|
|
1.IntroductionDiffuse optical imaging has received heightened attention in the last decade because of its capability to provide functional images of the tissue under investigation using nonionizing near-infrared light (600–1000 nm).1, 2 Specifically, imaging of brain and breast has been the primary applications of diffuse optical tomography.1, 2, 3 The critical step in obtaining these images is estimating internal distribution of optical properties of the tissue using the measurements made on the tissue boundary.4, 5 Because the scattering is the dominant mechanism for near-infrared light (NIR) interaction with tissue, the estimation problem, also known as the inverse problem, is nonlinear, ill-posed, and some times underdetermined.4 Thus, solving an inverse problem necessitates the use of computationally intensive models. The computed data using these models are matched with the experimental data iteratively in the least-squares sense to obtain the optical properties of the tissue.4 The major challenge in terms of obtaining these optical images in real time is the computational cost associated with these advanced computational models, because these are used repeatedly.2, 4, 6 These iterative techniques used in the inverse problem relies on the calculation of modeled data and Jacobian (or its variant) at each iteration to obtain an update of optical properties.4, 6, 7 Depending on the model used, these calculations can span up to several hours, especially in three-dimensional (3-D) cases.6, 8 There were attempts earlier to accelerate these calculations by using parallel computers, which have been shown to give a speedup of factor n, with n being the number of parallel processors used.6 The limitation with the use of parallel computers is the cost associated with achieving higher speedups and also the complex approach in parallelizing these computational models. Here, we aim to take advantage of general purpose graphics processing units (GPUs) in massively parallelizing these calculations. This work specifically aims to present accelerating the frequency domain diffuse optical image reconstruction using a cost-effective (∼$1200) programmable GPU (NVIDIA Tesla C 1060). This is achieved using an open-source GPU-accelerated linear algebra library (CULA) that utilizes the NVIDIA's compute unified device architecture (CUDA).9 Earlier works have shown the parallel computation capability of GPUs in performing high-speed Monte Carlo simulation of photon propagation in tissue and proven to give a speedup of ∼100 for simple cases10, 11 and ∼10 in heterogeneous tissues12 when compared to the implementation on a modern central processing unit (CPU). Also, the standard filtered backprojection (Radon transform-based) algorithm used in computed tomography (CT) has been shown to give a speedups of 100 compared to standard CPUs.13 These kinds of massively parallel programmable GPUs have been used in the context of optical projection tomography and shown to give 300-fold acceleration compared to traditional CPUs.14 For iterative algorithms that are used in CT reconstruction, such as simultaneous algebraic reconstruction, the reported speedups are 12-fold per iteration, comparing GPU to CPU.15 The image reconstruction algorithms that use Fourier transform or its variant have been shown to give speedups on the order of 100 or more using GPUs, and the reconstruction algorithms that mainly involve matrix computations have been shown to give speedups on the order of 10 or more.14, 15 The main aim of this work is to demonstrate the parallel computing power of these GPUs for performing diffuse optical tomographic image reconstruction. Here, we used a finite element (FE) method (FEM)–based computational model that solves the frequency-domain diffusion equation (DE) requiring complex arithmetic. The number of unknowns in the inverse problem will be equal to 2N N (N N absorption coefficients and N N-diffusion coefficients), with N N being the number of finite element nodes used in the FE mesh to obtain the modeled data (described later). To obtain the modeled data, Jacobian, and update of optical properties at every iteration, a set of linear system of equations (order of 2N N) needs to be solved along with the matrix-matrix multiplications (order of 2N N), which will take typically O[(2N N)3] operations. We will show that these operations could be done in parallel using a GPU (NVIDIA Tesla C 1060) to accelerate the diffuse optical tomographic image reconstruction procedure. As diffuse optical image reconstruction relies on numerical calculations, specifically system solve (i.e., solving x in Ax = b) and matrix multiplications. Several packages,16, 17, 18, 19, 20, 21, 22, 23, 24 listed in Table 1, exist that provide options for carrying out the calculations on GPUs. The FEM-based forward models based on frequency-domain DE results in sparse symmetric linear system of equations (complex type). This, in turn, limits the use of sparse numerical computations on GPUs in these cases (more discussion to follow). The only package that is capable of dealing with sparse system solve for the real type is Cusp,19 allowing only matrix-vector computations. It will be shown that even with the use of the full (nonsparse) matrices, the GPUs are capable of given an acceleration of up to 7 (for completing a start-to-end single iteration of the diffuse optical image reconstruction) compared to CPU sparse computations. Note that the Linux-based platform is used to carry out the computations performed in this work. Table 1Comparison of features of available CUDA-based numerical linear algebra packages along with their source.
2.Methods2.1.Diffuse Optical Tomographic Image ReconstructionDiffuse optical tomographic image reconstruction is typically performed using Newton-type algorithms, where the modeled data [G(μ) with μ representing the set of optical properties] is matched to the experimental data (y), iteratively, in the least-square sense.4, 7 The most popular technique among the full Newton-type algorithms is Levenberg–Marquardt minimization, described in detail in Ref. 7. The FEM-based frequency-domain diffusion model for the calculation of G(μ) is described in Refs. 25, 26; here, it is only briefly reviewed. The frequency-domain DE is given by25 Eq. 1[TeX:] \documentclass[12pt]{minimal}\begin{document}\begin{eqnarray} \qquad&& -\nabla D(r)\nabla \Phi (r,\omega)\, + \,\left({\mu _a (r)\, + \,\frac{{i\omega}}{c}} \right)\,\Phi (r,\omega)\nonumber\\ \qquad&&\quad= \,q_o (r,\omega), \end{eqnarray}\end{document}Eq. 2[TeX:] \documentclass[12pt]{minimal}\begin{document}\begin{equation} D(r)\, = \,\frac{1}{{3[\mu _{\rm a} (r)\, + \,\mu '_{\rm s}(r)]}}, \end{equation}\end{document}In the FEM framework, the imaging domain is discretized into linear triangular elements (for two dimensions) or linear tetrahedral elements (for three dimensions) connected at N N vertex nodes. Then the computational (forward) model for solving the diffusion equation reduces to25, 26 Eq. 3[TeX:] \documentclass[12pt]{minimal}\begin{document}\begin{equation} K\Phi \, = \,q, \end{equation}\end{document}Eq. 4[TeX:] \documentclass[12pt]{minimal}\begin{document}\begin{equation} \,P^{- 1} K\Phi \, = \,\,P^{- 1} q, \end{equation}\end{document}Eq. 5[TeX:] \documentclass[12pt]{minimal}\begin{document}\begin{equation} P_{i,j}^{- 1} \, = \left\{{\begin{array}{@{}l@{\quad}ll} \displaystyle\frac{1}{{K_{i,i}}}&{\rm if}&i = j \\ 0 & {\rm if}&i \ne j. \\ \end{array}} \right. \end{equation}\end{document}Because the complex sparse arithmetic is not yet supported in NVIDIA's CUDA, the GPU implementation of the same is performed by converting K to a full (nonsparse) matrix. The most important steps pertaining to the image reconstruction procedure are given as a flowchart in Fig. 1. The iterative image reconstruction procedure starts with an initial guess for the optical properties (μ0) typically obtained using the calibration procedure of experimental data (y).31, 32 Using this μ0, the forward model is solved to obtain G(μ) and, more importantly, the Jacobian (J = ∂G(μ)/∂μ), which gives the rate of change in the modeled data with respect to optical properties. J is typically obtained using the adjoint formulation,25 and the most important computations pertained to the calculation of J are given in Fig. 2. The computation times for each step (in percent) in calculation of J with adjoint formulation is also given in Fig. 2. It could be seen that calculation of J needs both Φ and Φ* (adjoint fluence, obtained by interchanging the source and detector positions) and typically takes ∼35% of time in the preconditioning {i.e., calculation of P −1 K with the order of calculations as O[(2N N)3]}, as both P −1 and K are complex [for the real case, it will be O(N N 3)].29 Obtaining Φ in Eq. 4 is performed using LU decomposition, and the number of operations required are O(2*(2N N)3/3),29, 30 contributing to ∼25% of total computation time (Fig. 2) taken for calculation of J. Fig. 1Flowchart presenting the important steps involved in reconstructing optical properties in diffuse optical tomography along with associated computation time (in percent of total time per iteration). The steps that used GPU-based computations are indicated with a double arrow. 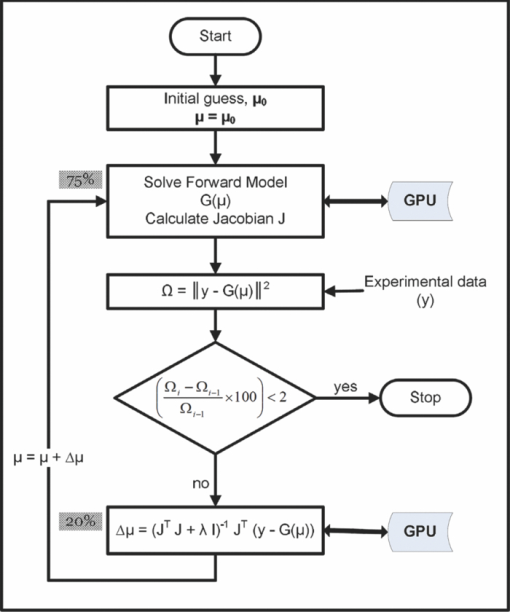 Fig. 2Flowchart showing the important computations for calculation of the Jacobian (J). The computation time in terms of percentage of total time taken in calculation of J at each step is also indicated. Note that the calculation of J accounts for 75% of the total time taken in completing one iteration of diffuse optical image reconstruction (also indicated in Fig. 1).  As the Rytov approximation is used in the work, the frequency domain data becomes, y = [ln(A); θ], where ln(A) is the natural logarithm of amplitude (A) and θ is the phase of the frequency domain signal, making the J a real valued matrix (dimension of 2N M×2N N, where N M is the number of measurements).4, 7 The procedure involved in the calculation of real valued J using complex Φ is given in Ref. 25. Note that the calculation of J [and G(μ)] consumes ∼75% of total computation time taken for completing a single iteration (see Fig. 1). To obtain an update of the optical properties (Δμ), the Levenberg–Marquardt (LM) minimization33, 34 is used and the objective function for this minimization scheme is given as33, 34 Eq. 6[TeX:] \documentclass[12pt]{minimal}\begin{document}\begin{equation} \Omega = \,\parallel \!y - G(\mu)\!\parallel^{2}. \end{equation}\end{document}The objective of the LM minimization scheme is to match y with G(μ) in the least-squares sense, by changing μ. The details of LM minimization scheme are discussed in Ref. 7. The update equation (for getting Δμ) for the LM minimization becomes7 Eq. 7[TeX:] \documentclass[12pt]{minimal}\begin{document}\begin{equation} [{J^{\rm T} J + \lambda I}]\,\Delta \mu = J^{\rm T} [{y -G(\mu)}], \end{equation}\end{document}2.2.GPU-Based Diffuse Optical Tomographic Image ReconstructionThe GPU typically consists of 100–200 stream processors that employ single-instruction multiple thread execution to give massive parallel computation power.35 The stream processing uses multiple processors to execute the same kernel of code in parallel on a large set of data.35 As this processing of large data sets is executed in parallel, the GPU accelerates the computation in cases such as large matrix computations, including multiplications and decompositions. In this work, a NVIDIA GPU, which is CUDA enabled, is used in parallelizing these matrix computations. The specification of this GPU card along with the CPU that is compared against it is given in Table 2. As indicated earlier, the matrix computations are carried out using CULA. The NIRFAST package26 is used for the frequency-domain diffuse optical tomographic image reconstruction. Because NIRFAST is built on Matlab-based routines,26 the CULA functions (written in C, which use CUDA libraries9) are wrapped to form Matlab executable (mex) files (details are given in Ref. 36). The routines that are used in this work along with examples of its usage are given in Table 3. Note that in this work only the basic package of CULA, which is open source, is used. This basic version of the CULA package is limited to single precision, which is sufficient for the diffuse optical tomographic image reconstruction (also shown later). Table 2Specifications of GPU and CPU used in this work along with its limitation on finite element mesh size (based on the available memory) in terms of number of nodes (N N).
Table 3GPU-based Matlab executable (mex) CULA functions that are used in this work.
As indicated in Fig. 1, the developed GPU routines (given in Table 3) based on CULA are used at six instances in a single iteration of the image reconstruction procedure. These instances are listed as follows:
Note that only in the case of matrix-matrix multiplications, CULA functions (CulaSgemm and CulaCgemm) are used. In the cases of matrix-vector multiplications, these functions resulted in an insignificant acceleration (speedup of ∼1.1). More over, in the case of GPU implementation, even though K with K* (along with P −1) are sparse, these matrices were converted to full before using them at the first four instances, because complex sparse arithmetic is not yet supported in NVIDIA CUDA (see Table 1). The main limitation in carrying out these large matrix-matrix computations on the GPU has been the available onboard memory,35 and this limitation puts a constraint on the FE mesh size (in turn, on the number of optical properties) that could be used for the GPU-based calculations. These sizes in terms of number of nodes (N N) are given in the last column of Table 2. 2.3.GPU and CPU ComputationsInitially, because single-precision CULA functions are used, a set of simulations were carried out using both single- and double-precision diffuse optical tomographic image reconstruction to show that single-precision calculations are sufficient. The 2-D FE circular mesh centered around the origin was used for this purpose. This 2-D traingular mesh had 4903 nodes connected to 9576 linear triangular elements. The circular domain had a radius of 43 mm, and 16 source/detector equidistant fibers were placed on the boundary of the domain. When one fiber is used as source, the other fibers act as detectors, resulting in N M of 240 (16×15). The source was placed at one mean transport length inside the boundary and had a Gaussian profile with full width at half maximum of 3 mm to mimic the experimental conditions.37 The background optical properties of the mesh were μa = 0.01 mm–1, μ′ s = 1 mm–1 with uniform refractive index of 1.33. An absorption anomaly with a contrast of 2:1 (i.e., μa = 0.02 mm–1) of diameter 15 mm was placed close to the boundary [at (30,0)] to mimic the tumor.38 Synthetic experimental data were generated using this mesh, and noise of 1% was added to this data to generate y. The background optical properties were used as μ0 (Fig. 1). This image reconstruction procedure is carried out over 15 iterations using the LM minimization procedure. The difference in the L2-norm of the data-model misfit [δ = y − G(μ)] between the single and double precision procedures along with the difference in the L2-norm of the estimated μ is compiled for the comparative purposes. The GPU-based matrix computations used full matrices (represented by GPU-Full for these type of computations). The CPU-based matrix computations (involving instances 1–4 in Section 2.2) were carried out using both sparse (represented by CPU-Sparse) and full (represented by CPU-Full) matrices for comparison purposes. Using these three different strategies, namely GPU-Full, CPU-Full, and CPU-Sparse, the optical image reconstruction procedure with varying sizes of FE meshes is implemented in both 2-D and 3-D cases. For the 2-D (circular geometry) case, the variation in N N is from 1933 to 10249 and for the 3-D case (cylindrical geometry), it is from 4378 to 12695. The 2-D computations are carried out on the circular imaging domain with similar specification given earlier for varying N N. In the 3-D case, similar to 2-D, a ring of equispaced 16 source/detector fibers placed at the center of the Z coordinate are used for boundary data collection, leading 240 (16×15) number of measurements (N M). The optical properties similar to the 2-D test object were used, with the shape of the tumor as a sphere (similar to a circular one in two dimensions). The 2-D meshes were created using Matlab PDE tool box and the 3-D tetrhedral meshes using NETGEN.39 The computation time per single iteration (start to end) is noted for all three strategies (GPU-Full, CPU-Full, and CPU-Sparse) for comparative purposes in both 2-D and 3-D cases, separately. Because the NVIDIA Tesla C 1060 GPU card onboard memory is limited to 4 GB (Table 2), imposing the upper limit of N N as 12,695 in the case of cylindrical FE meshes considered here (constructing a FE mesh of the exact size with N N as 13,377 was not plausible). 3.Results and DiscussionThere are many potential CUDA (GPU)-based numerical linear algebra packages that could be used to accelerate the diffuse optical image reconstruction procedure, some of these along with their features are listed in Table 1. With the need of matrix-matrix computations and system solve leads to the choice of CULA and Jacket from the ones listed in Table 1. Because CULA basic16 has an advantage of being open-source, the same is used in the presented work. Note that CULA is also part of Jacket.20 Also, from the Table 1, it is evident that the complex sparse arithmetic is not supported in CUDA. The difference (in percent) in the L2-norms of the data-model misfit (δ) and estimate optical properties (μ) with respect to iteration number between single- and double-precision computations using CPU are plotted in Fig. 3 for the test object described in Sec. 2.3. It is evident from Fig. 3 that the maximum difference in terms of percentage in either δ or estimated μ is <0.005%, asserting the fact that single precision is sufficient for carrying out the diffuse optical tomographic image reconstruction computations. Because Rytov approximated data are used in this work [i.e., using ln(A) rather than A] the single-precision calculations were adequate. The main advantages of single- over double-precision computations are twofold. First, given the limitation on the available GPU/CPU memory, diffuse optical tomographic image reconstruction could be performed on meshes twice as large. Second, the basic version of CULA (open source), which has only single-precision capability, could be used to perform the GPU computations. Fig. 3Difference (in percent) in the L2 norm between the single- and double-precision computation over the iterations in (a) data-model misfit (δ) and (b) estimated optical properties (μ). 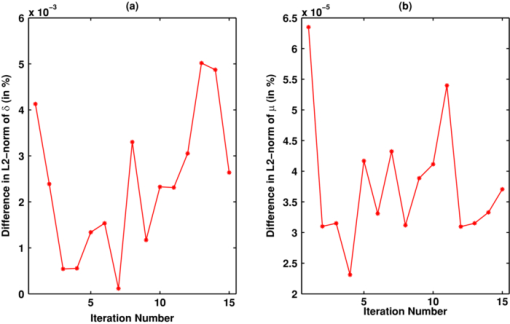 The optical images obtained with integration of CULA- based functions (Table 3 with the instances given in Sec. 2.2) into the diffuse optical image reconstruction procedure using the test case described earlier were identical (visually and numerically) compared to images obtained using single-precision CPU-based implementation. The observed difference (in percent) in the L2-norms of the data-model misfit (δ) and estimate optical properties (μ) between the GPU implementation (with CULA basic) and traditional CPU implementation (with double precision) are same as the observed values plotted in Fig. 3. A 2-D test case reconstruction results along with the target distribution using the three strategies (CPU-Full, CPU-Sparse, and GPU-Full, discussed in Sec. 2.3) are given in Fig. 4. The number of iterations to converge to a solution for each strategy is equal to 10. Obtained μa and μ′ s distributions were correlated to find a similarity measure among the three strategies, resulting in a correlation coefficient of 1 (with in the limits of single precision), asserting that the reconstructed results are identical. Fig. 4Two-dimensional reconstruction results obtained with 1% noisy data using three strategies—CPU-Full, CPU-Sparse, and GPU-Full— discussed in this work. The target 2-D distribution is given in column 1 for (a) μa images and (b) μ′ s images. The number of nodes along with computational time taken for each strategy in a given iteration is given in Table 4.  The time taken per iteration [start to end, including data transfer between CPU (host) and GPU (device) and vice versa] on GPU (NVIDIA Tesla C 1060) and CPU (Intel Xeon E 5410) for different mesh sizes both in two and three dimensions were plotted in Fig. 5 for the three strategies (GPU-Full, CPU-Full, and CPU-Sparse, discussed in Section. 2.3). In the 2-D case, it is clear from Fig. 5 that the computation time per iteration increases with increasing N N for all three strategies, with CPU-Full taking the maximum time. Among the three strategies, GPU-Full took the shortest computation time. As stated earlier, because sparse complex arithmetic is not supported in NVIDIA CUDA, the GPU-Sparse (GPU-based computation using sparse representation of matrices for instances 1–4, described Section 2.2) strategy was not attempted. Comparing the three strategies in this 2-D case, the speedup (acceleration) with the use of GPU was up to 7.5 comparing CPU-Full to GPU-Full. The maximum speedup has been lowered to 2 in comparing CPU-sparse to GPU-Full. For the 3-D case [Fig. 5], a similar trend in the 2-D case was observed with the computation time per iteration being higher than 2-D case for the same N N. In the 3-D case, the speedup (acceleration) with the use of GPU was up to 40 in comparing CPU-Full to GPU-Full, and was lowered to 7 in comparing CPU-sparse to GPU-Full. Fig. 5Plots showing the computation time taken for each strategy [indicated in the legend of (a)] per single iteration versus mesh size (node number, N N) for (a) the 2-D case and (b) 3-D case. The strategies are explained at the end of Section 2.3. The specifications of CPU and GPU are given in Table 2. 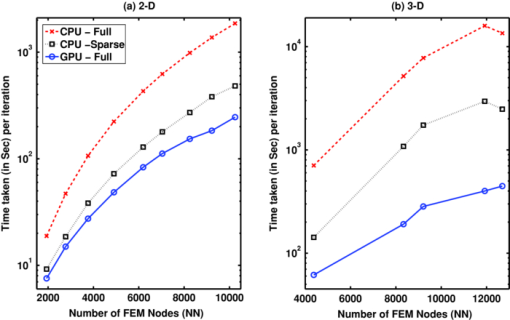 The main emphasis of this work is to prove that the GPU computing offers a considerable speedup compared to CPU computing and the comparison has been only performed by the time taken per iteration. The diffuse optical image reconstruction typically requires about 10 iterations, and for a typical three-dimensional problem (Table 4), the least total time taken by the CPU (CPU-sparse) to converge to a solution is 290 min, and 48 min for GPU. The gain in terms of total computation time is significant in nature (comparing ∼5 h to 0.75 h), making the GPU computing very attractive to be used in the real time, especially in multiwavelength cases (the problem gets scaled by the number of wavelengths), where the aim is to obtain functional images. Table 4Comparison of computation time (in seconds) taken at each instance (as listed in Section 2.2) between 2-D and 3-D meshes of similar size using both CPU (Intel Xeon E 5410 with 4 GB memory) and GPU (NVIDIA Tesla C 1060) in a given iteration using the three strategies discussed in Section 2.3. The last row indicates the total time taken for the iteration. Because J is full matrix, the CPU-Sparse calculations are not applicable (NA) in instances 5 and 6.
Even though it is not possible to attempt sparse complex arithmetic on NVIDIA CUDA-enable GPUs, we have attempted to test the capabilities of parallelizing sparse real computations (with the aid of Cusp, refer to Table 1) using a steady-state diffusion equation (where K and Φ are real) to estimate μa using intensity-based measurements in both 2-D and 3-D cases. Even though the GPU is able to provide speedup (up to 2) for calculation of Φ or Φ* in solving sparse system of linear equations, the overall computation time taken per iteration from start to end (including the overhead of transferring sparse matrices from the CPU memory to GPU memory) is increased (at least by 70%) in comparison of GPU to CPU in these sparse cases (similar to observed trend in Refs. 40, 41). The sparse cases have large overhead for GPU calculations in comparison to the CPU ones because the GPU compute kernels are not fully optimized. This overhead can become negligable when the problem size (number of nonzero entries) becomes bigger (typically >40,000). For fully bandwidth-optmized FEM meshes, the number of nonzero entries for the GPU-limited mesh (with N N = 13,377) is ∼30,000, making the GPU-sparse calculations not attractive compared to CPU-saprse calculations. Thus, only CPU-sparse implementation is attempted in this work. Also, note that sparse computations on currently available GPUs were mainly centered around matrix-vector computations (encountered in iterative solvers to deduce x in Ax = b), which are not as straightforward to implement and integrate into the existing domain-specific applications, such as optical image reconstruction.42, 43 Hopefully, the next generation of GPUs will be able to perform sparse computations with similar ease as current real computations and support sparse complex computations (including matrix-matrix computations). Although it is not attempted for the fair comparison of the CPU and GPU computations, the optimal computations will utilize both CPU (for sparse matrix computations) and GPU (for full matrix computations). Table 4 gives the computational time taken in a given iteration for the mesh of similar size (N N ≈ 9200) both in 2-D and 3-D cases. Although the GPU calculations took similar time between 2-D and 3-D calculations, there is a factor-of-20 difference between the 2-D and 3-D calculations with CPU-Full strategy, especially in solving for Φ in Eq. 4. This is primarily due to the higher numerical condition number of 3-D mass matrix [K inEq. 3], which takes more iterations to converge to a solution [Φ in Eq. 4] compared to their counterparts in 2-D cases for the same size K.8, 44 This higher condition number in the 3-D case could result from the nonuniform nature of 3-D mesh. The tetrahedral elements are formed using 3-D delaunay, which makes a uniform mesh more complex to generate,45 as compared to the 2-D mesh. Also, the condition number of the mass matrix (K) depends on the number of source fibers and their placement. Within this work, where a ring type of data acquisition is performed, the regions far away from the fibers have negligible contribution as opposed to regions close to source/detector fibers. Therefore, the 3-D problem has a higher condition number, requiring more iterations. Hence, this leads to longer computation time, as compared to the 2-D problem, for decomposing P –1 K and then solving for Φ in Eq. 4. Because the decomposition primarily involves matrix-vector computations, with GPU offering massive parallelization, results in similar computation time for the same size matrices both in the 2-D and 3-D cases (Table 4) with GPU giving peak performance in the 3-D case. Note that with J being a full matrix, computations for the last two instances (5 and 6) in Table 4 were not applicable to use sparse computations. These computations were carried out using CPU-Full strategy, and the same computation time taken by the CPU-Full strategy were taken into account in calculating the total time taken per iteration for CPU-sparse strategy. The imaging domains considered in this work are circular for the 2-D case and cylindrical for the 3-D case, with a tumor-to-background contrast being 2. But the trends observed in terms of computational speedup observed between CPU and GPU computing should be similar to any test object (varying tumor-to-background contrast, size, and position of tumor). It is very well known that the contrast recovery is highly dependent on the spatial location of the tumor;38 the images obtained either using single or double precision will be identical (shown in Fig. 3). As mentioned earlier, due to the available onboard memory (4 GB), the FEM mesh that could be used in GPU calculations has to be <13,377 nodes on a NVIDIA Tesla C1060 GPU card. With the advent of newer GPU architectures and generations, this limit could soon disappear. The current Fermi-based NVIDIA Tesla C2050 GPU card offers 6 GB of onboard memory (with 448 CUDA cores),46 which can enable GPU calculation for mesh with the number of nodes as 16,384. Note that the choice of FEM mesh is dependent on the size of the imaging domain, number of measurements, signal-to-noise ratio of the data, and the required stability of the forward solver.47 Discussion of the same is beyond the scope the presented work. Because modern day CPUs have multiple cores, it is possible to take advantage of the parallel computing power of these multicore CPUs. Recent work by Borsic 48 for a similar problem (electrical impedance tomography) like diffuse optical image reconstruction, the achievable speedup using a dual quad-core Intel Xeon processor was 7.6. But this required rewriting/optimizing the routines used in the computing of Jacobian and forward problem, where as the GPU calculations performed in this work used already preexisiting routines. The speedup obtained using a multicore processor with out the optimization is only 4.6.48 Also, the typical cost of dual quad-core processor is at least twice expensive compared to the top of the line GPU used in this work (Table 2). It is important to note that traditional parallel computing uses large number of processors connected in parallel to form a cluster, with an overhead of cost and demanding large footprint in the lab settings. These bulky parallel computer systems are not very attractive in the clinical settings. The GPU boards are comparatively low cost and fit into a traditional desktop computer, resulting in an desktop parallel computer (with a very small footprint), could become very attractive in the clinic to perform image reconstruction/analysis tasks. Usage of these GPU cards to accelerate FE meshing using multimodal imaging data that could be used in the NIR imaging studies is currently being explored. The CULA-based mex programs used in this work along with necessary documentation for installation and compiling are provided as open source.36 4.ConclusionsBecause general purpose computing using GPUs is becoming attractive in many areas of medical image reconstruction/processing, this work demonstrated that with the use of GPUs the speedups for a single iteration (start to end) of diffuse optical tomographic image reconstruction could be up to 40 compared to traditional CPU calculations. Also, the GPU computations are carried out using an open-source package (CULA) and appropriate code wrappers were used to integrate these routines into the existing diffuse optical image reconstruction package (NIRFAST). The current limitation to use GPU on a daily basis for diffuse optical tomographic image reconstruction is the available onboard memory (currently it is limited to 4 GB), which puts a restriction on mesh size that could be used. Moreover, the sparse complex calculations are not supported in GPU computing, making it less attractive for use in FE-based models. Because there is tremendous growth in the capabilities of GPU computing, this limitation might vanish, making GPU computing capabilities similar to those of CPU in applications that require more memory and sparse computations. More importantly, along with the cost effectiveness, these GPU cards are integrated with traditional desktop computers, giving massive parallel computing power at the desktop. Specific to diffuse optical tomography, GPU computing holds the promise of making the image reconstruction procedure match the time line, with the data acquisition, leading to dynamic NIR imaging viable in the clinic. AcknowledgmentsP. K. Y. acknowledges the IISc start-up grant [Grant No. IISc-Part(2A) Eleventh plan, 23/SERC] and the Apple Lauerate award. The authors are thankful to the chairman of the Supercomputer Education and Research Centre, Indian Institute of Science, for providing the GPU cards (NVIDIA Tesla C 1060) for carrying out this work. ReferencesD. A. Boas,
D. H. Brooks,
E. L. Miller,
C. A. DiMarzio,
M. Kilmer,
R. J. Gaudette, and
Q. Zhang,
“Imaging the body with diffuse optical tomography,”
IEEE Sig. Process. Mag., 18 57
–75
(2001). https://doi.org/10.1109/79.962278 Google Scholar
A. Gibson,
J. C. Hebden, and
S. R. Arridge,
“Recent advances in diffuse optical tomography,”
Phys. Med. Biol., 50 R1
–R43
(2005). https://doi.org/10.1088/0031-9155/50/4/R01 Google Scholar
D. R. Leff,
O. J. Warren,
L. C. Enfield,
A. Gibson,
T. Athanasiou,
D. K. Patten1,
J. Hebden,
G. Z. Yang, and
A. Darzi1,
“Diffuse optical imaging of the healthy and diseased breast: a systematic review,”
Breast Cancer Res. Treat., 108 9
–22
(2008). https://doi.org/10.1007/s10549-007-9582-z Google Scholar
S. R. Arridge,
“Optical tomography in medical imaging,”
Inverse Problems, 15 R41
–R93
(1999). https://doi.org/10.1088/0266-5611/15/2/022 Google Scholar
S. R. Arridge and
J. C. Hebden,
“Optical imaging in medicine: II. modelling and reconstruction,”
Phys. Med. Biol., 42 841
–853
(1997). https://doi.org/10.1088/0031-9155/42/5/008 Google Scholar
A. H. Hielscher and
S. Bartel,
“Parallel Programming of gradient-based iterative image reconstruction schemes for optical tomography,”
Comp. Meth. Prog. Biomed., 73 101
–113
(2004). https://doi.org/10.1016/S0169-2607(03)00020-8 Google Scholar
P. K. Yalavarthy,
B. W. Pogue,
H. Dehghani, and
K. D. Paulsen,
“Weight-matrix structured regularization provides optimal generalized least-squares estimate in diffuse optical tomography,”
Med. Phys., 34 2085
–2098
(2007). https://doi.org/10.1118/1.2733803 Google Scholar
P. K. Yalavarthy,
D. R. Lynch,
B. W. Pogue,
H. Dehghani, and
K. D. Paulsen,
“Implementation of a computationally efficient least-squares algorithm for highly under-determined three-dimensional diffuse optical tomography problems,”
Med. Phys., 35 1682
–1697
(2008). https://doi.org/10.1118/1.2889778 Google Scholar
E. Alerstam,
T. Svensson, and
S. Andersson-Engels,
“Parallel computing with graphics processing units for high-speed Monte Carlo simulation of photon migration,”
J. Biomed. Opt., 13 060504
(2008). https://doi.org/10.1117/1.3041496 Google Scholar
Q. Fang and
D. A. Boas,
“Monte Carlo simulation of photon migration in 3D turbid media accelerated by graphics processing units,”
Opt. Express, 17 20178
–20190
(2009). https://doi.org/10.1364/OE.17.020178 Google Scholar
N. Ren,
J. Liang,
X. Qu,
J. Li,
B. Lu, and
J. Tian,
“GPU-based Monte Carlo simulation for light propagation in complex heterogeneous tissues,”
Opt. Express, 18 6811
–6823
(2010). https://doi.org/10.1364/OE.18.006811 Google Scholar
G. C. Sharp,
N. Kandasamy,
H. Singh, and
M. Folkert,
“GPU-based streaming architectures for fast cone-beam CT image reconstruction and demons deformable registration,”
Phys. Med. Biol., 52 5771
–5783
(2007). https://doi.org/10.1088/0031-9155/52/19/003 Google Scholar
C. Vinegoni,
L. Fexon,
P. F. Feruglio,
M. Pivovarov,
J. Figueiredo, M. Nahrendorf,
A. Pozzo,
A. Sbarbati, and
R. Weissleder,
“High throughput transmission optical projection tomography using low cost graphics processing unit,”
Opt. Express, 17 22320
–22332
(2009). https://doi.org/10.1364/OE.17.022320 Google Scholar
F. Xu and
K. Mueller,
“Accelerating popular tomographic reconstruction algorithms on commodity PC graphics hardware,”
IEEE Trans. Nucl. Sci., 52 654
–663
(2005). https://doi.org/10.1109/TNS.2005.852703 Google Scholar
S. R. Arridge and
M. Schweiger,
“Photon-measurement density functions. Part 2: finite-element-method calculations,”
Appl. Opt., 34 8026
–8037
(1995). https://doi.org/10.1364/AO.34.008026 Google Scholar
H. Dehghani,
M. E. Eames,
P. K. Yalavarthy,
S. C. Davis,
S. Srinivasan,
C. M. Carpenter,
B. W. Pogue, and
K. D. Paulsen,
“Near infrared optical tomography using NIRFAST: algorithms for numerical model and image reconstruction algorithms,”
Commun. Numer. Methods Eng., 25 711
–732
(2009). https://doi.org/10.1002/cnm.1162 Google Scholar
M. Schweiger,
S. R. Arridge,
M. Hiroaka, and
D. T. Delpy,
“The finite element model for the propagation of light in scattering media: boundary and source conditions,”
Med. Phys., 22 1779
–1792
(1995). https://doi.org/10.1118/1.597634 Google Scholar
S. Ahn,
A. J. Chaudhari,
F. Darvas,
C. A. Bouman, and
R. M. Leahy,
“Fast iterative image reconstruction methods for fully 3D multispectral bioluminescence tomography,”
Phys. Med. Biol., 53 3921
–3942
(2008). https://doi.org/10.1088/0031-9155/53/14/013 Google Scholar
W. H. Press,
S. A. Teukolsky,
W. T. Vetterling, and
B. P. Flannery, Numerical Recipes: The Art of Scientific Computing, 3rd ed.Cambridge University Press, New York
(2007). Google Scholar
J. R. Westlake, A Handbook of Numerical Matrix Inversion and Solution of Linear Equations, 132 Wiley, Hoboken, NJ
(1968). Google Scholar
B. W. Pogue,
K. D. Paulsen,
H. Kaufman, and
C. Abele,
“Calibration of near infrared frequency-domain tissue spectroscopy for absolute absorption coefcient quantitation in neonatal head-simulating phantoms,”
J. Biomed. Opt., 5 185
–193
(2000). https://doi.org/10.1117/1.429985 Google Scholar
S. Jiang,
B. W. Pogue,
T. O. McBride,
M. M. Doyley,
S. P. Poplack, and
K. D. Paulsen,
“Near-infrared breast tomography calibration with opto-elastic tissue simulating phantoms,”
J. Electron. Imaging, 12 613
–620
(2003). https://doi.org/10.1117/1.1587153 Google Scholar
K. Levenberg,
“A method for the solution of certain nonlinear problems in least squares,”
Quart. Appl. Math., 2 164
–168
(1944). Google Scholar
D. W. Marquardt,
“An algorithm for least squares estimation of nonlinear parameters,”
J. Soc. Ind. Appl. Math., 11 431
–441
(1963). https://doi.org/10.1137/0111030 Google Scholar
J. D. Owens,
M. Houston,
D. Luebke,
S. Green,
J. E. Stone, J. C. Phillips,
“GPU computing,”
Proc. of IEEE, 96 879
–898
(2008). https://doi.org/10.1109/JPROC.2008.917757 Google Scholar
T. O. Mcbride,
B. W. Pogue,
S. Jiang,
U. L. Osterberg, K. D. Paulsen,
“A parallel-detection frequency-domain near-infrared tomography system for hemoglobin imaging of the breast in vivo,”
Rev. Sci. Instrum., 72 1817
–1824
(2001). https://doi.org/10.1063/1.1344180 Google Scholar
P. K. Yalavarthy,
H. Dehghani,
B. W. Pogue, and
K. D. Paulsen,
“Critical computational aspects of near infrared circular tomographic imaging: analysis of measurement number, mesh resolution and reconstruction basis,”
Opt. Express, 14 6113
–6127
(2006). https://doi.org/10.1364/OE.14.006113 Google Scholar
J. Schoberl,
“NETGEN–an automatic 3D tetrahedral mesh generator,”
(2010) http://www.hpfem.jku.at/netgen/ Google Scholar
K. Liu,
X. B. Wang,
Y. Zhang, and
C. Liao,
“Acceleration of time-domain finite element method (TD-FEM) using Graphics Processor Units (GPU),”
(2006). Google Scholar
M. Christen,
O. Schenk, and
H. Burkhart,
“General-purpose sparse matrix building blocks using the NVIDIA CUDA technology platform,”
(2007). Google Scholar
J. Kruger and
R. Westermann,
“Linear algebra operators for GPU implementation of numerical algorithms,”
ACM Trans. Graphics, 22 908
–916
(2003). https://doi.org/10.1145/882262.882363 Google Scholar
M. Schweiger and
S. R. Arridge,
“Comparison of two- and three-dimensional reconstruction methods in optical tomography,”
Appl. Opt., 37 7419
–7428
(1998). https://doi.org/10.1364/AO.37.007419 Google Scholar
M. Bern,
D. Eppstein, and
J. R. Gilbert,
“Provably good mesh generation,”
J. Comput. Syst. Sci., 48 384
–409
(1994). https://doi.org/10.1016/S0022-0000(05)80059-5 Google Scholar
M. Schweiger and
S. R. Arridge,
“Application of temporal filters to time resolved data in optical tomography,”
Phys. Med. Biol., 44 1699
–1717
(1999). https://doi.org/10.1088/0031-9155/44/7/310 Google Scholar
A. Borsic,
A. Hartov,
K. D. Paulsen, and
P. Manwaring,
“3D Electrical impedance Tomography Reconstruction on multi Core computing computing platform,”
1175
–1177
(2008). Google Scholar
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||

