|
|
1.IntroductionParticle tracking has drawn a lot of attention as a useful tool for rheology and micro-rheology studies. 1, 2 It is a popular way to evaluate the viscoelastic properties of micro-environments such as the cell cytoplasm, cell membrane, polymer solutions, etc. For cellular studies the probing beads are inserted into the cells or attached to the membrane, under the general assumption that they do not alter the normal physiology of the cell. Alternatively, fluorescence markers can be used for particle tracking. However, the fluorescence signal is usually weak, suffers from photobleaching, and may produce phototoxicity under long term observation. Therefore, it is valuable to develop label-free methods for tracking intrinsic particles within cells. Since the cells are usually transparent, or termed as phase object, a method based on phase information is required for imaging. Quantitative phase imaging (QPI) of cells and tissues has recently become a dynamic area of study (for a recent review, see Ref. 3). Compared to other qualitative phase imaging methods such as phase contrast and differential interference contrast, QPI has the ability to quantify the optical path-length map of a light field passing through the specimen. Due to its ability to render label-free, quantitative, and nanoscale information from live cells, a rapidly increasing number of QPI methods have been developed within the past decade or so. 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17 Despite all these developments, the range of QPI applications in biology has been largely limited to red blood cell imaging 18, 19, 20, 21 or assessment of global cell parameters such as dry mass, 12, 22, 23, 24, 25 average refractive index, 26, 27 and statistical parameters of tissue slices.14 In our opinion, this current limitation is due to two main reasons. First, because of the speckle generated by the high coherence of the illuminating light (typically lasers), the contrast to noise ratio in QPI images has never matched that exhibited in white light techniques such as phase contrast microscopy. Second, like all interferometric methods, the experimental arrangements tend to be rather complex, of high maintenance, which limits their in-depth biological applicability. A number of QPI-based tracking methods were reported recently. 28, 29, 30 However, in these previous reports, large beads or the cells themselves were tracked. On the other hand, qualitative methods such as phase contrast or differential interference contrast (DIC) provide better contrast to noise ratio, but their application to intrinsic particle tracking is limited as well. Toward this end, we have recently developed spatial light interference microscopy (SLIM) as a novel, highly sensitive QPI method, which is advantageous for enabling quantitative assessment of specimen structure and cell dynamics.31 SLIM combines Zernike's white light phase contrast microscopy which reveals the intrinsic contrast of transparent samples32 with Gabor's holography,33 by rendering quantitative phase maps across the specimen. In this paper, we exploit the exquisite spatial sensitivity of SLIM and demonstrate that the Laplace operator can be used to reveal a high-detail quantitative phase image without gradient artifacts common to DIC microscopy. In particular, this imaging approach, termed Laplace phase microscopy (LPM), allows us to quantify intracellular transport without the typical need for exogenous contrast agents. This opens up new avenues for particle tracking, which has been largely limited to fluorescently labeled tracers.1 We show that LPM can study transport in live cells over very broad time scales, unlimited by photobleaching or photoxicity, as demonstrated by tracking organelles in hippocampal neurons and cardiomyocytes. Our measurements indicate a diffusive regime for the particle motion, from which the diffusion coefficient can be extracted quantitatively. 2.Experimental SetupThe experimental setup is described in detail elsewhere.34 Here we briefly review its principle of operation (Fig. 1). The SLIM module is attached to a commercial phase contrast microscope (Axio Observer Z1, Zeiss, in this case). The image outputted by the microscope is Fourier transformed by lens L 1, which essentially images the objective phase contrast ring onto the surface of a liquid crystal phase modulator (LCPM). This device can modulate the phase difference Δφ between the scattered and unscattered light in increments of π/2, i.e., Δφ = 0, π/2, π, and 3π/2. The Fourier lens L 2 completes a (folded) 4f system, such that the conjugate image plane is reconstructed at the CCD plane. By recording one intensity image for each of the four phase shifts introduced by LCPM, we render the final, quantitative phase image associated with the sample. Fig. 1Schematic setup for SLIM. The black dots on the LCPM show the cross section through the ring along which the unscattered light is distributed. 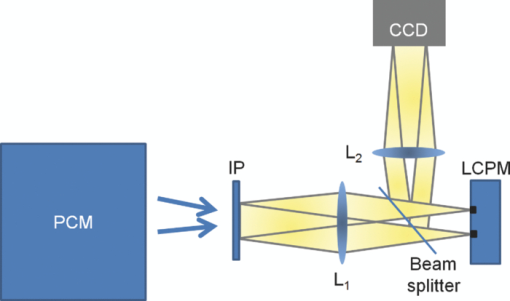 SLIM provides several advantages over existing techniques, including speckle-free images, which allows for spatially sensitive optical path-length measurement (0.3 nm), common path geometry that enables temporally sensitive optical path-length measurement (0.03 nm), and 3D tomographic rendering of transparent structures.31 Further, due to the broadband illumination spectrum, SLIM grants immediate potential for spectroscopic (i.e., phase dispersion) imaging. SLIM is likely to make a broad impact by implementation with existing phase contrast microscopes and overlaying with fluorescence imaging for multimodal, in-depth biological studies. 3.Data AnalysisFigure 2a (1) shows an example of a quantitative phase image, ϕ(x, y), of cardiomyocytes, which were obtained from 2-day old neonatal Sprague-Dawley rats (Harlan Laboratories, Inc.) using an approved protocol by the Institutional Animal Care and Use Committee. Briefly, whole hearts were excised35 in ice-cold Hanks balanced salt solution (HBSS) buffer, the atria were removed, and the remaining ventricles were quartered and digested in 0.05% (w/v) trypsin (Worthington Biochemicals) with gentle rotation (4°C, 16 h). To inhibit trypsin digestion, growth media (DMEM with 10% fetal bovine serum) was added for 5 min at 37°C. After discarding the supernatant, 0.1% (w/v) purified type II collagenase (Worthington Biochemicals) was added for 45 min while rotating at 37°C. The cardiac tissue was gently triturated to mechanically loosen the cells, filtered through a cell strainer, and centrifuged at 150×g for 10 min. The cell pellet was resuspended in 37°C growth media and pre-plated for 1 h to enrich the suspension for cardiac myocytes. Video 1SLIM and LPM imaging of cardiomyocytes in culture. Cells are beating simultaneously. Particles are revealed and traced by LPM (QuickTime, 7.6 MB). 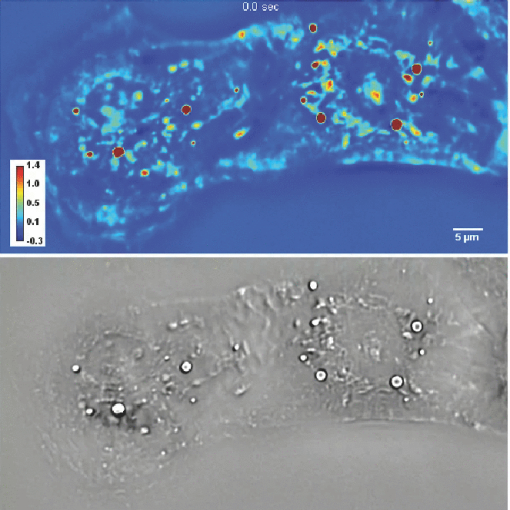 We computed the LPM image [Fig. 2b and 1] via the Laplace operator Eq. 1[TeX:] \documentclass[12pt]{minimal}\begin{document}\begin{equation} \nabla ^2 \phi (x,y) = \frac{{\partial ^2 \phi }}{{\partial x^2 }} + \frac{{\partial ^2 \phi }}{{\partial y^2 }}. \end{equation}\end{document}
[TeX:]
\documentclass[12pt]{minimal}\begin{document}\begin{eqnarray*}
\left[ {\begin{array}{*{20}r}
{\rm 0} &\quad {{\rm - 1}} &\quad {\rm 0} \\[2pt]
{{\rm - 1}} &\quad {\rm 4} &\quad {{\rm - 1}} \\[2pt]
{\rm 0} &\quad {{\rm - 1}} &\quad {\rm 0} \\
\end{array}} \right].
\end{eqnarray*}\end{document}
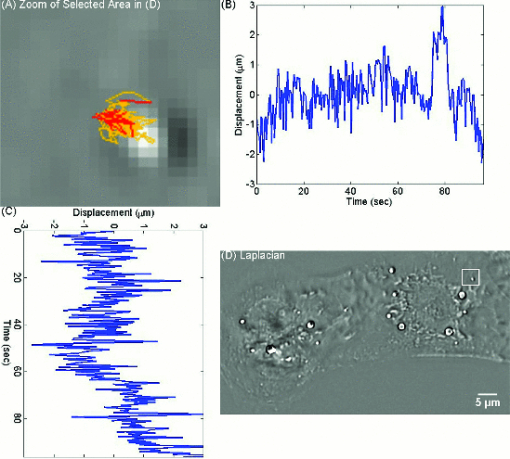
As evident from this image, the Laplace operator is able to clearly define the organelle structures within the cell. It is known that heart cells are very active, i.e., energy-consuming; therefore this type of cell is rich in mitochondria, which are responsible for the energy supply of the cell metabolism.37 Mitochondria are most likely the predominant type of visible organelles, especially in the area surrounding the cell nucleus [indicated by the circular region in Fig. 2a]. For comparison, we also computed a “synthetic” differential interference contrast (DIC or Nomarski) image [Fig. 2c], via the gradient of the quantitative phase image, Eq. 2[TeX:] \documentclass[12pt]{minimal}\begin{document}\begin{equation} \overline \nabla \phi (x,y) = \frac{{\partial \phi }}{{\partial x}}{\kern 1pt} \;\widehat{\bf x} + \frac{{\partial \phi }}{{\partial x}}\;\widehat{\bf y}, \end{equation}\end{document}Figure 3 demonstrates the procedure of quantifying intracellular diffusion via LPM. LPM images of a pair of beating cardiomyocytes in culture were recorded for more than a minute at a rate of 2.6 frames/s. The speed is currently limited by our acquisition camera which has a frame limit of 11 frames/s at full resolution. The organelle diffusion coefficient D is extracted from the measured mean squared displacement (MSD) of the particles, and is defined as2 Eq. 3[TeX:] \documentclass[12pt]{minimal}\begin{document}\begin{equation} \Delta r^2 (\tau) = \left\langle {\left[ {x(t + \tau) - x(t)} \right]^2 + \left[ {y(t + \tau) - y(t)} \right]^2 } \right\rangle.\end{equation}\end{document}Fig. 2(a) SLIM image of two cardiac myocytes in culture; color bar indicates phase shift in radians (1). The dashed circles show the cell nuclei. The objective is ZEISS Plan-Neofluar 40X/0.75. (b) Laplacian of (a) 1. (c) Simulated DIC, as gradient in X direction of (a). (d) Phase contrast image. (e) Laplacian of (d). (f) Gradient in X direction of (d). 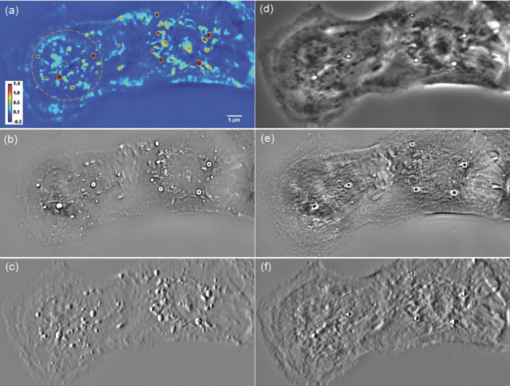 Fig. 3MSD measurement for particles within the cardiac myocytes in Fig. 2. (a) Zoom into the selected area shown in (d); (b) Displacement in Y direction; (c) Displacement in X direction. (d) Laplacian of the phase map [same as Fig. 2b]; (e) MSD for the particle shown in (a). (f) MSD ensemble-averaged over 15 particles in (d). The respective movie can be found in the supplemental material (2). 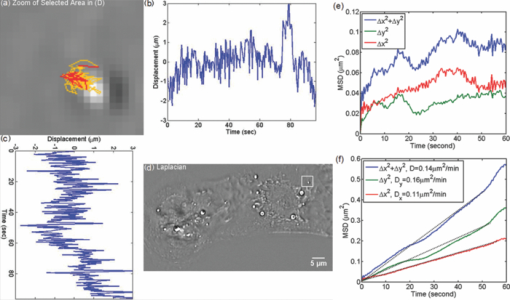 In Eq. 3, the angular bracket stands for temporal and ensemble average. For a diffusion process, the mean squared displacement grows linearly in time, Δr 2(τ) = 2nDτ, where n indicates the dimensionality, i.e., n = 1, 2, and 3 for diffusion in 1, 2, and 3 dimensions, respectively.2 Thus, the slope of Δr 2 reveals the diffusion coefficient. Figures 3a, 3b, 3c, 3d illustrate the tracking of a single particle within the cell. Figure 3a shows the magnified particle in the selected area of Fig. 3d. Particles are traced using an automatic algorithm implemented in Java. The tracking algorithm may lose the particle at certain frames (e.g., due to the particle going out of focus) and find the particle again in later frames. If the algorithm identifies the same particles and relinks the traces, the link (an indication of skipped frames) is in red and the trace is in yellow [e.g., red in Fig. 3a and 3]. This will not cause any problem in the calculation of MSD because it is a time and ensemble average and is not sensitive to the individual frame loss. Figures 3b and 3c show the displacement of the particle in the Y and X directions, respectively. The MSD for the specific particle shown in Fig. 3e shows no specific trend at all, as expected. It can be seen that the high frequency component of the signal is due to the cell beating. However, if we analyze all 15 particles in Fig. 3d and perform an ensemble average, the MSD is linear which clearly shows a diffusive movement, as summarized in Fig. 3f. The results demonstrate that the high-frequency beating signal is averaged out and that a linear dependence is obtained. The resulting diffusion along y is slightly smaller than along x, which might be explained by the beating signal propagation between the two cells, almost parallel to the x-axis. Note that the overall value, D = 0.14 μm2/min, obtained for the diffusion coefficient, D = (D x+D y)/2, is approximately 188 times smaller than that predicted by the Stokes–Einstein equation for a 1-μm particle diffusing in water at room temperature.39 This reduced diffusion of organelles within the cells can be understood by realizing that organelles occupy a crowded space, populated by cytoskeletal network and macromolecules, which makes the effective viscosity significantly higher than that of water.40 For comparison, we also captured the cardiomyocytes in culture with diffraction phase microscopy (DPM),41 an established laser technique for quantitative phase imaging (3). As evident in the movie, although the acquisition rate is faster, no particles can be resolved at all. It is clear that by simply applying a Laplacian operator to a laser-based quantitative phase image is not useful for tracking intrinsic particles. Video 3DPM imaging of the cardiomyocytes in culture. No particles can be resolved. (a) Phase map, (b) Laplacian of phase map, (c) gradient in vertical direction, (d) gradient in horizontal direction. The field of view is 32 μm × 32 μm (QuickTime, 5.9 MB). 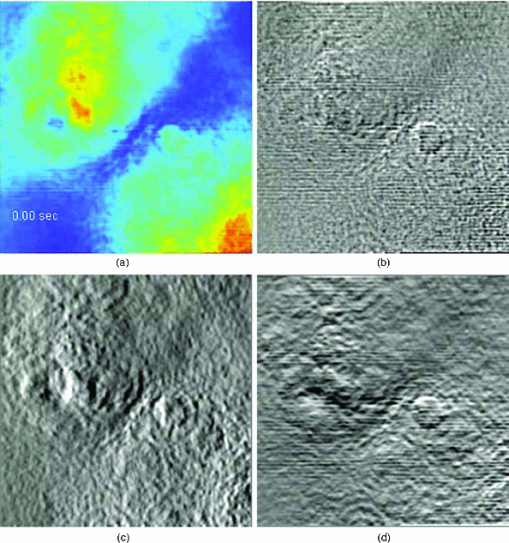 Further, we applied the LPM method to vesicles in hippocampal neurites. Recently, tracking of synaptic vesicles with far-field optical nanoscopy has been demonstrated.42 Primary neuronal cultures from the postnatal rat were generated on glass bottomed dishes for imaging analysis based on the previously defined protocol. Briefly, postnatal hippocampal neurons were isolated from postnatal rats through enzymatic digestion, tissue rinse, dissociation, and centrifugation, followed by resuspension and plating in supplemented neurobasal media until imaging. Prior to imaging, neuron media was replaced with supplemented, CO2 independent media Hibernate-A.43 With the high sensitivity of SLIM, the vesicles in neurites can be resolved, as seen from Fig. 4 (4). In our samples, SLIM images were acquired at 4 frames/min. The phase map shown [Fig. 4] for the dashed line includes five temporal traces [Figs. 4a and 4b ], trace 1 is the background, 2 and 4 are two large processes, and 3 and 5 are two small processes. The optical path length changes for individual traces are shown in Fig. 4b, where the background stays very stable, and the peaks in the figure correspond to the particles in transport. Since the optical path length change corresponds to the dry mass, the heights of the bumps are indicators of the protein mass under transport. Evident from Fig. 4b (4), SLIM is sensitive enough to display the optical 10 to 20 nm path-length changes associated with particles in transport. Figure 4c is the Laplacian of the selected area in Fig. 4a. The particles with the traces are shown in Fig. 4d. Video 4SLIM and LPM imaging of live hippocampal neuron. Neurons and putative glia in primary cell culture. Vesicle transports in hippocampal neurites are emphasized and traced using LPM. The contrast of LPM is inverted for better viewing purpose (QuickTime, 3.0 MB). 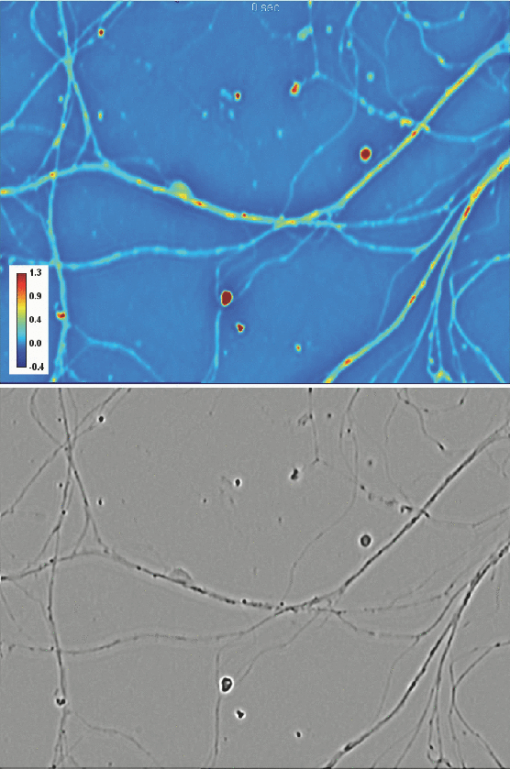 Neuronal processes demonstrate bidirectional transport of cargo (i.e., synaptic vesicle precursors, mitochondria, piccolo-bassoon transport vesicles, signaling endosomal organelles, and translation machinery) to and from the soma and distal tips. 44, 45, 46 As seen in the movie, a subset of particles exhibit a fast, directed (nondiffusive) motion. However, for our mean squared displacement analysis, we retain only particle trajectories that survive throughout the entire measurement window of 18 min. These long-time trajectories exhibit again diffusive motion, as indicated by the very strong linear dependence shown in the inset of Fig. 4e. To our knowledge, these are the first label-free diffusion measurements in neurons. As expected, the displacements parallel to the y-axis are approximately 2 orders of magnitude smaller than the x-axis displacements because the neurites are largely parallel to the x-axis. Because neurons have numerous long neurites, the surface area of a neuron far exceeds that of other cells by as much as 10 to 1000 times.47 Thus, the structure and function of the neuron has high demands on cellular resources, and the active transport of cellular resources is needed to maintain these demands. Most axonal transport velocities range between 0.5 to 5.0 μm/s.45 From our data, the directional transport velocity is about 0.5 μm/s and the diffusion coefficients D x = 6.50 μm2/min and D y = 0.046 μm2/min. Thus, the diffusion process in dendrites is largely one-dimensional, as obvious from the movie. Note that the diffusion coefficient in neurons is significantly larger than that in the heart cells, D = 0.14 μm2/min, which indicates that the transport in neurons is more active. This result is consistent with the neuronal function that involves mass transport over large distances. Fig. 4Particle transport in neurites of a hippocampal neuron processor network. The objective is ZEISS Plan-Neofluar 40X/0.75. (a) Phase map of the neuron network. The arrows 1 to 5 show the time-traces of the corresponding points along the dashed line. The whole field of view is 100 μm×75 μm. The objective used is Zeiss Plan-Neofluar 40×/0.75. (b) Optical path length change in time for the five points indicated in (a). Peaks in the point traces correspond to phase shifts associated with (fast) organelle traffic. (c) Laplacian of the selected area in (a). The scale bar is 5 μm. (d) Phase map of the same area as in (c), with some particle traces shown in fine lines. (e) Log–log plot of the MSD for 70 individual particles in (d). Since the particles are confined in the Y direction, the diffusion coefficient for this direction is 2 orders of magnitude smaller than for the other direction. The inset shows the same MSD curves in linear representation and two Y axes. See 4 for details. 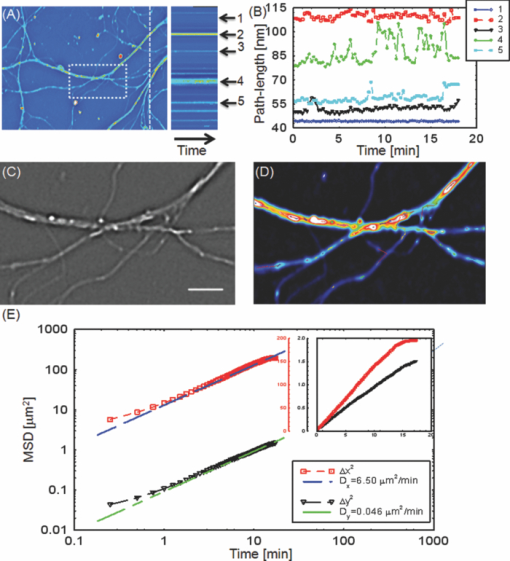 4.ConclusionIn summary, we demonstrated LPM as an efficient method for revealing and tracking organelles within unstained cells such as cardiomyocytes and neurons. To our knowledge, these are the first label-free diffusion measurements in such cells. Based on the advance of spatial light interference microscopy,31 this approach may help unravel important open questions regarding intracellular transport, which modulates cell development and cell signaling. It will also facilitate the study of the viscoelastic properties of the cells. In principle, the Laplacian operator can be applied to any quantitative phase image. However, we note that this operation enhances the high-frequency noise, e.g., noise due to speckles, which may reduce the overall utility of this operation (see 3). Laplace phase microscopy, Fourier phase microscopy,9 Hilbert phase microscopy,15 and Fresnel particle tracking28 are examples of optical techniques that exploit the ability of mathematical operators (from which they borrow their names) to enhance optical imaging. AcknowledgmentsThis research was supported in part by NSF CAREER 08-46660 (to G.P.) and Grainger Foundation (to G.P.). M.U.G, R.B., and G.P. are supported by the NSF-STC CBET-0939511. M.U.G. is supported by NIH grants HL 086870 and MH 085220. L.M. was supported by NIH/HD007333 Developmental Psychobiology and Neurobiology Training Grant. V.C. was supported by the NSF-STC CBET-0939511 and by a cooperative agreement that was awarded and administered by the US Army Medical Research & Materiel Command (USAMRMC) and the Telemedicine & Advanced Technology Research Center (TATRC), under Contract No. W81XWH0810701 (to R.B.). ReferencesD. Wirtz,
“Particle-Tracking Microrheology of Living Cells: Principles and Applications,”
Ann. Rev. Biophys., 38 301
–326
(2009). https://doi.org/10.1146/annurev.biophys.050708.133724 Google Scholar
T. A. Waigh,
“Microrheology of complex fluids,”
Rep. Prog. Phys., 68 685
–742
(2005). https://doi.org/10.1088/0034-4885/68/3/R04 Google Scholar
G. Popescu,
“Quantitative phase imaging of nanoscale cell structure and dynamics,”
Methods in Cell Biology, 87 Ed.Elsevier, Burlington, MA
(2008). Google Scholar
M. A. Choma, A. K. Ellerbee, C. H. Yang, T. L. Creazzo, and
J. A. Izatt,
“Spectral-domain phase microscopy,”
Opt. Lett., 30 1162
–1164
(2005). https://doi.org/10.1364/OL.30.001162 Google Scholar
C. Joo, T. Akkin, B. Cense, B. H. Park, and
J. E. de Boer,
“Spectral-domain optical coherence phase microscopy for quantitative phase-contrast imaging,”
Opt. Lett., 30 2131
–2133
(2005). https://doi.org/10.1364/OL.30.002131 Google Scholar
D. Paganin and
K. A. Nugent,
“Noninterferometric phase imaging with partially coherent light,”
Phys. Rev. Lett., 80 2586
–2589
(1998). https://doi.org/10.1103/PhysRevLett.80.2586 Google Scholar
A. Barty, K. A. Nugent, D. Paganin, and
A. Roberts,
“Quantitative optical phase microscopy,”
Opt. Lett., 23 817
–819
(1998). https://doi.org/10.1364/OL.23.000817 Google Scholar
E. D. Barone-Nugent, A. Barty, and
K. A. Nugent,
“Quantitative phase-amplitude microscopy I: optical microscopy,”
J. Microscopy, 206 194
–203
(2002). https://doi.org/10.1046/j.1365-2818.2002.01027.x Google Scholar
G. Popescu, L. P. Deflores, J. C. Vaughan, K. Badizadegan, H. Iwai, R. R. Dasari, and
M. S. Feld,
“Fourier phase microscopy for investigation of biological structures and dynamics,”
Opt. Lett., 29 2503
–2505
(2004). https://doi.org/10.1364/OL.29.002503 Google Scholar
W. S. Rockward, A. L. Thomas, B. Zhao, and
C. A. DiMarzio,
“Quantitative phase measurements using optical quadrature microscopy,”
Appl. Opt., 47 1684
–1696
(2008). https://doi.org/10.1364/AO.47.001684 Google Scholar
Z. Wang, L. J. Millet, M. U. Gillette, and
G. Popescu,
“Jones phase microscopy of transparent and anisotropic samples,”
Opt. Lett., 33 1270
–1272
(2008). https://doi.org/10.1364/OL.33.001270 Google Scholar
D. Zicha and
G. A. Dunn,
“An Image-Processing System For Cell Behavior Studies In Subconfluent Cultures,”
J. Microscopy, 179 11
–21
(1995). Google Scholar
P. Marquet, B. Rappaz, P. J. Magistretti, E. Cuche, Y. Emery, T. Colomb, and
C. Depeursinge,
“Digital holographic microscopy: a noninvasive contrast imaging technique allowing quantitative visualization of living cells with subwavelength axial accuracy,”
Opt. Lett., 30 468
–470
(2005). https://doi.org/10.1364/OL.30.000468 Google Scholar
H. Ding, Z. Wang, F. Nguyen, S. A. Boppart, and
G. Popescu,
“Fourier transform light scattering of inhomogeneous and dynamic structures,”
Phys. Rev. Lett., 101 238102
(2008). https://doi.org/10.1103/PhysRevLett.101.238102 Google Scholar
T. Ikeda, G. Popescu, R. R. Dasari, and
M. S. Feld,
“Hilbert phase microscopy for investigating fast dynamics in transparent systems,”
Opt. Lett., 30 1165
–1168
(2005). https://doi.org/10.1364/OL.30.001165 Google Scholar
J. G. Wu, Z. Yaqoob, X. Heng, L. M. Lee, X. Q. Cui, and
C. H. Yang,
“Full field phase imaging using a harmonically matched diffraction grating pair based homodyne quadrature interferometer,”
Appl. Phys. Lett., 90 151123
(2007). https://doi.org/10.1063/1.2722685 Google Scholar
N. T. Shaked, Y. Z. Zhu, N. Badie, N. Bursac, and
A. Wax,
“Reflective interferometric chamber for quantitative phase imaging of biological sample dynamics,”
J. Biomed, Opt., 15 030503
(2010). https://doi.org/10.1117/1.3420179 Google Scholar
G. Popescu, T. Ikeda, K. Goda, C. A. Best-Popescu, M. Laposata, S. Manley, R. R. Dasari, K. Badizadegan, and
M. S. Feld,
“Optical measurement of cell membrane tension,”
Phys. Rev. Lett., 97 218101
(2006). https://doi.org/10.1103/PhysRevLett.97.218101 Google Scholar
Y. K. Park, C. A. Best, K. Badizadegan, R. R. Dasari, M. S. Feld, T. Kuriabova, M. L. Henle, A. J. Levine, and
G. Popescu,
“Measurement of red blood cell mechanics during morphological changes,”
Proc. Natl. Acad. Sci. U.S.A., 107 6731
–6736
(2010). https://doi.org/10.1073/pnas.0909533107 Google Scholar
Y. K. Park, C. A. Best, T. Auth, N. Gov, S. A. Safran, G. Popescu, S. Suresh, and
M. S. Feld,
“Metabolic remodeling of the human red blood cell membran,”
Proc. Natl. Acad. Sci. U.S.A., 107 1289
(2010). https://doi.org/10.1073/pnas.0910785107 Google Scholar
Y. K. Park, M. Diez-Silva, G. Popescu, G. Lykotrafitis, W. Choi, M. S. Feld, and
S. Suresh,
“Refractive index maps and membrane dynamics of human red blood cells parasitized by Plasmodium falciparum,”
Proc. Natl. Acad. Sci. U.S.A., 105 13730
–13735
(2008). https://doi.org/10.1073/pnas.0806100105 Google Scholar
G. Popescu, Y. Park, N. Lue, C. Best-Popescu, L. Deflores, R. R. Dasari, M. S. Feld, and
K. Badizadegan,
“Optical imaging of cell mass and growth dynamics,”
Am. J. Physiol. Cell. Physiol., 295 C538
–C544
(2008). https://doi.org/10.1152/ajpcell.00121.2008 Google Scholar
H. G. Davies and
M. H. Wilkins,
“Interference microscopy and mass determination,”
Nature (London), 169 541
–541
(1952). https://doi.org/10.1038/169541a0 Google Scholar
R. Barer,
“Interference microscopy and mass determination,”
Nature (London), 169 366
–367
(1952). https://doi.org/10.1038/169366b0 Google Scholar
B. Rappaz, E. Cano, T. Colomb, J. Kuhn, C. Depeursinge, V. Simanis, P. J. Magistretti, and
P. Marquet,
“Noninvasive characterization of the fission yeast cell cycle by monitoring dry mass with digital holographic microscopy,”
J. Biomed. Opt., 14 034049
(2009). https://doi.org/10.1117/1.3147385 Google Scholar
N. Lue, G. Popescu, T. Ikeda, R. R. Dasari, K. Badizadegan, and
M. S. Feld,
“Live cell refractometry using microfluidic devices,”
Opt. Lett., 31 2759
–2761
(2006). https://doi.org/10.1364/OL.31.002759 Google Scholar
C. L. Curl, C. J. Bellair, T. Harris, B. E. Allman, P. J. Harris, A. G. Stewart, A. Roberts, K. A. Nugent, and
L. M. D. Delbridge,
“Refractive index measurement in viable cells using quantitative phase-amplitude microscopy and confocal microscopy,”
Cytometry, Part A, 65A 88
–92
(2005). https://doi.org/10.1002/cyto.a.20134 Google Scholar
Y. K. Park, G. Popescu, R. R. Dasari, K. Badizadegan, and
M. S. Feld,
“Fresnel particle tracking in three dimensions using diffraction phase microscopy,”
Opt. Lett., 32 811
–813
(2007). https://doi.org/10.1364/OL.32.000811 Google Scholar
P. Langehanenberg, L. Ivanova, I. Bernhardt, S. Ketelhut, A. Vollmer, D. Dirksen, G. Georgiev, G. von Bally, and
B. Kemper,
“Automated three-dimensional tracking of living cells by digital holographic microscopy,”
Biomed. Opt., 14 014018
(2009). https://doi.org/10.1117/1.3080133 Google Scholar
J. Sheng, E. Malkiel, J. Katz, J. Adolf, R. Belas, and
A. R. Place,
“Digital holographic microscopy reveals prey-induced changes in swimming behavior of predatory dinoflagellates,”
Proc. Natl. Acad. Sci. U.S.A., 104 17512
–17517
(2007). https://doi.org/10.1073/pnas.0704658104 Google Scholar
Z. Wang, K. Tangella, and
G. Popescu,
“Tissue refractive index as marker for disease,”
Nat. Photonics, Google Scholar
F. Zernike,
“How I discovered phase contrast,”
Science, 121 345
–349
(1955). https://doi.org/10.1126/science.121.3141.345 Google Scholar
D. Gabor,
“A new microscopic principle,”
Nature (London), 161 777
–778
(1948). https://doi.org/10.1038/161777a0 Google Scholar
Z. Wang, L. Millet, M. Mir, H. Ding, S. Unarunotai, J. Rogers, M. Gillette, and
G. Popescu,
“Spatial light interference microscopy (SLIM),”
Opt. Express, 19 1016
–1026
(2011). https://doi.org/10.1364/OE.19.001016 Google Scholar
A. H. Maass and
M. Buvoli,
“Cardiomyocyte Preparation, Culture, and Gene Transfer,”
Methods Mol. Biol., 366 321
–330
(2007). https://doi.org/10.1007/978-1-59745-030-0 Google Scholar
R. C. Gonzales and
R. E. Woods, Digital Image Processing, 2ndPrentice Hall, Englewood Cliffs, NJ
(2002). Google Scholar
B. Alberts, Essential Cell Biology: An Introduction to the Molecular Biology of the Cell, Garland Publishers, New York
(2004). Google Scholar
J. B. Pawley, Handbook of Biological Confocal Microscopy, Springer, New York
(2006). Google Scholar
A. Einstein,
“Investigations on the theory of the Brownian movement,”
Ann. Physik., 17 549
–560
(1905). https://doi.org/10.1002/andp.19053220806 Google Scholar
M. T. Valentine, P. D. Kaplan, D. Thota, J. C. Crocker, T. Gisler, R. K. Prud'homme, M. Beck, and
D. A. Weitz,
“Investigating the microenvironments of inhomogeneous soft materials with multiple particle tracking,”
Phys. Rev. E, 6406 061506
(2001). https://doi.org/10.1103/PhysRevE.64.061506 Google Scholar
G. Popescu, T. Ikeda, R. R. Dasari, and
M. S. Feld,
“Diffraction phase microscopy for quantifying cell structure and dynamics,”
Opt. Lett., 31 775
–777
(2006). https://doi.org/10.1364/OL.31.000775 Google Scholar
V. Westphal, S. O. Rizzoli, M. A. Lauterbach, D. Kamin, R. Jahn, and
S. W. Hell,
“Video-rate far-field optical nanoscopy dissects synaptic vesicle movement,”
Science, 320 246
–249
(2008). https://doi.org/10.1126/science.1154228 Google Scholar
L. J. Millet, M. E. Stewart, J. V. Sweedler, R. G. Nuzzo, and
M. U. Gillette,
“Microfluidic devices for culturing primary mammalian neurons at low densities,”
Lab Chip, 7 987
–994
(2007). https://doi.org/10.1039/b705266a Google Scholar
A. Y. N. Goldstein, X. N. Wang, and
T. L. Schwarz,
“Axonal transport and the delivery of pre-synaptic components,”
Curr. Opin. Neurobiol., 18 495
–503
(2008). https://doi.org/10.1016/j.conb.2008.10.003 Google Scholar
C. F. Ibanez,
“Message in a bottle: long-range retrograde signaling nervous system,”
Trends Cell Biol., 17 519
–528
(2007). https://doi.org/10.1016/j.tcb.2007.09.003 Google Scholar
J. R. Sotelo-Silveira, A. Calliari, A. Kun, E. Koenig, and
J. R. Sotelo,
“RNA trafficking in axons,”
Traffic, 7 508
–515
(2006). https://doi.org/10.1111/j.1600-0854.2006.00405.x Google Scholar
S. Sann, Z. P. Wang, H. Brown, and
Y. S. Jin,
“Roles of endosomal trafficking in neurite outgrowth and guidance,”
Trends Cell Biol., 19 317
–324
(2009). https://doi.org/10.1016/j.tcb.2009.05.001 Google Scholar
|