|
|
1.IntroductionInfrared (IR) and Raman spectroscopy have proven to be valuable tools for the study of biological samples due to the inherent chemical specificity of vibrational frequencies in the spectrum. Spectroscopic imaging techniques that couple a spectrometer and microscopy allow the measurement of a specific area within a sample thereby supplying spatially resolved molecular structure information. These techniques have, in the last 10 to 15 years, been developed into powerful biophysical approaches used by this laboratory, and others, for the studies of skin, brain, bone, and other biological samples.1, 2, 3, 4, 5, 6 Spatially resolved spectroscopic images provide, in essence, a histological image of the distribution of endogenous biochemical components (proteins, lipids, DNA) within a tissue section without the use of stains or probes. In addition, the vast amount of data inherent in spectral imaging permit sophisticated multivariate analyses such as cluster analysis, principal component analysis, and factor analysis, which enhance the ability to detect altered spatial regions in a heterogeneous sample. Combining univariate analysis and multivariate techniques provide a unique spectral-structure correlated characterization of biological samples. Human hair is a complex biological material consisting of distinct morphological components each composed of several chemical species. The cuticle, the outermost layer of hair, consists of 5 to 10 layers of thin, flat, but circumferentially curved, overlapping cuticle cells and provides a chemically resistant region surrounding the cortex. The cortex, the major component of the hair mass, contains elongated keratinized cortical cells and the cell membrane complex. Cortical cells are filled with spindle-shaped macrofilaments composed of intermediate filaments embedded in matrix materials. The medulla is innermost and generally occupies only a small percentage of the mass. In some fine hair, the medulla is completely absent. To date, complete chemical analysis of the medulla of human hair fibers has not been reported. As demonstrated in this paper, Fourier transform infrared (FTIR) spectroscopic imaging is uniquely able to characterize the medulla in situ in hair sections. Both FTIR and Raman spectroscopy have been extensively used to probe hair chemical composition and keratin structural changes under different treatments but without directly supplying spatial resolved information.7, 8, 9, 10, 11 The inherent distinct sensitivities of these two techniques to molecular vibrations provide complementary structure and chemistry information. IR spectroscopic imaging studies of hair cross sections have been reported with synchrotron12 and micro attenuated total reflection infrared microscopy.13 But images corresponding to relevant features of hair chemistry, anatomy, and organization were not completely disclosed. The current study focuses on the utility of transmission infrared and Raman spectroscopic imaging for the study of hair fibers. We demonstrate the ability of these techniques to generate spatially resolved images of hair chemistry, anatomy, lipid and protein distribution, lipid conformation, and protein secondary structure changes, with both univariate and multivariate data reduction approaches. 2.MethodsBoth European dark brown and white hair tresses were purchased from International Hair Importers and Products Inc. (Glendale, New York). Hair tresses were washed with a 10% (w/w) sodium lauryl ether sulfate solution followed by extensive water rinsing to remove any surface contamination. 2.1.Sample Preparation for IR ImagingA 1-cm long hair bundle was cut from the middle of European dark hair tresses and mounted on the top of a sample holder by embedding it in ice. The hair bundle was then microtomed at −30ºC into 5-μm thick cross sections with a Leica CM 1850 cryostat (Leica Microsystems Inc., Bannockburn, Illinois). Hair cross sections were collected onto CaF2 windows for IR imaging. This preparation technique avoids any possibility of contamination with embedding or fixing medium. 2.2.Sample Preparation for Raman ImagingThe high melanin granule content in pigmented (black) hair results in the absorption of laser light and subsequent sample destruction and/or fluorescence. Thus, Raman spectroscopic analysis is limited to hair fibers with very little color. In the current study, European white hair fibers were mounted on the surface of a copper plate. The hair fibers were covered with a glass cover slip to avoid any contamination from oil used by the oil-immersion objective. The working distance of the objective is up to ∼80 μm in the current setup. 2.3.Infrared MicroscopyHair cross sections were imaged with a Perkin Elmer Spotlight system that couples a FTIR spectrometer with an optical microscope, the system consists of a linear array of mercury-cadmium-telluride detectors and an automated high precision XY sample stage. Images were acquired with a 6.25-μm step size, eight scans for each spectrum, and 8 cm−1 spectral resolution. 2.4.Raman MicroscopyRaman spectra were acquired with a Kaiser Optical Systems Raman Microprobe. Excitation was achieved with a solid-state diode laser generating 4-mW laser power at 785 nm using a 100× objective. Spectral coverage is 100 to 3450 cm−1 with a spectral resolution of 4 cm−1. Confocal Raman lines were acquired from the hair surface in toward the medulla, i.e., perpendicular to longitudinal axis of the hair fiber. Each spectrum was collected with a 60-s exposure time, two accumulations, and cosmic ray correction. The spectral data from each confocal line were converted into MAT-file format and imported into ISys 3.1 software (Marlven Instrument Ltd., United Kingdom) for further analysis. 2.5.Data AnalysisBoth IR and Raman spectra were analyzed and a variety of image planes were generated using ISys 3.1 software. Spectral data were baseline-corrected before peak heights and peak areas were determined. As appropriate, factor analysis was used to detect relationships and patterns between observed variables. Factor analysis was performed using the ISys score segregation routine and its details have been described in previous studies.3 Briefly, the algorithm starts with a principal component analysis (PCA). PCA is inherently variance-oriented. It describes the maximum variance within the data set. The loading vectors from PCA calculation are not generally pure component spectra. It is difficult to use PCA loadings to explain the original spectra. PCA produces the data matrix: X = Σj S j L j, where the S j are the scores and the L j are the corresponding loadings. The data matrix can be factorized in terms of concentrations and spectra. Factor analysis seeks transformations between the L j and the e m (e m is the molar extinction coefficient of species m). The corresponding score images will then represent the concentration profiles of the various components. At the end of factor analysis, one set of factor loadings and score images are generated. Factor loadings are assumed to represent spectra from the location at which they are determined while scores indicate the relative contributions of various factors from particular locations. 3.Results and Discussions3.1.IR Spectral Imaging of Chemical Composition Across Hair Fiber SectionsA visual micrograph of microtomed hair cross sections is presented in Fig. 1a with the corresponding IR image of the hair section showing the protein Amide I band (∼1652 cm−1) intensity as a pixelwise pattern in Fig. 1b. Each pixel contains a full IR spectrum covering the spectral region from 700 to 4000 cm−1. Three small white squares, one each located in regions of the medulla, cortex, and cuticle in pixilated IR image, denote the pixels (6.25 × 6.25 μm) from which the three IR spectra shown to the right in Fig. 1c were acquired. Spectral regions from 1000 to 1800 cm−1 and 2800 to 3700 cm−1 are displayed. Several strong bands characteristic of proteins and lipids are observed in the spectra. Fig. 1(a) Optical image of microtomed hair cross sections (scale bar: 20 μm). (b) IR image of the band intensity at 1650 cm−1. Each pixel (each square) contains a full IR spectrum (700 to 4000 cm−1). Three small white boxes mark the pixels from which the corresponding spectra shown in (c) were obtained. (c) Representative IR spectra obtained from the medulla, cuticle, and cortex in the spectral regions 1000 to 1800 cm−1 and 2800 to 3700 cm−1. Each spectrum was obtained from a single pixel. Amides I and II are marked as AI and AII. (d) Spectra shown in (c) are plotted with second derivative spectra in the Amide region (1480 to 1700 cm−1). Spectra obtained from the cortex and cuticle are presented. The second derivative spectra are used to resolve peaks overlapped in raw spectra. Bands characteristic of different protein conformations emerge in the second derivative spectra: Amide I at 1652 cm−1 and Amide II at 1548 cm−1 for α-helix, Amide I at 1630 cm−1 in conjunction with 1695 cm−1 and Amide II at 1516 cm−1 for anti-parallel for β-sheet, band 1680 cm−1 for unidentified conformational structure. (e) IR image of β-sheet distribution relative to α-helical conformation across hair sections. The image was obtained from the intensity ratio of 1516/1548 cm−1. (f) IR image of lipid distribution. The image was obtained from the ratio of the peak areas at 2850 to 2960 cm−1. These two bands arise from C–H stretching of CH2 of lipid chains and CH3 of protein terminal groups. 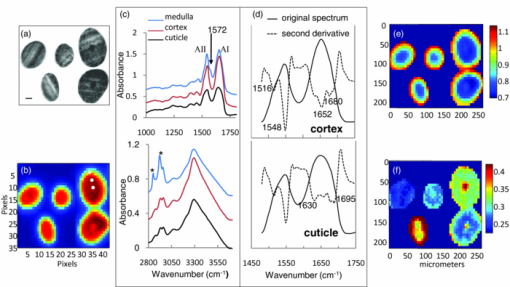 The Amide I and II modes associated with protein backbone vibrations are located in the 1480 to 1750 cm−1 spectral region. Both are very sensitive protein conformation and correlations between the frequencies of Amides I and II and protein secondary structures are well documented for different types of keratin.14 The current work extends the investigation of keratin to different hair regions by adding spatial resolution within the hair. Thus, in the spectrum obtained from cortex shown in Fig. 1c, Amide I at ∼1652 cm−1 and Amide II at ∼1548 cm−1 (marked as AI and AII) reflect that fibril proteins from the cortex are predominately α-helical in structure. However, both of the Amide I and II bands in the spectrum of the cuticle exhibit obvious broadening indicating that cuticle proteins have different secondary structure from those of the cortex. It has been reported from amino acid analysis that human cuticle cells contain a higher percentage of amino acids that are not usually found in α-helical peptides.15, 16 Further examination of these spectra was conducted using second derivative spectra as displayed in Fig. 1d. In addition to a weaker Amide I at 1652 cm−1, two other Amide I bands are clearly visible in the second derivative spectrum of the cuticle at 1630 and 1695 cm−1. These two bands also appear in the spectrum of cortex but they are much weaker. It has been reported that this pattern, Amide I at 1630 cm−1 in conjunction with a peak 1695 cm−1, is diagnostic of an anti-parallel β-sheet keratin structure.3, 17 The band assignment of ∼1680 cm−1 appearing in the second derivative spectra of both cortex and cuticle has not been previously reported. This may indicate that other unidentified conformations are present in hair proteins. The Amide II frequency at 1548 cm−1 is ascribed to an α-helical conformation, while the frequency of Amide II for β-sheet conformation varies from 1515 to 1525 cm−1 with different types of proteins.18 Previous studies have shown that the anti-parallel β-sheet in human keratin has an increased intensity for Amide II at ∼1516 cm−1.3, 17 Therefore, imaging the intensity ratio of 1516 cm/1548 cm−1 provides a measurement of β-sheet spatial distribution across hair sections relative to helical conformations [Fig. 1e]. It is evident from this image that the outer layer of hair (cuticle region) has a higher level of β-sheets than the inner hair fiber. The medulla exhibits the same frequencies for Amides I and II as the cortex, indicating that proteins in the medulla are also helical [Fig. 1c]. This result agrees with a previous investigation using transmission electron microscopy, which showed the medulla had a microfibrillary structure similar to that found in the cortex.19 The band at 1572 cm−1 shown in the spectrum of medulla is irrelevant to protein conformation and is assigned to the asymmetric stretching mode of COO- groups from amino acid side chains or fatty acids. In the spectral region of 2800 to 3700 cm−1 [Fig. 1c], the medulla presents strong bands at 2850 and 2924 cm−1 (marked with asterisk). These bands are attributed to the symmetric and asymmetric C–H stretching of lipid CH2 groups, respectively. The band at 2960 cm−1 arises from C–H asymmetric stretching of the CH3 moiety, mostly from amino acid side chains. The peak area ratio of 2850 to 2960 cm−1, shown in Fig. 1f, reveals relative lipid distribution across hair sections. It is observed that lipid concentrations vary among different hair fibers. However, the medulla and the cuticle always have relatively higher concentration of lipids. 3.2.Confocal Raman Microscopic Imaging of Hair FibersRaman spectroscopy complements the molecular vibration information available from IR spectroscopy and likewise contains detailed information about the structure, chemistry, and interactions of molecules of biomedical interest. Raman imaging has a higher spatial resolution and is suitable for the characterization of localized biochemical processes. In addition, Raman microscopy has the benefit of permitting the confocal measurement of molecular structure or chemistry. Thus, the physical sectioning of hair fibers is not required to obtain molecular chemistry and structure information within the fiber. This provides opportunities for the nondestructive examination of hair fibers under many different conditions. A series of Raman spectra were collected as a function of depth with a 2-μm step size from the hair cuticle surface. Due to the ∼80 μm working distance of the oil-immersion objective used in the current setup, the measurement was not able to completely reach through the fiber from one cuticle surface to the opposing cuticle surface. Nevertheless, a total of 38 spectra were collected, the first 19 spectra covering the hair cuticle, cortex, and medulla. Spectra in the regions of 400 to 1750 cm−1 and 2800 to 3020 cm−1 are presented in Figs. 2a, 2b, respectively. No data pretreatment is applied to the raw spectral data except a linear baseline correction between the spectral end points. Representative spectra (1200 to 1750 cm−1) of these three hair regions acquired at depths of 2, 20, and 38 μm, respectively, are displayed in the Fig. 2c. As expected, spatial variations in spectral features are apparent. Amide I located in the spectral region from 1500 to 1720 cm−1 is centered at different frequencies depending on the secondary structural conformation. As shown in Fig. 2c, all Amide I bands acquired at different depths present a broadened contour with several visible overlapping bands. This indicates that a variety of protein conformations exist in hair fibers. Distinct from IR spectra, the Amide I band of β-sheet confirmations in Raman spectra occurs at 1668 cm−1. The bands on the higher frequency side are mostly related to protein random structures, for example, the band at ∼1685 cm−1 has been assigned to nonhydrogen bonded disordered structures.20 It is notable that random structures are present across the hair fibers. However, the confocal Raman spectra clearly show that the cuticle contains less α-helical structure than the cortex and the medulla. The Raman confocal results are consistent with those obtained from the IR imaging of hair cross sections. Fig. 2Raman spectra of hair fibers in spectral regions (a) 400 to 1750 cm−1 and (b) 2800 to 3020 cm−1. Spectra were confocally obtained in 2 μm increments. Nineteen spectra are presented covering the cuticle, cortex, and medulla. (c) Raman spectra of Amide region (1200 to 1750 cm−1) at depths of 2, 20, and 38 μm below the hair fiber surface.  Raman spectra also contain detailed information regarding lipid distribution and the conformational order (fluidity) of lipid chains. As confocal Raman data acquisition proceeds from the hair surface to the medulla [Fig. 3a], bands at 1080, 1125,1295, 2848, and 2878 cm−1 increase in intensity [Figs. 2a, 2b], these bands are all assigned to the vibrational modes of lipids. The bands at 1295, 2848, and 2878 cm−1 are assigned to C–H twisting, symmetric stretching, and asymmetric stretching vibrations of CH2 group, respectively, while the band at 2934 cm−1 is assigned to symmetric stretching of the (mostly amino acid side chains) CH3 moiety. As shown in Fig. 3b, the intensity ratio of 2848/2934 cm−1 reflects lipid to protein distribution across the hair fiber. Consistent with the IR imaging analysis of sections, it is evident in the confocal Raman spectra that the medulla has a relatively high concentration of lipids. Fig. 3(a) Schematic diagram of confocal Raman acquisition from the hair fiber. (b) Raman image of lipid distribution. The map was constructed from the intensity ratio of 2848/2934 cm−1. The bands at 2848 and 2934 cm−1 are attributed to the C–H stretching modes of methylene and methyl groups, respectively. (c) Raman image of ordered lipid distribution. The map was obtained from the intensity ratio of 1125/1080 cm−1 of lipid chains. Both the 1125 and 1080 cm−1 peaks are assigned to C–C skeletal stretching of lipids, the former band from all-trans acyl chains, and the latter from random conformations. (d) Raman image of S–S cross links distribution. The map was created by integrating the peak areas of the band at 509 cm−1 arising from the S-S stretching mode of hair keratin fibers. Before peak integration, Raman spectra were normalized by band intensity of 1448 cm−1. The white dashed lines mark the position of the medulla. The color coding is red> orange> yellow> green> blue> purple and is separately scaled for image (b), (c), and (d). 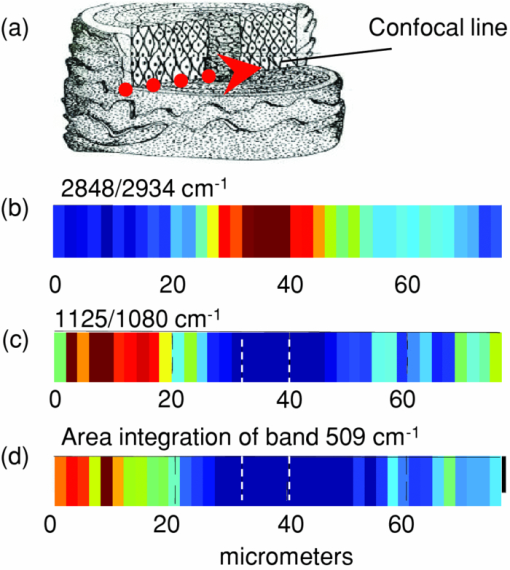 It is worthwhile mentioning that complete chemical analysis of the medulla of human hair fibers is not reported in the literature due to its poor solubility and the difficulty of isolating the medulla. The exact function of the medulla remains unknown, interestingly, however, some diseases are associated with abnormalities of the medulla.21 As demonstrated above, both IR imaging and confocal Raman microspectroscopy are capable of providing direct chemical and molecular structure-based information of this region in the hair fiber without any sample destruction or chemical treatment. It is quite reasonable to speculate that these techniques will greatly facilitate the process of the medical diagnosis of medulla related diseases. The lipid bands at 1125 and 1080 cm−1 are both assigned to C–C skeletal stretching. The former band in particular is indicative of all-trans acyl chain conformation and has been extensively used for monitoring lipid conformational order (fluidity).22, 23 The peak intensity ratio of 1125/1080 cm−1, shown in Fig. 3c, indicates that the lipids in the cuticle region are considerably more conformationally ordered than the lipids in the medulla. Raman microscopy is also capable of probing the conformation of disulfide linkages in hair fibers, a parameter not accessible by IR spectroscopy. The importance of disulfide linkages for the stabilization of hair protein structures is well known. In the Raman spectra shown in Fig. 2a, the band near 509 cm−1 is assigned to the S–S stretching mode. It has been reported that the position of this band varies with different disulfide conformers and that less stable disulfide conformers contribute to hair brittleness.10, 24 To evaluate the spatial variation of S–S cross-linking across hair fibers, spectra obtained from the confocal lines were normalized to the peak area of the ∼1448 cm−1 band (the C–H scissoring mode from both CH2 and CH3 groups). The map of the integrated area of band 509 cm−1 is shown in Fig. 3d and indicates that proteins in the cuticle region contain a higher level of S–S cross-links, which is consistent with the cuticle containing a higher content of sulfur than the cortex. It is noticeable that the S–S cross linkages are unevenly distributed through different layers of the cuticle. 3.3.IR Imaging of Spectra-structure Correlations of Hair Sections by Factor AnalysisIR imaging techniques provide the capability to sample large areas in short time periods with high spectral quality. IR images acquired from multiple hair sections collected in a single experiment can include hundreds to thousands of spectra with significantly different spectral features due to the heterogeneity of the hair sections. To efficiently extract the information inherent in the large arrays of spectral data, a multivariate algorithm was introduced to condense the information into a small set of dimensions (factors) with a minimum loss of information. Factor analysis was chosen to analyze the data in this work. The details of factor analysis have been briefly described in Sec. 2. The efficiency of using factor analysis to characterize the microanatomy of hair sections is demonstrated below. An IR image was acquired of 18 microtomed virgin hair sections. A corresponding optical micrograph is shown in Fig. 4a. The IR image was analyzed by factor analysis performed in two separate spectral regions: 1500 to 1700 cm−1 and 2832 to 3700 cm−1. Four factor loadings from each spectral region are shown in Fig. 4b and labeled f1 to f4. Representative score images corresponding to the four factor loadings in the spectral region 1480 to 1700 cm−1 are presented in Fig. 4c with the color bar marked from deep red for the highest score to dark blue for the lowest score. Score images of factors loadings from the remaining spectral region of 2832 to 3700 cm−1 show essentially the same results (not shown). As described in Sec. 2, score distributions depict correlations between factor loadings and raw spectra directly obtained from these measurements. Factor loadings generated from factor analysis are assumed to represent spectral features of the raw spectra acquired from the areas with the higher score. As shown in Fig. 4c, the score distribution images clearly delineate the known anatomical regions of hair, i.e., f1 corresponds to the medulla, f2 to the cortex, f4 to the cuticle. The remaining factor (f3) highlights the transition region between the cortex and the cuticle. In the same way, the factor loadings contain spectral features specific to each hair region. Briefly, in the factor loadings of the spectral region from 2830 to 3700 cm−1, f1 presents strong lipid bands at 2850 and 2924 cm−1 indicating generally higher lipid content in the medulla area. The band centered at 3400 cm−1 is assigned to O–H stretching and the increased intensity at 3400 cm−1 shown in f4 reflects relatively higher content of O–H groups in the epi-cuticle area. The high level of O–H groups may be attributed to water absorbed on the hair surface and/or a high level of serine residues in the cuticle.16 In the factor loadings of the spectral region from 1480 to 1750 cm−1, the frequencies of Amides I and II shown in f1 and f2 indicate predominantly an α-helical structural conformation in the cortex and medulla regions. The broadening contour of Amide I and the relatively increased intensities at ∼1630 cm−1 (Amide I) and 1516 cm−1 (Amide II) shown in f3 and f4 indicate more β-sheet contribution in the cuticle area. Fig. 4(a) Optical micrograph of a microtomed 5-μm thick hair cross section. Factor analysis was conducted on the IR image acquired from the hair cross sections. (b) Four distinct factor loadings in the regions 1480 to 1700 cm−1 and 2800 to 3700 cm−1 generated by the ISys score segregation algorithm (see Sec. 2) are offset and labeled as f1 to f4. (c) Spatial distribution of factor scores for each of the loadings in the spectral region 1480 to 1700 cm−1. Dark blue indicates the lowest score with green, yellow, orange, and red indicative of progressively higher scores. Factor loadings and score images have been assigned to different micro regions in hair as described in the text. 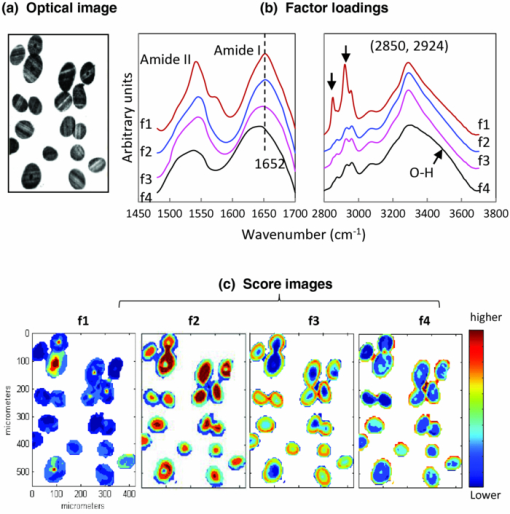 As demonstrated above, although the factor analysis condenses a large number of spectra to several factor loadings, these do represent the molecular structural features of the raw spectra from the location at which they are measured. Score images spatially delineate microregions within hair sections according to their underlying molecular structure. It is feasible to utilize such a multivariate algorithm to efficiently examine the altered spatial regions of hair samples of biomedical interest. 4.ConclusionsThis study demonstrates the unique utility of IR and Raman microscopic imaging for hair fiber studies through the acquisition of spatially resolved spectra of hair cross sections or the confocally acquired spectra of intact hair fibers. Using multivariate and univariate data analysis, the full informational content of spectra can be spatially correlated to hair composition, component concentration, lipid organization, and protein conformation. It is possible, therefore, to generate images corresponding to any relevant feature of hair chemistry, anatomy, and organization. An understanding of the spatially resolved spectroscopy of hair components paves the way to the investigation of molecular alteration of hair proteins and lipids resulting from environmental stresses and diseases status. AcknowledgmentsThe authors thank Professor Richard Mendelsohn and Dr. Carol Flach of Rutgers University for instrument support with the IR and Raman imaging data acquisition. ReferencesG. Zhang, D. J. Moore, R. Mendelsohn, and
C. R. Flach,
“Vibrational microspectroscopy and imaging of molecular composition and structure during human corneocyte maturation,”
J. Invest. Dermatol., 126
(5), 1088
–1094
(2006). https://doi.org/10.1038/sj.jid.5700225 Google Scholar
K. L. A. Chan, G. Zhang, M. Tomic-Canic, O. Stojadinovic, B. Lee, C. R. Flach, and
R. Mendelsohn,
“A coordinated approach to cutaneous wound healing: vibrational microscopy and molecular biology,”
J. Cell. Mol. Med., 12
(5B), 2145
–2154
(2008). https://doi.org/10.1111/j.1582-4934.2008.00459.x Google Scholar
G. Zhang, D. J. Moore, C. R. Flach, and
R. Mendelsohn,
“Vibrational microscopy and imaging of skin: from single cells to intact tissue,”
Anal. Bioanal. Chem., 387
(5), 1591
–1599
(2007). https://doi.org/10.1007/s00216-006-0852-0 Google Scholar
P. J. Caspers, G. W. Lucassen, E. A. Carter, H. A. Bruining, and
G. J. Puppels,
“In vivo confocal Raman microspectroscopy of the skin: noninvasive determination of molecular concentration profiles,”
J. Invest. Dermatol., 116
(3), 434
–442
(2001). https://doi.org/10.1046/j.1523-1747.2001.01258.x Google Scholar
X. Bi, X. Yang, M. P. Bostrom, and
N. P. Camacho,
“Fourier transform infrared imaging spectroscopy investigations in the pathogenesis and repair of cartilage,”
Biochim. Biophys. Acta, 1758
(7), 934
–941
(2006). https://doi.org/10.1016/j.bbamem.2006.05.014 Google Scholar
S. Koljenović, T. C. Bakker Schut, R. Wolthuis,
“Raman spectroscopic characterization of porcine brain tissue using a single fiber-optic probe,”
Anal. Chem., 79 557
–564
(2007). https://doi.org/10.1021/ac0616512 Google Scholar
A. Kuzuhara,
“Analysis of structural changes in permanent waved human hair using Raman spectroscopy,”
Biopolymers, 85
(3), 274
–283
(2006). https://doi.org/10.1002/bip.20646 Google Scholar
A. Kuzuhara,
“Analysis of structural change in keratin fibers resulting from chemical treatments using Raman spectroscopy,”
Biopolymers, 77
(6), 335
–344
(2005). https://doi.org/10.1002/bip.20221 Google Scholar
J. Strassburger and
M. M. Breuer,
“Quantitative Fourier transform infrared spectroscopy of oxidized hair,”
J. Soc. Comet. Chem., 36
(1), 61
–74
(1985). Google Scholar
S. Schlucker, K. R. Strehle, J. J. DiGiovanna, K. H. Kraemer, and
I. W. Levin,
“Conformational differences in protein disulfide linkages between normal hair and hair from subjects with trichothildystrophy: a quantitative analysis by Raman microspectroscopy,”
Biopolymers, 82
(6), 615
–622
(2006). https://doi.org/10.1002/bip.20515 Google Scholar
V. F. Kalasinski,
“Biomedical applications of infrared and Raman microscopy,”
Appl. Spectrosc. Rev., 31
(3), 193
–249
(1996). https://doi.org/10.1080/05704929608000570 Google Scholar
K. L. A. Chan, S. G. Kazarian, A. Mavraki, and
D. R. Williams,
“Fourier transform infrared imaging of human hair with a high spatial resolution without the use of a synchrotron,”
Appl. Spectrosc., 59
(2), 149
–155
(2005). https://doi.org/10.1366/0003702053085070 Google Scholar
P. Dumas and
L. Miller,
“The use of synchrotron infrared microspectroscopy in biological and biomedical investigations,”
Vib. Spectrosc., 32
(1), 3
–21
(2003). https://doi.org/10.1016/S0924-2031(03)00043-2 Google Scholar
R. P. Oertel,
“Protein conformational changes induced in human stratum corneum by organic sulfoxides: an infrared spectroscopic investigation,”
Biopolymers, 16
(10), 2329
–2345
(1977). https://doi.org/10.1002/bip.1977.360161017 Google Scholar
E. R. Blout, C. de Lozé, S. M. Bloom, and
G. D. Fasman,
“The dependence of the conformations of synthetic polypeptides on amino acid composition,”
J. Am. Chem. Soc., 82
(14), 3787
–3789
(1960). https://doi.org/10.1021/ja01499a080 Google Scholar
R. R. Clarence,
“Chemical composition,”
Chemical and Physical Behavior of Human Hair, 39
–64 2nd ed.Springer-Verlag, New York
(1988). Google Scholar
R. Mendelsohn and
H. H. Mantsch,
“Fourier transform infrared studies of lipid -protein interaction,”
Progress in Protein-Lipid Interaction, 2 103
–146 Elsevier, Amsterdam
(1986). Google Scholar
M. Ishida, M. Takai, H. Okabayashi, H. Masuda, E. Nishio, and
C. J. O’Connor,
“FTIR evidence for antiparallel -sheet structures of long oligomeric N-acetyl-l-glutamic acid benzyl esters,”
Vib. Spectrosc., 27
(2), 135
–184
(2001). https://doi.org/10.1016/S0924-2031(01)00130-8 Google Scholar
J. I. Clement, A. Le Pareux, and
P. F. Ceccaldi,
“The specificity of the ultrastructure of human hair medulla,”
J. Forensic Sci. Soc., 22
(4), 396
–398
(1982). https://doi.org/10.1016/S0015-7368(82)71518-X Google Scholar
P. Yager and
B. P. Gaber,
“Membranes,”
Biological Applications of Raman Spectroscopy, 203
–261 Wiley, New York
(1987). Google Scholar
W. H. Irwin Mclean,
“Close shave for a keratin disorder—k6hf polymorphism linked to pseudofolliculitis barbae,”
J. Invest. Dermatol., 122 xi
–xiii
(2004). https://doi.org/10.1111/j.0022-202X.2004.22351.x Google Scholar
R. Mendelsohn,
“Laser-Raman spectroscopic study of egg lecithin and egg lecithin-cholesterol mixtures,”
Biochim. Biophys. Acta, 290 15
–21
(1972). https://doi.org/10.1016/0005-2736(72)90047-8 Google Scholar
C. Xiao, C. R. Flach, C. Marcott, and
R. Mendelsohn,
“Uncertainties in depth determination and comparison of multivariate with univariate analysis in confocal Raman studies of a laminated polymer and skin,”
Appl. Spectrosc., 58
(4), 382
–389
(2004). https://doi.org/10.1366/000370204773580202 Google Scholar
H. Sugeta, A. Go, and
T. Miyazawa,
“Vibrational spectra and molecular conformations of dialkyl disulfides,”
Bull. Chem. Soc. Jpn., 46 3407
–3411
(1973). https://doi.org/10.1246/bcsj.46.3407 Google Scholar
|

