|
|
|
In surgical oncology, the selective excision of tumors with minimal damage to the surrounding normal tissue is critical. Tumor removal is guided by examining pathology that is prepared during surgery from the excisions. Preparation of pathology is labor intensive and time consuming. Typical preparation time is hours for frozen pathology during Mohs surgery and days for fixed pathology in other surgical settings such as head-and-neck and breast.1 This often results in insufficient sampling of tissue and incomplete removal of a tumor such that 20% to 70% of the patients must subsequently undergo further resection, radiotherapy and/or chemotherapy.2, 3 Confocal mosaicing microscopy potentially offers an approach for detecting cancer margins rapidly and with the necessary sub-cellular resolution.1, 4 In this process, merging individual images creates mosaics that display large areas of tissue. The feasibility of imaging5 and mosaicing6 skin cancer margins in vivo in reflectance contrast has been demonstrated. More recently, feasibility for mosaicing in endoscopy and intraorally was reported.7, 8 However, at present, mosaicing in vivo has been demonstrated either on relatively small areas (∼mm2) or along linear paths (∼mm) with long acquisition times (∼minutes), whereas surgeons need to examine much larger areas (∼cm2) in shorter times (∼1 min). To address these issues, we designed a new method called strip-mosaicing. We previously reported a confocal mosaicing microscope that images a 12×12 mm2 area of Mohs surgical excisions by acquiring 36×36 images and merging them with custom software.9, 10 Fluorescence contrast using acridine orange to stain nuclei was shown to be superior to reflectance contrast for the detection of basal cell carcinomas. In a blinded examination of 45 fluorescence mosaics by two Mohs surgeons, basal cell carcinomas were detected with sensitivity of 96.6% and specificity of 89.2%. The time for image acquisition and two-dimensional mosaicing was 9 min. While this showed initial feasibility, the acquisition and stitching time required for larger excisions (∼cm2) that are routinely taken in other surgical settings make this impracticable. In this report, we eliminate one of the stitching dimensions by acquiring long image strips instead of the standard “square” images. Instead of merging a two-dimensional array of images, a single one-dimensional array of strips is stitched together (see Fig. 1). The benefit of this is threefold: the acquisition time, merging time, and the artifacts due to the illumination variations are all reduced by half. The system described uses a combination of optical and mechanical scanning to generate the images. Preliminary data shows that the system can produce a 10×10 mm2 strip mosaic in about 3 min. Fig. 1(a) A two-dimensional mosaic acquisition pattern. The individual images have radial illumination falloff (as shown in the diagram) that must be corrected and each image must be “stitched” to two neighbors (on average). (b) In strip mosaicing the optics acquire a single line in the X direction while a stage scans the sample in the Y direction (straight arrows). The intensity fall-off is in the horizontal direction only and each image must be stitched to a single neighbor. 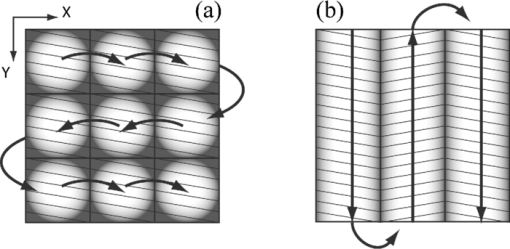 Our system is based on a point-scanning confocal microscope with a rotating polygonal mirror and a galvanometrically driven mirror (Vivascope 2000, Lucid Inc.)9, 10 A laser (Ar+ 488 nm) is scanned in the fast (X) direction at 6.8 kHz over 408 μm at the sample through a 30×0.9 NA, water immersion objective lens (Stableview, Lucid Inc.) The captured field of view is 330 μm to limit the intensity falloff at the edges of the scan. The fluorescence signal is captured with a digital acquisition card (DAQ PCI-6110, National Instruments). A custom tissue fixture is used for mounting large skin excisions from Mohs surgery with precise orientation and alignment to the objective lens.11 It provides tip-tilt adjustments of the glass slide and samples relative to the objective lens to make the imaging and scanning planes parallel, ensuring that the data is captured at a constant depth from the glass-tissue interface. The theoretical lateral and axial resolutions, as per the Rayleigh criteria, are 0.33 μm radially (Airy radius) and 1.61-μm axially, assuming a planar wave and a circular aperture. We approximate this by overfilling the back aperture of the objective lens and using the central portion of the Gaussian beam. The Rayleigh criterion requires sampling at half the Airy radius. To increase the scanning speed we undersample the data by a factor of ∼6 which results in a ∼1 μm pixel size. As reported in our earlier studies9, 10 the undersampled images are adequate for interpretation by surgeons and pathologists. To obtain square pixels and equal spatial sampling rates in X and Y, the stage speed must be ∼6.8 mm/s (1 μm pixel×6.8 kHz line rate). Thus, it takes ∼1.5 s to scan a 10 mm strip plus 0.5 s to move the stage laterally before starting to acquire the adjacent strip. This is about 2 s/strip. A 31 strip (∼10-mm wide) mosaic takes approximately 1 min to capture. To acquire a strip image the galvanometric mirror is locked to its center position and the fast (X) scanner is started. The Y stage motor is started and monitored by a hardware counter. On reaching a constant speed after N steps, the DAQ starts acquiring image lines on the next valid horizontal line trigger from the scanner. This ensures that the strips do not have more than a single line of misalignment. M lines are acquired and then the stage is stopped. The X stage motor moves the sample laterally by 330 μm (80% of the imaged field) and the process starts anew. Thirty-one strips were acquired in the example shown in this paper (Fig. 2). After completing the acquisition, the images are loaded into the MosaicJ open source mosaicing software,12 together with information on their relative positions. The program automatically places the images in their acquired positions and merges the images with corrections for any remnant misalignments and variations in signal level. The merging time depends on the power of the processing computer. On an Mac Pro (2×2.93 GHz Quad-Core Xeon, 16 GB RAM) it took 35 s to load 31 images and 92 s of processing by MosaicJ to form Fig. 2. Although our system is designed to accurately synchronize the scanner and stage to acquire strip images that are adjacent to each other with a vertical misalignment of no more than one line, there remain some misalignments that are entirely due to inadequate precision in our stages. This, of course, can be easily corrected with newer and better quality hardware. Fig. 2A 10×10 mm2 mosaic consisting of 31 fluorescence image strips of excised tissue from Mohs surgery. The tissue was stained with 0.6 mM acridine orange for 20 s (Refs. 9, 10). Nests of basal cell carcinomas (A) are observed, showing nuclear detail such as increased density, pleomorphism, and palisading. Typical normal features such as hair follicles and sebaceous glands (B) and eccrine ducts (C) can be seen. The mosaic dimensions are 11,415×10,291 (pixels wide × high) with 8 bits/pixel. Note that the magnified areas are digital zooms obtained from the original image showing the detail and resolution of the mosaic. The features in the mosaic compare well to the pathology (Fig. 3), in terms of location, shape, size, nuclear detail and overall morphology of both basal cell carcinomas and normal features. 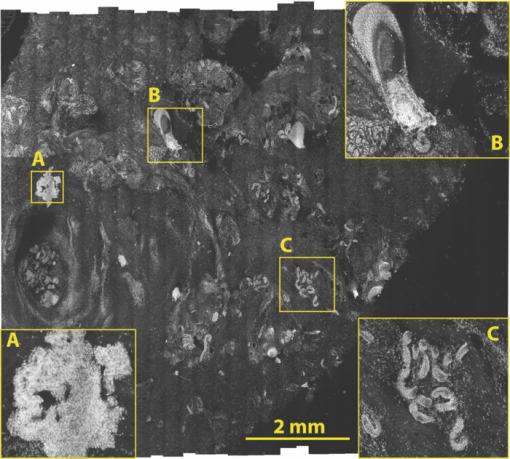 Figure 2 shows a strip mosaic of a skin excision from Mohs surgery. The total time for acquisition and merging of the 31 image strips was about 3.1 min. The mosaic shown is in fluorescence contrast. Acridine orange was used to stain because its excitation matches the 488-nm wavelength in our system. Any contrast agent can be used, such as methylene blue,4 with a suitable excitation source. The mosaic shows the morphology of a basal cell carcinoma and typical normal features (Fig. 2). The nuclear morphology is not readily visible in small figures but is clearly seen on a large monitor. To show the detail as seen on a monitor, we present the magnified inserts A, B, and C. The morphologic features in the mosaics compare well to the corresponding pathology of Fig. 3. As detailed in earlier reports9, 10 an exact correlation between features in the mosaics to those in the pathology is not expected because the H&E preparation is from the cut adjacent to the section imaged by the confocal microscope (Fig. 2). Further, the tissue sections are pliant and are inevitably distorted during fixing for mosaicing and hematoxylin and eosin (H&E) preparation. Fig. 3Frozen H&E-stained pathology of excised tissue from Mohs surgery. The wide-field microscopy images correspond to a tissue slice adjacent to the one shown in Fig. 2. The features corresponding to areas A, B, and C can be identified across the two figures. 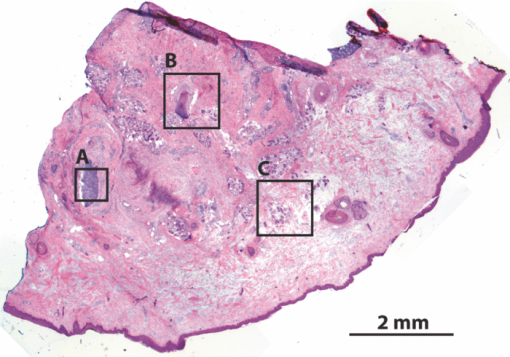 Our results demonstrate that strip mosaicing is three times as fast as the previously reported two-dimensional mosaicing method with square images. Starting the merging process as the strips are acquired through the integration of the acquisition and merging programs will further reduce the turnaround time to produce mosaics. This, together with more precise stages, will allow us to generate 10×10 mm2 mosaics in less than the 3.1 min reported here. The projected acquisition time for a 20×20 mm2 mosaic would be 3.5 min and we anticipate a linear increase of acquisition time with sample area. AcknowledgmentsThe authors gratefully acknowledge support from NIH Grant No. R01EB012466 from NIBIB's Image Guided Interventions Program (Program Director Dr. John Haller). We thank Dr. Kishwer Nehal and Dr. Erica Lee for supplying discarded tissue from Mohs surgery (under an IRB-approved protocol), and William Fox, Scott Grodevant, and Zachary Eastman at Lucid Inc. for technical support. ReferencesM. Rajadhyaksha, G. Menaker, T. Flotte, P. J. Dwyer, and
S. Gonzalez,
“Confocal examination of nonmelanoma cancers in thick skin excisions to potentially guide mohs micrographic surgery without frozen histopathology,”
J. Invest. Dermatol., 117
(5), 1137
–1143
(2001). https://doi.org/10.1046/j.0022-202x.2001.01524.x Google Scholar
R. Haque, R. Contreras, M. P. McNicoll, E. C. Eckberg, and
D. B. Petitti,
“Surgical margins and survival after head and neck cancer surgery,”
BMC Ear Nose Throat Disord, 6
(2)),
(2006). https://doi.org/10.1186/1472-6815-6-2 Google Scholar
L. Jacobs,
“Positive margins: The challenge continues for breast surgeons,”
Ann. Surg. Oncol., 15
(5), 1271
–1272
(2008). https://doi.org/10.1245/s10434-007-9766-0 Google Scholar
A. N. Yaroslavsky, J. Barbosa, V. Neel, C. DiMarzio, and
R. R. Anderson,
“Combining multispectral polarized light imaging and confocal microscopy for localization of nonmelanoma skin cancer,”
J. Biomed. Opt., 10
(1), 014011
(2005). https://doi.org/10.1117/1.1854173 Google Scholar
Z. Tannous, A. Torres and
S. Gonzalez,
“In vivo real-time confocal reflectance microscopy: A noninvasive guide for mohs micrographic surgery facilitated by aluminum chloride, an excellent contrast enhancer,”
Dermatol. Surg., 29
(8), 839
–846
(2003). https://doi.org/10.1046/j.1524-4725.2003.29219.x Google Scholar
A. Scope, U. Mahmood, D. S. Gareau, M. Kenkre, J. A. Lieb, K. S. Nehal, and
M. Rajadhyaksha,
“In vivo reflectance confocal microscopy of shave biopsy wounds: feasibility of intraoperative mapping of cancer margins,”
Br. J. Dermatol., 163
(6), 1218
–1228
(2010). https://doi.org/10.1111/j.1365-2133.2010.10063.x Google Scholar
V. Becker, T. Vercauteren, C. H. von Weyhern, C. Prinz, R. M. Schmid, and
A. Meining,
“High-resolution miniprobe-based confocal microscopy in combination with video mosaicing (with video),”
Gastrointest. Endosc., 66 1001
–1007
(2007). https://doi.org/10.1016/j.gie.2007.04.015 Google Scholar
K. E. Loewke, D. B. Camarillo, C. A. Jobst, and
J. K. Salisbury,
“Real-time image mosaicing for medical applications,”
Studies in Health Technology and Informatics, 125 304
–309
(2007). Google Scholar
J. K. Karen, D. S. Gareau, S. W. Dusza, M. Tudisco, M. Rajadhyaksha, and
K. S. Nehal,
“Detection of basal cell carcinomas in Mohs excisions with fluorescence confocal mosaicing microscopy,”
Br. J. Dermatol., 160
(6), 1242
–1250
(2009). https://doi.org/10.1111/j.1365-2133.2009.09141.x Google Scholar
D. S. Gareau, J. K. Karen, S. W. Dusza, M. Tudisco, K. S. Nehal, and
M. Rajadhyaksha,
“Sensitivity and specificity for detecting basal cell carcinomas in Mohs excisions with confocal fluorescence mosaicing microscopy,”
J. Biomed. Opt., 14
(3), 034012
(2009). https://doi.org/10.1117/1.3130331 Google Scholar
D. S. Gareau, Y. B. Li, B. Huang, Z. Eastman, K. S. Nehal, and
M. Rajadhyaksha,
“Confocal mosaicing microscopy in Mohs skin excisions: feasibility of rapid surgical pathology,”
J. Biomed. Opt., 13
(5), 054001
(2008). https://doi.org/10.1117/1.2981828 Google Scholar
P. Thevenaz and
M. Unser,
“User-friendly semiautomated assembly of accurate image mosaics in microscopy,”
Microsc. Res. Tech., 70
(2), 135
–146
(2007). https://doi.org/10.1002/jemt.20393 Google Scholar
|

