|
|
1.IntroductionEpithelial ovarian cancer has the highest mortality rate of any of the gynecology cancers and spreads beyond the ovary in 90% of the women diagnosed with ovarian cancer. Ovarian cancer goes undetected in both developed and developing countries because of inadequate technology to detect pre-invasive or early-stage disease. There are approximately 25,600 new cases of ovarian cancer in the United States per year, and there was an estimated 16,000 deaths from ovarian cancer in 2004.1 High prevalence of late-stage disease and the poor prognosis associated with these later stages are the major factors that give ovarian cancer patients such a dismal prognosis.1, 2, 3 In recent years, the need for new methods for early detection at a pre-cancer level has stimulated the rapid development of a reliable, objective, and minimally invasive optical tools. The optical (spectroscopic) methods are ideal for this, because in these techniques, spectral changes can be measured with very high sensitivity and they can be used to monitor biochemical alterations, which are the precursors for many diseases including cancer.4, 5 Laser-induced spectroscopic techniques4 examine different types of light-tissue interactions and noninvasively provide biochemical and morphological information at the molecular, cellular, and tissue levels. They are extremely sensitive and usually cause no discomfort. Laser-induced spectroscopy techniques, 4, 5, 6, 7, 8, 9, 10, 11, 12 such as fluorescence, 4, 5, 6, 7, 8, 9, 10 Raman,11 and photoacoustic12 (PA), have been widely investigated as nondestructive optical tools for medical diagnostics. 4, 5, 6, 7, 8, 9, 10, 11, 12, 13 Pulsed photoacoustic spectroscopy is one of the blossoming fields of science and, in recent years, the technique has become increasingly popular as a tool for biomedical imaging and detection.13, 14 The technique is based on the detection of an acoustic signal induced by photons and is different than the optical techniques detecting optical signals. The propagation of acoustic signals within biological tissues are less susceptible to attenuation and scattering compared to the optical signals and the information contained within them is much less likely to be lost. Because of this advantage, the photoacoustic technique projects itself as a robust and attractive modality for imaging beyond the possible range that exists for all-optical techniques. The technique of pulsed photoacoustic generation in liquid is well established.14, 15 Scientific applications of this technique have been widely reported in a range of areas including semiconductor research,16 physical processes in liquids,14, 15 trace gas monitoring,17 analyte detection in liquids,15, 16, 17, 18, 19 depth-resolved analysis of tissue models,20 photoacoustic imaging,21, 22, 23, 24, 25, 26 and volumetric analysis of protein.18, 19 A broad range of potential applications have emerged including detection of breast,22 skin,20 and oral cancers27 and vascular applications such as imaging superficial blood vessels28, 29 and the characterization of arterial tissues.29 In pulsed photoacoustic spectroscopy for biological samples, when a short laser pulse interacts with a tissue, during the laser pulse duration, some of the inherent biomolecules whose concentration depends upon the condition of the tissue having resonance absorption at the excitation absorb the incident photon energy and get excited. The excited biomolecules are then relaxed through nonradiative relaxations releasing the absorbed energy in the form of heat in the absorbing volume of the sample. Subsequently, the thermal expansion of the instantaneously heated sample followed by its contraction due to the periodically applied excitation causes a pressure variation in the irradiated volume. This pressure variation is nothing but the acoustic waves that travel outward through the medium and can be detected at the surface.30 If the tissue is placed in a cuvette forming the PA cell,13 the acoustic waves propagating outward from the excitation volume get reflected from the cuvette walls and superpose to form standing waves of different modes depending upon the tissue structure through which they are originated. A few superposed acoustic waves with slight damping will result in complex and long duration time domain photoacoustic transients.31 The goal of the present work is to explore whether photoacoustic spectroscopy could eventually be adapted to improve early diagnosis of ovarian neoplasia. With this idea, in our laboratory, we have recorded the photoacoustic spectra of normal as well as different stages of ovarian cancer tissue samples in vitro and principal component analysis (PCA) (Refs. 7, 8, 9, 10 and 32) based k-nearest neighbor (k-NN),6, 8, 9 artificial neural network (ANN),7, 10 and support vector machine (SVM) (Refs. 33 and 34) classification algorithms are developed. This multi-algorithm approach, comprised of computational methods such as ANN, k-NN, and SVM, was used to improve the reliability in classification and hence to determine their relative diagnostic performance. 2.Materials and Methods2.1.Sample Collection and HandlingBiopsied or surgically resected ovarian normal, benign, and malignant tissue samples of nearly 5×5 to 5×6 mm2 size were obtained from the Department of Obstetrics and Gynecology, Kasturba Hospital, Manipal University, Manipal, India. Tissues from uninvolved areas from the same subjects are used as healthy controls. A mirror image of each sample was fixed in 10% neutral buffered formalin and was sent for histopathological certification. The samples were kept moist with saline (pH = 7.4), and spectra were recorded within half an hour of tissue removal. The sample details are given in Table 1. According to the histopathology report, samples used in this study were stratified into three main categories: normal ovary (no structures except stroma, epithelium, corpus albicans, and corpus luteum), benign neoplasm (abnormal growth without invasive areas) and endometriosis (growth of both endometrial glands and stroma on the ovary), and malignant (invasion of carcinoma into the ovary, ovarian tumor, hemorrhage, cystic, Papillary cystadenoma). Fig. 7ROC graph showing relative diagnostic performance of the three discrete classifiers, k-NN, ANN, and SVM analyses. 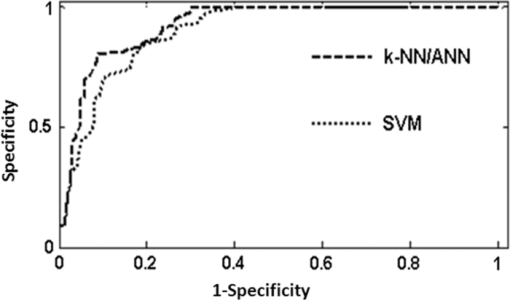 Table 1Sample details.
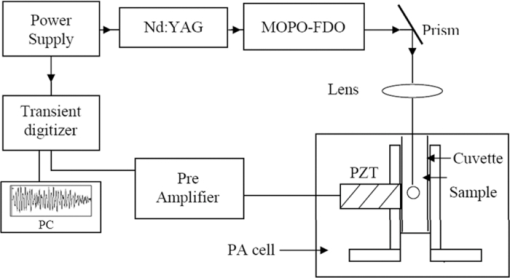
2.2.Experimental SetupThe block diagram of the experimental set up used to record the ovarian tissue photoacoustic spectra is shown in Fig. 1. The samples were excited using 325-nm pulsed laser radiation obtained from an Nd-YAG/MOPO/FDO source (Spectra Physics, Quanta Ray, Model: PRO 230 10, MOPO SL) at 10 Hz repetition of 5 ns pulses with energy 100 to 200 μJ. An indigenously developed PA cell consisting of the “sample holder” and the “PZT mounting” was utilized to generate the PA signal.12, 13 The laser light was focused onto the moist tissue samples kept in a quartz cuvette positioned in the sample holder of the PA cell. The focusing of the laser beam was achieved using a 10 cm focal length convex lens vertically placed above the cuvette. The acoustic transient signal generated upon laser excitation of the samples was detected with a PZT detector (3 mm diameter and 4 mm thickness, PI-ceramic, Germany) mounted in the photoacoustic cell12, 13, 14 held in contact with the cuvette sidewall. After suitable amplification using a home-made pre-amplifier, the transient signals were recorded in time domain with transient digitizer (EG&G Models 9826 and 9846) personnel computer combination. By changing the incidence position of the laser beam on the sample under study more than two PA transients were recorded from each tissue sample. In the present study, 102 PA transients (34 normal, 38 benign, and 30 malignant) were recorded involving ten normal, 13 benign, and nine malignant tissue samples. All of the PA transients were recorded with maximum care using fixed excitation geometry, as well as with consistent recording procedures. 2.3.Fast Fourier Transform of PA spectraSince the PA cell is essentially a resonant cavity, it is most informative to Fourier transform of the acoustic waves providing the frequency response of the cell and its resonant frequencies for the different types of samples. By examining the signal in the frequency domain one can easily observe the modal frequencies that developed upon excitation of the sample under study. Thus, the acoustic waves (PA signals) originated from different pathological samples, such as normal, benign, and malignant, have slightly different modal frequencies giving rise to slightly different spectral profiles that are directly related to their pathological states. Each pathological sample, thus, produces its own characteristic spectral profile slightly different from one another depending upon the state of the tissue. The spectral features, if extracted from each type of samples, will thus carry significantly different optical properties representing the sample. In the present study, various spectral features extracted from different pathological samples are used for discrimination analysis of the samples. Before extracting spectral features, all of the recorded time domain PA transients were smoothened and Fourier transformed using MATLAB@R6 (Ref. 35) tools, Medfilt1, and FFT (fast Fourier transform), respectively, and the frequency region 0 to 4×105 Hz of the corresponding FFT pattern was windowed. Subsequently using MATLAB@R6 algorithms, six different features from each of 102 FFT spectra were extracted. These features were mean, median, spectral residual, energy, standard deviation, and maximum intensity, and involving them, a feature space matrix is formed. The process of feature extraction, thus, transformed the frequency domain spectra into a feature space matrix, reducing the number of computations needed for further data analysis. Different spectral features extracted from each of 102 spectra are described below.
First, the four features, mean, median, standard deviation, and maximum intensity, were obtained using the inbuilt tool, “Data Statistics,” the feature Energy using the function ‘SUM (X), and the spectral residuals using the Basic Fitting tool of the MATLAB software. Thus, six features extracted from a total of 102 spectra produced an original data matrix of dimension (6×102). 2.4.Data AnalysisIn many of the earlier studies with fluorescence6, 7, 10 and Raman spectroscopy,11 when a number of spectra are recorded from different sites of a tissue sample, the mean of all of the spectra was taken as representative for that sample. In the present study, all of the 102 PA transients recorded are treated as independent data. This, we believe, is a better approach because often a tissue specimen can have normal and neoplastic regions adjacent to each other showing variations in the spectra from site to site.10, 36 The analysis of the spectral data for their classification between different groups has been carried out using PCA based k-NN, ANN, and SVM algorithms. 2.4.1.Principal component analysisPCA can be used to reduce the dimensionality of a dataset, while still retaining as much of the variability of the dataset as possible.7, 10, 32 It is a classical statistical method that transforms attributes of a dataset into a new set of uncorrelated attributes called principal components (PCs). Applying PCA to the feature space matrix, the original data gets transformed into a set of PC scores. The contribution of each PC to the total variance of spectral data is proportional to its eigenvalue. Higher-order PCs often account for less than 1% of the total variation and represent mostly noise.7, 10, 32 PCs that have variance more than 1% in the spectral data are considered as informative PCs. In PCA, first the calibration sets for each class of samples are prepared and an appropriate number of principal components is selected. In the present study, calibration sets are formed using 45 spectra, 15 randomly selected from each group of pathologically certified normal, benign, and malignant samples. PCA is then performed on the data matrix comprised of the six spectral features extracted from each of the 45 calibration samples. The calibration sets are then optimized by cluster analysis removing outliers and an optimum number of factors (PCs) containing maximum diagnostic information is selected.7, 10, 32 Outlier samples usually arise from incorrect measurements, whether it is in the concentration data (i.e., errors in the primary calibration technique) or the spectral data (i.e., sample handling procedures, environmental control such as temperature, humidity, etc). The outlier samples, if present in a calibration set, will tend to “pull” the model in their direction, causing the predicted concentrations of valid samples to be less accurate (or even erroneous) than if the sample was completely eliminated from the training set. The cluster plot for detecting outliers in the calibration model is generally plotted between the PC scores of the first two principal components because the first two principal components represent the plane of best fit through the data. In the present study, when this graph was plotted with arbitrarily chosen spectra of three classes, data points of some spectra were found inconsistent with other samples of the same class in the calibration model and their scores were far from their respective classes. Those spectra with inconsistent scores were then replaced by other spectra of the same class. This process was repeated for a class until spectra with appropriate scores have been identified and found consistent with other scores in the calibration model. Thus, we have optimized the calibration models by including only those spectra that have consistent scores with other samples in the same class and an appropriate number of PCs is identified for each. After the PCs with diagnostic information are identified, the scores of the selected PCs are utilized and the k-NN, ANN, and SVM algorithms are trained. These trained algorithms are then used to classify unknown spectra. The algorithms for k-NN, ANN, and SVM classification are constructed using only those PCs that have significantly different projection scores for the normal, malignant, and benign spectra.*** 2.4.2.Classification algorithm: k-nearest neighborNearest neighbor methods provide an important data classification tool for recognizing object classes in the pattern recognition domain.6, 8, 9 In this method, an unknown sample is classified to that class having the most “similar” or “nearest” sample point in the training set of data. In the present study, for classification of ovarian data, the single nearest neighbor method has been used. For classification, a prototype sample is computed from the reference (calibration) set and a given test sample is classified as belonging to the class of the closest prototype. That means, when an input pattern is presented to a k-NN classifier, the classifier computes the k nearest prototypes to it using a distance (Euclidean distances) measure. Then the classifier assigns the class label using a majority vote among the labels of the k nearest prototypes. This prototype vector is known as the centroid vector. For any test sample, the three spectral distances (i.e., Euclidean distances), corresponding to “normal centroid,” “malignant centroid,” and “benign centroid” are estimated first and are used in the classification. Details of the k-NN algorithm and steps involved in the classification analysis are explained elsewhere.6, 9 2.4.3.Classification algorithm: artificial neural networkIn the present study, ANN with back propagation was used for classification of the ovarian normal, benign, and malignant samples. The back propagation ANN is a potential method for finding a relationship between different input variables and binomial output variables.7, 10, 36, 37 The back propagation algorithm consists of fitting the parameters (weights) of the model by a criterion function, usually mean squared error (MSE) or maximum likelihood, using a gradient optimization method. In back propagation artificial neural network, the error (the difference between the predicted outcome and the true outcome) is propagated back from the output to the connection weights in order to adjust the weights in the direction of minimum error. The design of ANN with back propagation algorithm is explained elsewhere.10 In the classification using ANN, two output neurons are used and the performance goal was fixed at an accuracy of 0.01. The input passes through two processing elements to reach the output. Since the activation function is a sigmoid function, the network (when trained) can represent a nonlinear classifier of any order. The fact that each component of input data reaches each of the output terminals through several parallel paths provides the network considerable flexibility in deciding the nonlinear input-output relationship.37, 38 2.4.4.Support vector machine analysisSVM is basically a binary classifier.33, 34 It can be a one-class, two-class, or multiclass SVM.33, 34 In multiclass SVM, it assign labels to instances by using a support vector machine, where the labels are drawn from a finite set of several elements. This approach reduces the single multiclass problem into multiple binary problems. In the present study, we have used a multiclass SVM one-versus-rest kernel-radial basis function (rbf) based classifier.33, 34 A common approach, in case of SVMs, is the use of recursive feature elimination (RFE) and elimination of the least important features corresponding to the smallest ranking criterion and training the classifier with the remaining features. But, in the present study, we performed MATLAB@R6 based PCA on the training set data matrix comprising six different spectral features extracted from each spectrum as mentioned earlier and an appropriate number of PCs is selected. The scores of the selected PCs containing maximum diagnostic information are then used to train the classifier and the remaining samples are classified using their respective scores with the trained network. Details of SVM design and execution is explained elsewhere.33, 34 SVMmulticlass is an implementation of the multiclass support vector machine (SVM).33, 34 Given a training set of instance-label pairs (x i; y i), i = 1, . . . . . . ., l where x i ∈ R n and y∈ {1, −1.}.l, the SVMs require the solution of the following optimization problem:
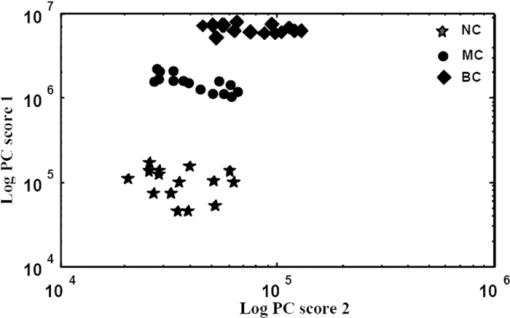
[TeX:]
\documentclass[12pt]{minimal}\begin{document}\begin{eqnarray*}
&&\mathop {\min }\limits_{w,b,\xi } \frac{1}{2}w^T w + C\sum\limits_{i = 1}^l {\xi _i} \\
&&\text{subject to } y_i ({w^T \phi (x_i) + b}) \ge 1 - \xi _i,\xi _i \ge 0,
\end{eqnarray*}\end{document}
where R is the radius of the ball containing the data, n is the dimension of the input space, l is the training set size, Φ (x
i) is the real valued function before taking threshold, ξ is the slack variable, w is the weight vector, b is the bias, and w
T is transpose of weight vector. The set of slack variables ξi allow for the class overlap, controlled by the penalty weight C > 0. This parameter C, called the regularization parameter, basically controls the trade-off between the largest margin and lowest number of errors. For, C = ∞, no class overlap is allowed. In the present study, for the optimal value C = 100000, the SVM classifier was trained with the training set.The steps followed in SVM:
2.4.5.Receiver operating characteristics analysisThe performances of the classifiers can be visualized through receiver operating characteristic (ROC) graphs and a particular classifier can also be selected with them.39, 40 The ROC graphs are plotted with the true positive rate (TPR) in the y-axis and false positive rate (FPR) in the x-axis.39, 40 Each point on the ROC graph stands for a pair of sensitivity/specificity values corresponding to a particular decision threshold. In case of multiple ROC curves, the area under the curves (AUC) is generally used to evaluate the performance of the curves. For ROC analysis and subsequently for determining the AUC, in the present study, Statistical Package for the Social Sciences (SPSS) 11.0 software41 was used. The AUC is equivalent to Mann-Whitney U-statistic39 (nonparametric test of difference between disease/nondisease test results). 3.Results and DiscussionTypical smoothened ovarian tissue photoacoustic transients of normal, malignant, and benign classes (left) and the corresponding FFT pattern (right) are shown in Fig. 2. From Fig. 2, noticeable differences among normal, benign, and malignant conditions have been observed. As we know, tissue is basically inhomogeneous in nature containing different chromophores (e.g., water, oxy-hemoglobin, deoxy-hemoglobin, lipid, cytochrome oxidase, and melanin) and fluorophores (tryptophan, collagen, NADH, FAD, etc.) in it at different proportions.42, 43 A small change in its condition alters its biochemical composition leading to changes in its spectral profile. These are essentially the fingerprints that help to determine the condition of the tissue. The PA signal induced by photons depends on the optical energy deposition at the target tissue, which is the product of the tissue optical absorption coefficient and the local light fluence. The optical properties including the absorption coefficient, scattering coefficient, refractive index, and anisotropy factor of the target tissue and the background medium, are all highly dependent on the excitation wavelength.42 Thus, the complex PA transients originated from normal, benign, and malignant tissues carry different spectral information related to their pathological states. Fig. 2Typical time domain photoacoustic spectra (left) and the corresponding frequency domain FFT spectra (right) of ovarian tissues. (a) Normal, (b) benign, and (c) malignant. 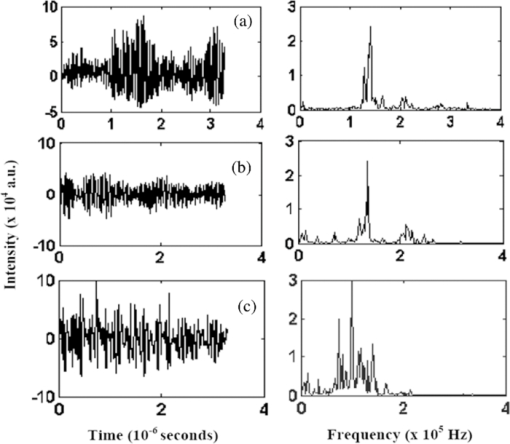 3.1.Principal Component AnalysisAs mentioned in Sec. 2.4, we have six different features extracted from each of 102 photoacoustic spectra (45 calibration plus 57 test) used to form a feature space matrix of dimension (6×102). On this data matrix, PCA is performed further reducing data dimensionality. In PCA, first, the calibration sets for normal, benign, and malignant classes are formed by randomly selecting 15 pathologically certified spectra from each class. Subsequently, the calibration sets are optimized using cluster analysis removing outliers and an appropriate number of factors (PCs) containing maximum diagnostic information is selected. Figures 3a and 3b respectively show the eigenvalues for the factors and total percentage variance (i.e., total percentage contribution to the variation spectra with increasing number of factors) of 45 calibration set spectra (15 normal, 15 malignant, and 15 benign). From Figs. 3a and 3b, it is clear that only three factors (PCs) are sufficient to explain the calibration set data. The first PC itself covers over 98.1% of variance and the first three PCs represent 99.92% of the total variance. This shows that the feature vector of length six could be reduced to three components using the PCA technique. As a result, the feature space matrix of dimension (6×102) was dramatically reduced to dimension (3×102) making classification computationally more efficient. Thus, we have considered the first three factors for further analysis of the ovarian data. Fig. 3Plots of calculated eigenvalues. (a) Total% variance against (b) factors for a PCA decomposition of 45 calibration spectra (15 normal, 15 malignant, and 15 benign). 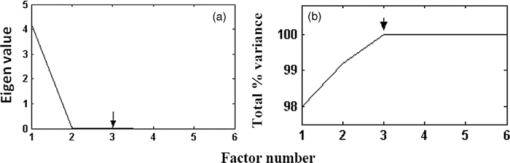 In PCA, the calibration sets are optimized using cluster analysis removing outliers.15, 16, 17 Figure 4 shows the cluster/scatter plot between the scores of the first two principal components of 45 calibration samples. It is clearly seen from the plot that all of the samples diagnosed as normal, malignant, and benign by pathological examination in the calibration set are clustered in three distinct regions without any overlap, indicating that there are no outliers in these sets. The calibration sets are thus optimized and three informative PCs representing these samples are selected. The scores of these first three factors (PCs) of the 45 (15 normal, 15 malignant, and 15 benign) calibration samples are then used as input feature space for training of k-NN, ANN, and SVM algorithms and the remaining 57 (19 normal, 15 malignant, and 23 benign) samples are tested against them using their respective scores. 3.2.k-Nearest Neighbor AnalysisFor predicting unknown new samples using k-NN, first the k-NN based programs are executed and then the classification of the unknown samples is carried out as per the algorithm protocol as mentioned in Ref. 6. This technique when used for the 45 calibration samples, all samples are classified to their respective classes. When the 57 test samples (19 normal, 15 malignant, and 23 benign) are tested against the trained algorithm, all normal are classified as normal; two out of 15 malignant are not classified as malignant but as normal; and out of 23 benign, 16 are classified as benign, two are classified as normal, and five are classified as malignant. The two malignant test samples that are classified as normal are the sample numbers 71 and 72 and the two benign test samples which are also classified as normal are the samples 89 and 90. The five benign test samples that are classified as malignant are the samples 95, 96, 97, 98, and 99, respectively. The specificity, sensitivity, and accuracy of this analysis are found to be 100, 86.76, and 91.17%, respectively. The classification results for the calibration and the test samples are shown in Table 2. Table 2Fifteen calibration/test normal/malignant/benign samples tested against normal/malignant/benign calibration/test set.
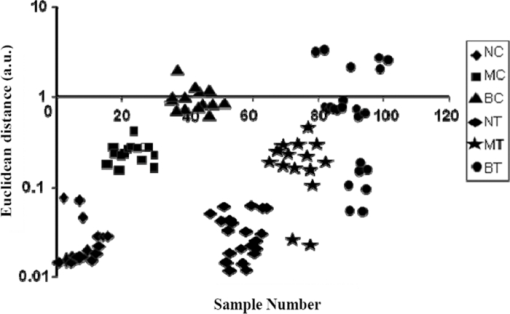
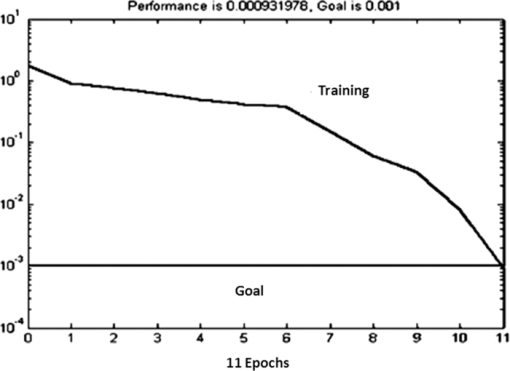
Figure 5 shows the plot of Euclidean distance against sample number for the 102 samples (15+19 normal, 15+15 malignant, and 15+23 benign). In Fig. 5, the Euclidean distances for all of the samples are plotted considering normal centroid as the reference point. The calibration set samples (normal, benign, and malignant) are clustered into three distinct groups without any overlap. The test normal and malignant samples are also clustered along with the corresponding calibration set samples, showing 100% discrimination for these samples. Two of the benign test samples are overlapped with the normal calibration set cluster and five are overlapped with the malignant cluster, showing their tendency toward the normal and malignant. To sum up, only a very small number of malignant/benign samples (two malignant + seven benign) fall outside the general range of malignant/benign species, indicating the probability of malignant/benign samples being in the respective cluster of about 93.33%/81.57% and finding them out of the cluster of about 6.67%/18.43%. The discrimination between different classes of samples is clearly visible in the plot. 3.3.Artificial Neural Network AnalysisAs mentioned in Sec. 2.4, the ANN algorithm was trained with the input feature vectors formed with the scores of three factors for 15 normal, 15 malignant, and 15 benign samples. Using these feature vectors (15×3 matrix for each case), MATLAB-based ANN programs are executed to train the network and subsequently to predict any new data. The performance goal for the training of ANN was met at 0.001 as shown in Fig. 6 and the convergence was achieved with 11 epochs. The network was given the instruction to show binary digit 1 1 for normal, 1 −1 benign, and −1 −1 for malignant conditions. When this classification analysis was used on the calibration samples, all 45 samples (15 normal, 15 malignant, and 15 benign) are classified to their respective groups. In this analysis, all of the 19 normal test spectra are classified as normal, two out of 15 malignant test spectra are not classified as malignant but as normal, and 16 out of 23 benign test spectra were classified as benign. Out of the seven remaining benign test spectra, two are classified as normal and five are classified as malignant. The classification results for the calibration and test samples are shown in Table 3. Similar to the k-NN analysis, in this case also, the same two malignant test samples 71 and 72 are classified as normal, the same two benign test samples 89 and 90 are classified as normal, and the same five test benign samples 95, 96, 97, 98, and 99 are classified as malignant. This shows that the ANN classification results are exactly the same type as that of the k-NN results. The specificity, sensitivity, and accuracy of this analysis are also found to be 100, 86.76, and 91.17%, respectively. Table 3Calibration/test set of normal/malignant/benign (45+57 spectra) tested against the trained neural network and results.
3.4.Support Vector Machine AnalysisAfter constructing the feature space matrix (15×3) for all three calibration sets comprising their PC scores, a MATLAB-based program was executed and an SVM classifier was trained to predict any new data. The classifier was trained with one-versus-rest (SVM kernel rbf = 10 C = 100,000), where C is the regularization parameter that trades off margin size and training error. The classifier was given the instruction to show binary digit 1 −1 −1 for normal, −1 1 −1 for malignant, and −1 −1 1 for benign conditions. When this classification technique was used on calibration set samples, all 45 samples (15 normal, 15 malignant, and 15 benign) are classified into their respective groups. In this case, all of the 19 normal test spectra are classified as normal, four out of 15 malignant test spectra are not classified as malignant but as normal, and 14 out of 23 benign test spectra are classified as benign. Out of the nine remaining benign test spectra, six are classified as normal and three are classified as malignant. The calibration and test sample results are given in Table 4. Similar to the k-NN/ANN analysis, in this case also, the same two malignant test spectra 71 and 72 are classified as normal and the same two benign test spectra 89 and 90 are classified as normal. Also, the same three benign test spectra 97, 98, and 99 that are classified as malignant by k-NN/ANN analysis are classified as malignant by SVM. But, the two benign test spectra 95, 96 that are classified as malignant in k-NN/ANN analysis are classified as normal in SVM analysis. In addition to this, two more benign test spectra, 86 and 87 that are classified as benign by the other two classifiers, are classified as normal by SVM. This shows that the SVM classification results are not exactly the same type as that of the k-NN/ANN results. The specificity, sensitivity, and accuracy of the SVM analysis are found to be 100, 80.18, and 90.09%, respectively. Table 4Calibration/test set of normal/malignant/benign (45+57 spectra) tested against the trained SVM classifier and results.
3.5.Receiver Operating Characteristic AnalysisAfter plotting the ROC (Refs. 39 and 40) curves for k-NN/ANN and SVM classifiers, their relative diagnostic performance was determined by measuring the AUC for each curve. The ROC curves plotted with FPR versus TPR for the two classifiers is shown in Fig. 7. The curves are drawn by showing each and every sensitivity/specificity pair resulting from the continuously varying decision threshold over the entire range of results observed in all three analyses. The calculated values of the AUCs for k-NN/ANN and SVM are found to be 0.86 and 0.82, respectively. 4.ConclusionThe pulsed laser induced photoacoustic spectroscopy study conducted on ovarian tissues and the subsequent statistical analysis using PCA-based kNN/ANN/SVM algorithms was motivated by the idea of developing an objective and sensitive photoacoustic-based technique for optical pathology. As a preliminary investigation, the current study reports the discrimination of a limited number of normal, benign, and malignant ovarian tissues in vitro. The photoacoustic spectroscopy in combination with PCA-based k-NN/ANN/SVM algorithms has properly and efficiently classified the ovarian tissues suggesting its possible potential application in the field as an alternative or complementary technique to the existing other conventional methods of disease diagnosis. The small time needed to acquire and analyze the photoacoustic spectra together with the high rates of success proves that the technique is very attractive for real time applications. Although, the technique in its current form may not be suitable for such applications, however, with proper instrumentation using a fiber optic probe and a thin film PZT detector coupled to an endoscope, the technique may be suitable. Of course, for clinical validation of the methodology, further studies with a sufficient number of subjects belonging to normal, benign, and malignant classes as well as on blind samples are extremely essential. Overall, our results indicate that a PCA-based multi-algorithm approach has great promise to classify high dimensional ovarian tissue photoacoustic data and appears to be highly capable to detect ovarian carcinoma, which would be a big improvement in guiding biopsies and diagnosing tissues in different pathological conditions. AcknowledgmentsThe authors are thankful to the Manipal University, Manipal and to Dr. K. Satyamoothy, Professor and Director, Manipal Life Sciences Centre for providing the necessary facilities to carry out this study. The authors are also thankful to Dr. Rani A. Bhat for providing some of the ovarian tissues used in the study. ReferencesM. Brewer, U. Utzinger, E. Silva, D. Gershenson, J. R. C. Bast, M. Follen, and
R. R. Kortum,
“Fluorescence spectroscopy for in vivo characterization of ovarian tissue,”
Lasers Surg. Med., 29 128
–135
(2001). https://doi.org/10.1002/lsm.1098 Google Scholar
M. Brewer, U. Utzinger, W. Satterfield, L. Hill, D. Gershenson, R. Bast, J. T. Wharton, R. R. Kortum, and
M. Follen,
“Biomarker modulation in a nonhuman Rhesus primate model for ovarian cancer chemoprevention,”
Cancer Epidemiol. Biomarkers Prev., 10 889
–893
(2001). Google Scholar
M. A. Brewer, U. Utzinger, J. K. Barton, J. B. Hoying, N. D. Kirkpatrick, W. R. Brands, J. R. Davis, K. Hunt, S. J. Stevens, and
A. F. Gmitro,
“Imaging of the ovary,”
Technol. Cancer Res. Treat., 3
(6), 617
–627
(2004). Google Scholar
N. Ramanujam,
“Fluorescence spectroscopy of neoplastic and non-neoplastic tissues,”
Neoplasia, 2 89
–117
(2000). https://doi.org/10.1038/sj.neo.7900077 Google Scholar
A. Pradhan, P. Pal, G. Durocher, L. Villeneuve, A. Balassy, F. Babai, L. Gaboury, and
L. Blanchard,
“Steady state and time resolved fluorescence properties of metastatic and non-metastatic malignant cells from different species,”
J. Photochem. Photobiol., B, 31 101
–112
(1995). https://doi.org/10.1016/1011-1344(95)07178-4 Google Scholar
S. D. Kamath and
K. K. Mahato,
“Optical pathology using oral tissue fluorescence spectra: classification by principal component analysis (PCA) and k-means nearest neighbour (k-NN) analysis,”
J. Biomed. Opt., 12
(1), 014028
(2007). https://doi.org/10.1117/1.2437738 Google Scholar
S. D. Kamath, C. D'souza, S. Mathew, S. George, S. Chadangil, and
K. K. Mahato,
“A pilot study on colonic mucosal tissues by fluorescence spectroscopy technique: discrimination by principal component analysis (PCA) and artificial neural network (ANN) analysis,”
J. Chemom., 22 408
–418
(2008). https://doi.org/10.1002/cem.1154 Google Scholar
S. D. Kamath, R. A. Bhat, S. Ray, and
K. K. Mahato,
“Autofluorescence of ovarian normal, benign, and malignant tissues: a pilot study,”
Photomed. Laser Surg., 27
(2), 325
–335
(2009). https://doi.org/10.1089/pho.2008.2261 Google Scholar
S. D. Kamath and
K. K. Mahato,
“Principal component analysis (PCA) based k nearest neighbour (k-NN) analysis of colonic mucosal tissue fluorescence spectra,”
Photomed. Laser Surg., 27
(4), 659
–668
(2009). Google Scholar
G. S. Nayak, S. D. Kamath, K. M. Pai, A. Sarkar, S. Ray, J. Kurien, L. D’Almeida, B. R. Krishnanand, S. Chadangil, V. B. Kartha, and
K. K. Mahato,
“Principal component analysis (PCA) and artificial neural network (ANN) analysis of oral tissue fluorescence spectra: classification of normal, pre-malignant, and malignant pathological conditions,”
Biopolymers, 82 152
–166
(2006). https://doi.org/10.1002/bip.20473 Google Scholar
C. M. Krishna, G. D. Sockalingum, J. Kurien, L. Rao, L. L. Venteo, M. Pluot, M. Manfait, and
V. B. Kartha,
“Micro-Raman spectroscopy for optical pathology of oral squamous cell carcinoma,”
Appl. Spectrosc., 58 107
–114
(2004). https://doi.org/10.1366/0003702041959460 Google Scholar
R. A. Bhat, S. D. Kamath, K. K. Mahato, S. Ray, and
V. B. Kartha,
“Photoacoustic spectroscopic studies of ovarian tissue in different pathological conditions: classification using cluster analysis,”
(2006). Google Scholar
S. D. Kamath, V. B. Kartha, and
K. K. Mahato,
“Dynamics of L-tryptophan in aqueous solution by simultaneous laser induced fluorescence (LIF) and photoacoustic spectroscopy (PAS),”
Spectrochim. Acta Part A, 70
(1), 187
–194
(2007). Google Scholar
C. K. N. Patel and
A. C. Tam,
“Pulsed optoacoustic spectroscopy condensed matter,”
Rev. Mod. Phys., 53 517
–550
(1981). https://doi.org/10.1103/RevModPhys.53.517 Google Scholar
M. W. Sigrist,
“Laser generation of acoustic waves in liquids and gases,”
J. Appl. Phys., 60 R83
–R121
(1986). https://doi.org/10.1063/1.337089 Google Scholar
A. Rosencwaig and
A. Gersho,
“Theory of the photoacoustic effect with solids,”
J. Appl. Phys., 47 64
–69
(1976). https://doi.org/10.1063/1.322296 Google Scholar
P. Hess,
“Photoacoustic, photothermal and photochemical processes in gases,”
Topics in Current Physics, 46 Springer-Verlag, Berlin
(1989). Google Scholar
S. E. Braslavsky,
“Photoacoustic and photothermal methods applied to the study of radiationless deactivation processes in biological system and in substances of biological interest,”
Photochem. Photobiol., 43 667
–679
(1986). https://doi.org/10.1111/j.1751-1097.1986.tb05645.x Google Scholar
T. Schmid,
“Photoacoustic spectroscopy for process analysis,”
Anal. Bioanal. Chem., 384 1071
–1086
(2006). https://doi.org/10.1007/s00216-005-3281-6 Google Scholar
G. Puccetti, F. Lahjomri, and
R. M. Leblanc,
“Pulsed photoacoustic spectroscopy applied to the diffusion of sunscreen chromophores in human skin: the weakly absorbent regime,”
J. Photochem. Photobiol. B, 39 110
–120
(1997). https://doi.org/10.1016/S1011-1344(96)00017-6 Google Scholar
E. Zhang, J. Laufer, and
P. Beard,
“Backward-mode multiwavelength photoacoustic scanner using a planar Fabry–Perot polymer film ultrasound sensor for high-resolution three-dimensional imaging of biological tissues,”
Appl. Opt., 47
(4), 561
–577
(2008). https://doi.org/10.1364/AO.47.000561 Google Scholar
S. Manohar, S. E. Vaartjes, C. G. Johan, V. Hespen, J. M. Klaase, M. E. Frank, W. Steenbergen, and
T. G. van Leeuwen,
“Initial results of in vivo non-invasive cancer imaging in the human breast using near-infrared photoacoustics,”
Opt. Express, 15
(19), 12277
–12285
(2007). https://doi.org/10.1364/OE.15.012277 Google Scholar
L. Xiang, D. Xing, H. Gu, D. Yang, S. Yang, L. Zeng, and
W. R. Chen,
“Real-time optoacoustic monitoring of vascular damage during photodynamic therapy treatment of tumor,”
J. Biomed. Opt., 12 014001
(2007). https://doi.org/10.1117/1.2437752 Google Scholar
D. W. Yang, D. Xing, Y. Tan, H. Gu, and
S. Yang,
“Integrative prototype B-scan photoacoustic tomography system based on a novel hybridized scanning head,”
Appl. Phys. Lett., 88 174101
(2006). https://doi.org/10.1063/1.2198488 Google Scholar
B. T. Cox, S. R. Arridge, and
P. C. Beard,
“Quantitative photoacoustic image reconstruction for molecular imaging,”
Proc. SPIE, 6086 60861M
(2006). https://doi.org/10.1117/12.644806 Google Scholar
L. Xiang and
F. Zhou,
“Photoacoustic imaging application in tumor diagnosis and monitoring,”
Key Eng. Mater., 364–366 1100
–1104
(2008). https://doi.org/10.4028/www.scientific.net/KEM.364-366.1100 Google Scholar
A. A. Oraevsky, A. A. Karabutov, E. B. Savateeva, B. Bell, M. Motamedi, S. L. Thomsen, and
P. Pasricha,
“Opto-acoustic imaging of oral cancer: feasibility studies in hamster model of squamous cell carcinoma,”
385
–396
(1999). https://doi.org/10.1117/12.356791 Google Scholar
J. Laufer, D. Delpy, C. Elwell, and
P. Beard,
“Quantitative spatially resolved measurement of tissue chromophore concentrations using photoacoustic spectroscopy: application to the measurement of blood oxygenation and hemoglobin concentration,”
Phys. Med. Biol., 52 141
–168
(2007). https://doi.org/10.1088/0031-9155/52/1/010 Google Scholar
P. C. Beard and
T. N. Mills,
“Characterization of post mortem arterial tissue using time-resolved photoacoustic spectroscopy at 436nm, 461nm and 532nm,”
Phys. Med. Biol., 42
(1), 177
–198
(1997). https://doi.org/10.1088/0031-9155/42/1/012 Google Scholar
Y. Su, F. Zhang, K. Xu, J. Yao, and
R. K. Wang,
“A photoacoustic tomography system for imaging of biological tissues,”
J. Phys. D: Appl. Phys., 38 2640
–2644
(2005). https://doi.org/10.1088/0022-3727/38/15/016 Google Scholar
D. A. Schurig, G. L. Klunder, M. A. Shannon, and
R. E. Russoa,
“Signal analysis of transients in pulsed photoacoustic spectroscopy,”
Rev. Sci. Instrum., 64
(2), 363
–373
(1993). https://doi.org/10.1063/1.1144258 Google Scholar
I. T. Joliffe,
“Principal component analysis,”
(1986) Google Scholar
S. S. Keerthi and
C. J. Lin,
“Asymptotic behaviors of support vector machines with Gaussian kernel,”
Neural Comput., 15
(7), 1667
–1689
(2003). https://doi.org/10.1162/089976603321891855 Google Scholar
C. C. Chang and
C. J. Lin,
“Training υ-support vector classifiers: theory and algorithms,”
Neural Computation, 13
(9), 2119
–2147
(2001). https://doi.org/10.1162/089976601750399335 Google Scholar
Introduction to MATLAB 6 for engineers,”
(1994) Google Scholar
M. F. Mitchell,
“Accuracy of colposcopy,”
Consult. Obstetrics Gynecol., 6 70
–73
(1994). Google Scholar
J. M. Zurada, Introduction to Artificial Neural Systems, 3rd ed.Jaico Publishing House, Mumbai, India
(2002). Google Scholar
S. Hykin, Neural Networks. A Comprehensive Foundation, 2nd ed.Pearson Education, Singapore
(2001). Google Scholar
T. Fawcett,
“An introduction to ROC analysis,”
Pattern Recogn. Lett., 27 861
–874
(2006). https://doi.org/10.1016/j.patrec.2005.10.010 Google Scholar
J. A. Hanley and
B. J. McNeil,
“The meaning and use of the area under a receiver operating characteristic (ROC) curve,”
Radiology, 143 29
–36
(1982). Google Scholar
V. Gupta,
“SPSS for-beginners,”
http://www.brothersoft.com/spss-11-for-beginners-2nd-edition-34060.html Google Scholar
J. R. Rajian, P. L. Carson, and
X. Wang,
“Quantitative photoacoustic measurement of tissue optical absorption spectrum aided by an optical contrast agent,”
Opt. Express, 17
(6), 4879
–4889
(2009). https://doi.org/10.1364/OE.17.004879 Google Scholar
G. Wagnieres, W. Star, and
B. C. Wilson,
“In vivo fluorescence spectroscopy and imaging for oncological applications,”
Photochem. Photobiol., 68
(5), 603
–632
(1998). Google Scholar
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||