fig. 1. An 830-nm diode laser (Micro Laser Systems, Inc., Garden Grove, California) was used to excite Raman scattering. This near-IR wavelength was selected to help avoid the fluorescence commonly observed in biological materials, as well as to prevent damage to the sample as these results will be used to study biofilms in situ in the future. The beam passed through a bandpass filter (Chroma Technology Corp., Bellows Falls, Vermont) and a spatial filter (10×objective, Newport Corp., Irvine, California; 10-μm pinhole). The beam was then reflected from a holographic notch filter (Semrock, Inc., Rochester, New York) at near-normal incidence before being directed into the upright microscope. A 50×, 0.8 numerical aperture (NA) air objective (Nikon Corp., Tokyo, Japan) focused the laser to a spot of diameter 1.5 μm, delivering ∼40 mW of laser light to the sample plane. Epidirected Raman scattered light was collected by the objective and the Raman-shifted light was transmitted through the notch filter. The signal was focused onto a circular optical fiber array (100-μm core fibers), where only the central fiber was used in confocal operation. The fibers were mapped to a linear array to deliver the light to the spectrometer (HoloSpec f/1.8, Kaiser Optical Systems Inc., Ann Arbor, Michigan). A thermoelectrically cooled, front-illuminated, open electrode charge-coupled device (CCD) array (DU420-OE, Andor Technology PLC, Belfast, Northern Ireland) was used to collect the Raman spectra. The CCD was operated using code written in-house within MATLAB® (Version 7.8.0, The MathWorksTM, Inc., Natick, Massachusetts). The system has a spectral resolution of ∼7 cm−1, as measured from neon gas emission lines. The axial sectioning depth is ∼5 μm, as determined from the derivative of the response curve when scanning into plastic, following the method described by Caspers 9 Fig. 1Schematic overview of confocal Raman microscope; see text for details. Abbreviations: BPF, bandpass filter; SF, spatial filter; HNF, holographic notch filter. 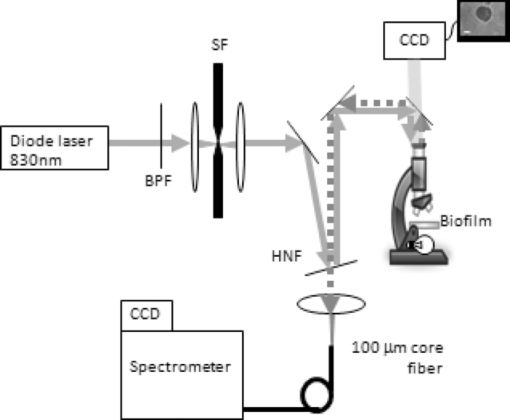 Samples were placed on an electronically controlled stage (x-y; Applied Scientific Instrumentation, Eugene, Oregon; z; Nikon). When performing lateral scans to assess the spatial resolution of our species identification, the stage was axially positioned to place the laser focus 2 μm below the air-sample boundary to probe a consistent depth. This was achieved through an autofocusing algorithm based on the image of the laser light reflected from the sample's surface, with code written in MATLAB®. 2.3.Data Acquisition and PreprocessingRegions for Raman analysis were either chosen randomly from areas thicker than ∼10 μm or as part of a gridlike scan across a dried biofilm sample. Six frames of 30-s exposures were recorded to generate one spectrum for each location studied. All spectra were submitted to preprocessing consisting of cosmic-ray and system background removal and spectral throughput correction. The fluorescent background was removed using a modified polynomial-fitting method described previously.8 After preprocessing separately, the frames were averaged. Due to spectrum-to-spectrum instabilities in the laser's excitation wavelength, all spectra were recalibrated to align the 1003 cm−1 phenylalanine peak10 and resampled to a common wavenumber axis. The wavenumber region of 706 to 1810 cm−1 from each spectrum was submitted for species identification. 2.4.Species Identification MethodsPrincipal component (PC) analysis was used for data reduction of the 651 included pixels from each spectrum. The number of PC scores to retain for subsequent calculations was selected using the smallest number that provided >95% classification accuracy in a leave-one-group-out cross-validation (LOGOCV). This subset of PC scores, along with their corresponding species labels, were then submitted to logistic regression 11 (LR) to create a model for species separation. Because the PC-LR method is spectroscopically abstract, a more intuitive classification scheme was sought to verify the hypothesis of Raman-based classification. The regression coefficients from the LR calibration b λ and the difference of the mean spectra from each species M λ (≡ [TeX:] \documentclass[12pt]{minimal}\begin{document}$\bar{s}^{\rm mutans}_{\lambda}$\end{document} − [TeX:] \documentclass[12pt]{minimal}\begin{document}$\bar{s}^{\rm sanguinis}_\lambda$\end{document} ), were used to construct the spectrum b λ M λ. By definition, b λ M λ assigns large values to spectral regions with the greatest predictive power. Generally, these regions coincided with slight but noticeable peak-height changes in the mean spectra of the two species. Areas under four such peaks were used as another method of data reduction, with these values also being submitted to LR for species classification. 3.ResultsFigure 2 shows white light transmission images of the biofilm samples from each species. The images appear similar, making it impossible to identify the species of origin based on conventional white light transmission imaging alone. Raman spectroscopy's chemical specificity offers the ability to distinguish between the species. Fig. 2White light transmission images of (a) S. mutans and (b) S. sanguinis show that the species can not be distinguished based on appearance alone. (Bar length = 10 μm.) 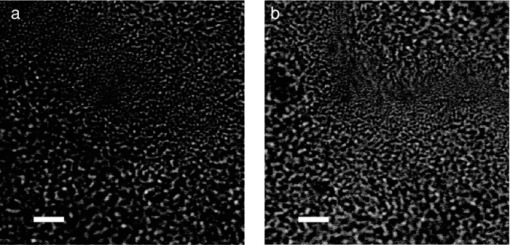 3.1.Calibration on Pure BiofilmsFigure 3 shows the mean spectra for 173 and 162 voxels from S. sanguinis and S. mutans, respectively, acquired from 37 biofilms over 6 months with a minimum of five spectra acquired from each biofilm. The mean spectrum of each species was calculated for each measurement day; the shaded backgrounds in Fig. 3 indicate the standard deviations of those mean spectra. These 335 spectra comprise the calibration set for discrimination of future samples. One acquired spectrum was not entered into the calibration set due to its abnormal background, which could not be properly fit in the background removal process. Twenty was the minimum number of PCs needed to achieve 95% LOGOCV accuracy and was therefore selected for the analysis here, although using 19 or more PCs gave fairly consistent LOGOCV results. To give a sense of scale, these PCs account for 93.4% of the variance in the calibration set. 3.2.Validation on Pure BiofilmsThe PC-LR model was applied to a validation set of 96 spectra acquired from 11 pure biofilms over 9 months, with all biofilms grown and examined after the completion of the calibration set. One additional acquired spectrum was omitted after being flagged by an automated outlier detection method based on that described by Haaland and Thomas.12 Table 1 shows the results of the PC-LR species identification for these spectra; an overall accuracy of 96% was achieved. Table 1Validation of PC-LR model on 96 new spectra.
The arrows in Fig. 3 indicate regions of spectral variation between species, as flagged by large values of b λ M λ in the PC-LR model, as already mentioned. The areas under a subset of these peaks (after baseline removal, indicated by solid arrows in Fig. 3), were then used as spectral markers for Raman feature-based classifications, submitting the peak areas to LR in place of PC scores. Using this approach, 95% of spectra were classified correctly by species. It is a satisfying observation that the classification process can be performed nearly as well using visible Raman features rather than PC decomposition, although no chemical interpretation for these peaks is offered at this time. Species-specific distinctions in Raman spectra are most likely a result of genomic differences between organisms, translating into diversity in expressed proteins. While some of these proteins reside within the cells, others are secreted and thereby influence the extracellular environment. Certain oral bacteria, in particular, produce glucosyltransferase enzymes that assemble glucose chains called glucans; these are essential components of the EPS matrix that typifies oral plaque's architecture. Streptococcus sanguinis and S. mutans are known to produce different types and relative quantities of glucans.13, 14, 15, 16 Since our sample volume size is larger than a single bacterium and the sample transferral process leads to some homogenization of the biofilm, presumably some EPS may be present in the Raman sampling volume along with (or even to the exclusion of) cells. In principle, the source of species-specific spectral signatures could be the EPS rather than the bacterial cells themselves. Cellular differences were most likely involved, however, because spectra of these bacteria grown without the nutrients necessary for EPS production could also be classified according to species (data not shown). This is also in agreement with previous results from our group, with bacteria in colonies grown on agar or centrifuged from planktonic culture.17, 18 3.3.Analysis of Mixed BiofilmsWith confidence in our calibration set as validated by the set of pure biofilm samples, species identifications were performed when both species were present in a mixed biofilm. To have a means of knowing the true species in a given location, one species was stained before stirring it together with a biofilm sample from the opposite species, creating pseudo-mixed biofilms like that shown in Fig. 4. The modified Gram staining procedure left the treated cells with spectral artifacts from the stain; however, it was still possible to scan over boundaries of stained-unstained cells and achieve good species identification in the unstained regions. In the unstained S. mutans scan in Fig. 4, for example, 110 of 115 unstained locations were classified as S. mutans. It should be noted that three of the locations classified as S. sanguinis occur at boundaries where one cannot rule out the possibility that the voxel was indeed dominated by unstained or minimally stained S. sanguinis, though this seems unlikely. Assuming all locations not flagged as containing stain are in fact the unstained species, for all six pseudo-mixed biofilms examined, 97% of 526 locations sampled were correctly identified. This good classification was achieved within ∼2 μm of the boundary between stained and unstained cells. Given that a single bacterium is ∼1 μm in diameter, this indicates that our system cannot image a single bacteria in a biofilm environment, but should be suitable for imaging larger scale structure in intact, multispecies biofilms. Fig. 4White light transmission image of pseudo-mixed biofilm (left) with a close-up of the scanned region (right). Darker regions are S. sanguinis stained with CV, while lighter regions are unstained S. mutans. Assignments in the scan: dark circles, S. mutans; light circles, S. sanguinis; x, spectral outlier (for example, due to the stain). Circle sizes represent laser focus. (Bar length = 10 μm.) 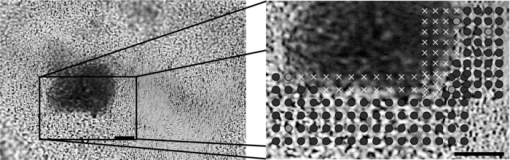 4.DiscussionMost literature on Raman analysis of bacteria focuses on either single cells or bulk samples, typically taken from colonies on an agar plate1, 2, 3, 17 or centrifuged from liquid suspensions.7, 18 When studying biofilms, however, the use of confocal Raman microscopy provides the additional opportunity to study spatial distributions, with the option of performing this study in situ.19 While confocal mapping of biofilms is still largely dominated by fluorescence techniques, Raman has been used to map distributions of water and biomass,20 pigmented EPS and cellular content,21 and chemical components such as sugars or proteins.10 Concerning biofilms including S. mutans, only diffusion of polyethylene glycol has been studied.22 Numerous fluorescence studies have been performed on this species; however, these are generally limited to resolving distributions of cells versus other materials and are incapable of distinguishing between species.23 Fluorescence methods capable of species discrimination include green fluorescence protein (GFP) incorporation or fluorescent in situ hybridization (FISH). These methods rely on genetically altered samples24 (GFP) or require invasive preparations of grown biofilms19 (FISH). Raman spectroscopy, in contrast, has the potential to spatially discriminate between unaltered species in intact biofilms. Pätzold mapped the distribution of two species in a wastewater-generated biofilm, although one species was unidentified. The mean spectra for these species were quite different, being easily distinguished by eye, due to their coming from a variety of genera.19 In the work presented here, species of the same genus with Raman spectra that are visually almost identical (see Fig. 3) were distinguished. Confocal Raman microscopy's spatial discrimination should enable the study of the interplay between species under different growth conditions. Observing the interaction of S. sanguinis and S. mutans may lead to insights for treatment methods to prevent tooth decay. 5.ConclusionConfocal Raman microscopy was used to discriminate between samples of S. sanguinis and S. mutans in biofilm form, both in isolation and in pseudo-mixed biofilms. For these two validation sets, 97% of 622 spectra were properly identified. The lateral resolution achieved in the scans of psuedomixed biofilms was ∼2 μm, which should be reasonable for the structural study of intact, multispecies biofilms. Future work will include the study of biofilms of S. sanguinis and S. mutans grown together throughout the preparation of the biofilm, under various growth conditions. The methodology used here is general in nature and should directly transfer to studies of other multibacterial biofilms. AcknowledgmentsThe authors would like to acknowledge Roberta Faustoferri and Ben Metcalf from the Center for Oral Biology for advice in microbiological preparations. Helpful conversations with Hyun Koo and Jin Xiao are gratefully acknowledged. This work was partially funded through National Institutes of Health (NIH) Grants 1-R21-DE016111-01A1 (AJB), DE0-17157 (RGQ), and DE0-13683 (RGQ), and by a graduate fellowship from the Department of Defense's (DoD’s) National Defense Science & Engineering (NDSEG) program (BDB). ReferencesK. Maquelin,
L.-P. Choo-Smith,
T. van Vreeswijk,
H. P. Endtz,
B. Smith,
R. Bennett,
H. A. Bruining, and
G. J. Puppels,
“Raman spectroscopic method for identification of clinically relevant microorganisms growing on solid culture medium,”
Anal. Chem., 72
(1), 12
–19
(2000). https://doi.org/10.1021/ac991011h Google Scholar
P. Rösch,
M. Schmitt,
W. Kiefer, and
J. Popp,
“The identification of microorganisms by micro-Raman spectroscopy,”
J. Molec. Struct., 661‐662 363
–369
(2003). Google Scholar
M. Harz,
P. Rösch,
K. D. Peschke,
O. Ronneberger,
H. Burkhardt, and
J. Popp,
“Micro-Raman spectroscopic identification of bacterial cells of the genus Staphylococcus and dependence on their cultivation conditions,”
Analyst, 130
(11), 1543
–1550
(2005). https://doi.org/10.1039/b507715j Google Scholar
S. S. Socransky and
S. D. Manganiello,
“The oral microbiota of man from birth to senility,”
J. Periodontol., 42
(8), 485
–496
(1971). Google Scholar
W. J. Loesche,
“Role of Streptococcus mutans in human dental decay,”
Microbiol. Rev., 50
(4), 353
–380
(1986). Google Scholar
P. D. Marsh,
“Microbiologic aspects of dental plaque and dental caries,”
Dent. Clin. North Am., 43
(4), 599
–614
(1999). Google Scholar
Q. Zhu,
R. G. Quivey, and
A. J. Berger,
“Raman spectroscopic measurement of relative concentrations in mixtures of oral bacteria,”
Appl. Spectrosc., 61
(11), 1233
–1237
(2007). Google Scholar
B. D. Beier and
A. J. Berger,
“Method for automated background subtraction from Raman spectra containing known contaminants,”
Analyst, 134
(6), 1198
–1202
(2009). https://doi.org/10.1039/b821856k Google Scholar
P. J. Caspers,
G. W. Lucassen,
H. A. Bruining, and
G. J. Puppels,
“Automated depth-scanning confocal Raman microspectrometer for rapid in vivo determination of water concentration profiles in human skin,”
J. Raman Spectros., 31
(8–9), 813
–818
(2000). https://doi.org/10.1002/1097-4555(200008/09)31:8/9813::AID-JRS5733.0.CO;2-7 Google Scholar
M. Wagner,
N. P. Ivleva,
C. Haisch,
R. Niessner, and
H. Horn,
“Combined use of confocal laser scanning microscopy (CLSM) and Raman microscopy (RM): investigations on EPS matrix,”
Water Res., 43
(1), 63
–76
(2009). https://doi.org/10.1016/j.watres.2008.10.034 Google Scholar
V. Bewick,
L. Cheek, and
J. Ball,
“Statistics review 14: logistic regression,”
Crit. Care, 9
(1), 112
–118
(2005). https://doi.org/10.1186/cc3045 Google Scholar
D. M. Haaland and
E. V. Thomas,
“Partial least-squares methods for spectral analyses. 1. Relation to other quantitative calibration methods and the extraction of qualitative information,”
Anal. Chem., 60
(11), 1193
–202
(1988). https://doi.org/10.1021/ac00162a020 Google Scholar
H. Aoki,
T. Shiroza,
M. Hayakawa,
S. Sato, and
H. K. Kuramitsu,
“Cloning of a,”
Streptococcus mutans glucosyltransferase gene coding for insoluble glucan synthesisInfect. Immun., 53
(3), 587
–594
(1986). Google Scholar
N. Hanada and
H. K. Kuramitsu,
“Isolation and characterization of the Streptococcus mutans gtfC gene, coding for synthesis of both soluble and insoluble glucans,”
Infect. Immun., 56
(8), 1999
–2005
(1988). Google Scholar
N. Hanada and
H. K. Kuramitsu,
“Isolation and characterization of the Streptococcus mutans gtfD gene, coding for primer-dependent soluble glucan synthesis,”
Infect. Immun., 57
(7), 2079
–2085
(1989). Google Scholar
L. K. Kopec,
A. M. V. Smith,
D. Wunder,
L. Ng-Evans, and
W. H. Bowen,
“Properties of Streptococcus sanguinis glucans formed under various conditions,”
Caries Res., 35
(1), 67
–74
(2001). Google Scholar
A. J. Berger and
Q. Zhu,
“Identification of oral bacteria by Raman microspectroscopy,”
J. Mod. Opt., 50
(15–17), 2375
–2380
(2003). Google Scholar
Q. Zhu,
R. G. Quivey, and
A. J. Berger,
“Measurement of bacterial concentration fractions in polymicrobial mixtures by Raman microspectroscopy,”
J. Biomed. Opt., 9
(6), 1182
–1186
(2004). https://doi.org/10.1117/1.1803844 Google Scholar
R. Pätzold,
M. Keuntje, and
A. Anders-von Ahlften,
“A new approach to nondestructive analysis of biofilms by confocal Raman microscopy,”
Anal. Bioanal. Chem., 386
(2), 286
–292
(2006). https://doi.org/10.1007/s00216-006-0663-3 Google Scholar
C. Sandt,
T. Smith-Palmer,
J. Comeau, and
D. Pink,
“Quantification of water and biomass in small colony variant PAO1 biofilms by confocal Raman microspectroscopy,”
Appl. Microbiol. Biotechnol., 83
(6), 1171
–1182
(2009). https://doi.org/10.1007/s00253-009-2072-y Google Scholar
C. Sandt,
T. Smith-Palmer,
J. Pink,
L. Brennan, and
D. Pink,
“Confocal Raman microspectroscopy as a tool for studying the chemical heterogeneities of biofilms in situ,”
J. Appl. Microbiol., 103 1808
–1820
(2007). https://doi.org/10.1111/j.1365-2672.2007.03413.x Google Scholar
L. Marcotte,
J. Barbeau, and
M. Lafleur,
“Characterization of the diffusion of polyethylene glycol in Streptococcus mutans biofilms by Raman microspectroscopy,”
Appl. Spectrosc., 58
(11), 1295
–1301
(2004). Google Scholar
M. I. Klein,
S. Duarte,
J. Xiao,
S. Mitra,
T. H. Foster, and
H. Koo,
“Structural and molecular basis of the role of starch and sucrose in Streptococcus mutans biofilm development,”
Appl. Environ. Microbiol., 75
(3), 837
–841
(2009). Google Scholar
M. B. Aspiras,
K. M. Kazmerzak,
P. E. Kolenbrander,
R. McNab,
N. Hardegen, and
H. F. Jenkinson,
“Expression of green fluorescent protein in Streptococcus gordonii DL1 and its use as a species-specific marker in coadhesion with Streptococcus oralis 34 in saliva-conditioned biofilms in vitro,”
Appl. Environ. Microbiol., 66
(9), 4074
–4083
(2000). https://doi.org/10.1128/AEM.66.9.4074-4083.2000 Google Scholar
|
|||||||||||||||