|
|
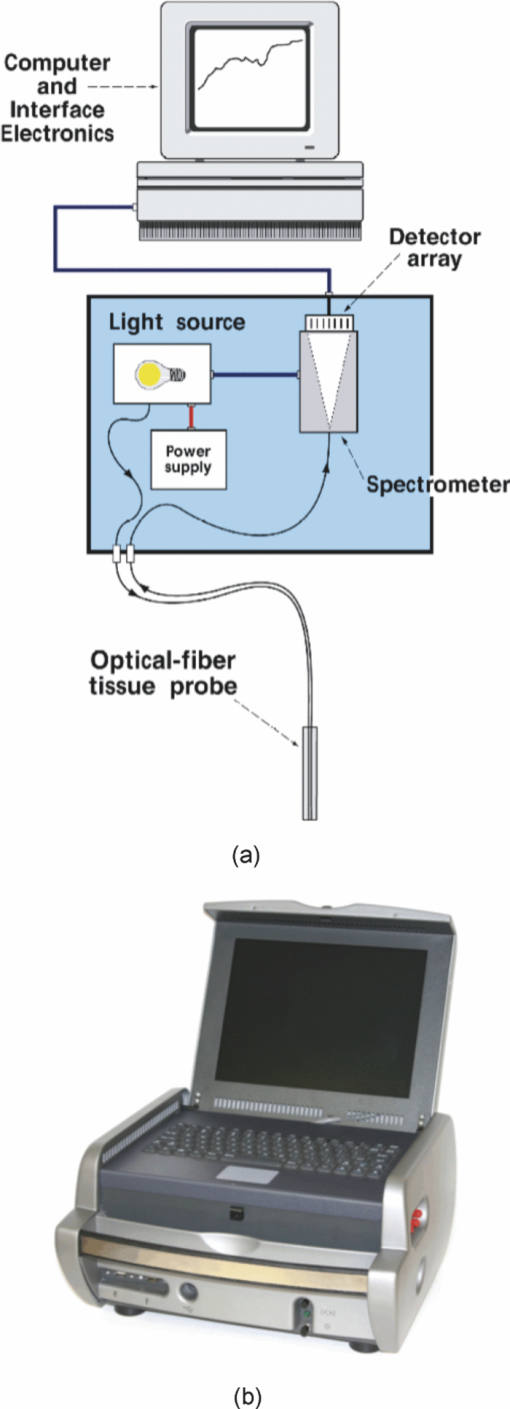
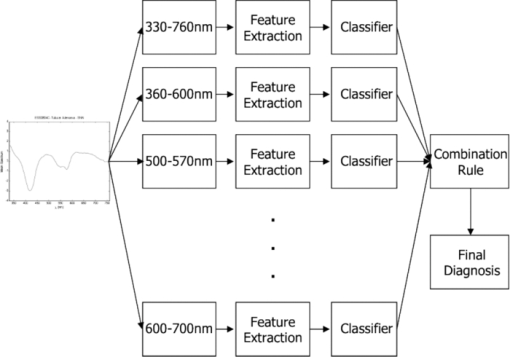
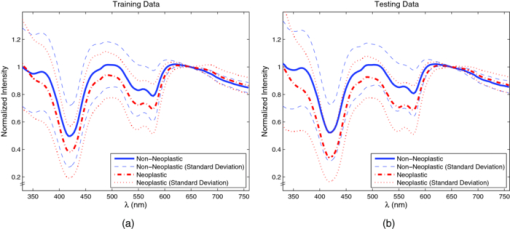
1.Introduction1.1.Spectral Classification for Diagnostic SpectroscopyThe promise of optically guided biopsy using elastic-scattering spectroscopy (ESS) in colorectal cancer screening relies on achieving a high accuracy for detecting neoplasms, because misdiagnoses can be potentially costly. Both statistical and model-based methods of spectral classification have been studied by a number of groups.1, 2, 3, 4 The present report focuses on improvements in pattern-recognition and machine-learning methods, as applied to ESS, with the complexities of biological variability in mind. The specific clinical application addressed here is the assessment of polyps during colonoscopy for the screening/management of colorectal cancer. Current algorithms, based on a single classifier, have shown the feasibility of ESS as a diagnostic tool,1, 2, 5, 6, 7 yet increased accuracy is desired for ESS to be widely adopted in clinical settings, particularly for the in situ classification of polyps during colorectal cancer screening.8 A different classifier framework, one where the decision is made based on the input from various classifiers, can enhance performance over a single-classifier scheme.9, 10, 11, 12 This field of combining multiple classifiers, or ensemble classifiers, has garnered increased attention over the past decade in the pattern-recognition and machine-learning community motivated by the prospect of improving classification performance. Ensemble classifiers are generally designed based on different approaches for generating the base classifiers (i.e., the set of classifiers that composes the ensemble).11 One approach, the data level, uses different subsets of the training data to design the base classifiers. Two common sampling methods to achieve this include bagging and boosting,11, 13 with AdaBoost11, 14, 15 being a well-known classifier employing the latter method. In a second approach, the feature level, the base classifiers are designed on different subsets of data features. This approach was used to classify autofluorescence spectra acquired from healthy and diseased mucosa in the oral cavity.10 Another approach is the classifier level, where each of the base classifiers is designed using a different classification method. Tumer 12 employed the classifier-level approach when classifying fluorescence spectra for cervical dysplasia by designing ensembles with different radial basis-function neural networks as base classifiers. An additional classification paradigm can also be used to improve current performance levels. The misclassification-rejection (MR) approach, known in the literature as error-rejection, is one where the classifier identifies samples at high risk of being misclassified, in most cases the ones lying near the decision boundary, and as such, refrains from making a decision on them. Thus, for a binary classification problem there is a third possible outcome: A sample is not assigned a class or label, and is considered as a rejected sample. The expected result is an improvement in performance relative to the one obtained with a regular classifier at the expense of having a percentage of the samples not classified. Rejected samples would then have to be examined by some other method (repeat optical measurements or biopsy) in order to obtain their classification. With this paradigm, decisions become a sequential process, suitable for colonoscopic cancer screening, where an initial real-time, in vivo examination of a polyp using ESS is followed by biopsy and histopathology if it could not be classified spectroscopically. The idea of misclassification-rejection has been mentioned for spectroscopic applications;16, 17 however, no formal framework has been presented. The concept of misclassification-rejection was first introduced by Chow18 for minimum probability of error classifiers, where the optimal decision rules take the form of a threshold for the posterior probabilities, assuming that the underlying probability distributions are known. Extensions to this work have been presented for cases where the underlying probability distributions are unknown and estimated;19, 20 artificial neural networks;21, 22 and support vector machines (SVM).23, 24, 25, 26, 27 In addition, reformulations of the SVM training problem, embedding the rejection option have been presented.28, 29, 30 In this framework, the decision boundary becomes a decision region that, in its simplest form, consists of a pair of parallel hyperplanes separated by some distance. Samples lying within this decision region are rejected. In this paradigm, the orientation of the decision regions as well as the distance between them are obtained during the training phase, in contrast to previous work that has focused on methods to threshold samples once the classifier has been designed. This has resulted in an improvement in error-reject trade-off when compared to simply thresholding the output after designing the classifier. The notion of misclassification-rejection for classifying spectra in “optical biopsy” schemes can be thought of as relating to the all-too-frequent histopathology designation of “indeterminate,” or to cases where multiple histopathology assessments of the same tissue lack agreement. A different type of “error” identification has been introduced by Zhu,31 wherein principal components that relate to variations in practitioner technique (e.g., probe pressure, angle) are ignored in the classification. These differences are often variations of overall intensity, and their removal can lead to improvement in classification performance.31, 32 Another type of error rejection is often referred to as “outlier rejection,” in which spectra that clearly invoked experimental error (e.g., broken probe, noncontact with the tissue, or dirt on the probe tip) are rejected. Both of these error types differ from our misclassification-risk reduction, which is more akin to identifying biological uncertainly. In our experimental approach, such outliers can be recognized and instantly rejected at the moment of the measurement. 1.2.Optical Biopsy in Management of Colorectal CancerIn the United States, colorectal cancer (CRC) is the second leading cause of cancer and cancer death for both sexes (behind lung cancer), with nearly 150,000 new cases and 50,000 deaths annually in recent years.33 Current recommendations from multiple professional societies advocate early detection by screening the entire average-risk population for CRC beginning at age 50.34 Accepted screening modalities include (i) stool occult blood or deoxyribose nucleic acid testing, (ii) flexible sigmoidoscopy every five years (iii) colonoscopy every 10 years, and (iv) radiological examination via double-contrast barium enema or computed tomography colonography every five years. Of these, colonoscopy emerged as the most effective method of screening because a high-quality examination permits simultaneous detection and removal of precancerous polyps, thus modifying CRC risk.35 The effectiveness of colonoscopic cancer prevention hinges on the complete removal of all polyps detected during a standard-definition white-light endoscopy (SDWLE).35, 36, 37 However, a large proportion (up to 50%) of polyps that are removed are, in fact, non-neoplastic by histopathology (i.e., they have negligible malignant potential). Thus, there is an inherent inefficiency: Because neoplastic and non-neoplastic polyps cannot be reliably distinguished by SDWLE alone, substantial resources are devoted to the removal of clinically inconsequential tissue, while the actual benefit derives from the identification and removal of premalignant lesions. Thus, there is great need for simple, rapid, and low-cost methods for “smart” tissue evaluation, because removing inconsequential tissue introduces additional unnecessary time, cost, and incremental risk of bleeding and perforation, to a high-demand, high-volume screening procedure. Optical spectroscopy has been suggested to assist endoscopists in classifying polyps in situ and in real time in a simple and cost-effective manner.1, 2, 3, 4 In particular, ESS has shown promise for detecting dysplasia and/or cancer in various epithelial-lined hollow organs, including the urinary bladder,38 esophagus,7, 39, 40, 41, 42 and colon,1, 2, 3 as well as in tissues such as breast and lymph nodes.5, 6 ESS, mediated by specific fiberoptic probes with specific optical geometries, is sensitive to the absorption spectra of major chromophores (e.g., oxy-/deoxy-hemoglobin) and, more importantly, the scattering spectra, which relate to micromorphological features of superficial tissue. ESS spectra derive from the wavelength-dependent optical scattering efficiency (and the effects of changes in the scattering angular probability) caused by optical index gradients exhibited by cellular and subcellular structures. Unlike Raman and fluorescence spectroscopy, ESS provides largely microstructural, not biochemical, information. ESS is sensitive to features such as nuclear size, crowding, and chromaticity, chromatin granularity, and mitochondrial and organellar size and density [Fig. 1a]. Because neoplasia is associated with changes in subcellular, nuclear, and organellar features, scattering signatures represent the spectroscopic equivalent of a histopathological interpretation. The ESS method, however, senses morphological changes semiquantitatively without actually rendering a microscopic image.43, 44 In practice, ESS is a point-source measurement obtained over a broad wavelength range (320–900 nm) that samples a tissue volume of ≤0.1 mm3. Probes are comprised of separate illuminating and collecting fibers [Fig. 1b] and require optical contact with tissue being interrogated. Collected light transmitted to the analyzing spectrometer must first undergo one or more scattering events through a small volume of the tissue before entering the collection fiber(s) in the “backward” direction. No light is collected from surface Fresnel reflection. The standard ESS catheter-type probe consists of a pair of fibers (each with a core diameter of 200 μm) with a center-to-center separation of ∼250 μm. Because of the small separation of the source and detector fibers, the collected light predominantly samples the mucosal layer, which is typically 300–400 μm thick in the GI tract. Fig. 1(a) Cartoon illustration of optical scattering from density gradients in cells, and (b) a diagram of the optical geometry for the fiber-optic tissue measurements. Fiber tips are in optical contact with the tissue surface. Only light that has scattered elastically within the epithelial layer is collected. 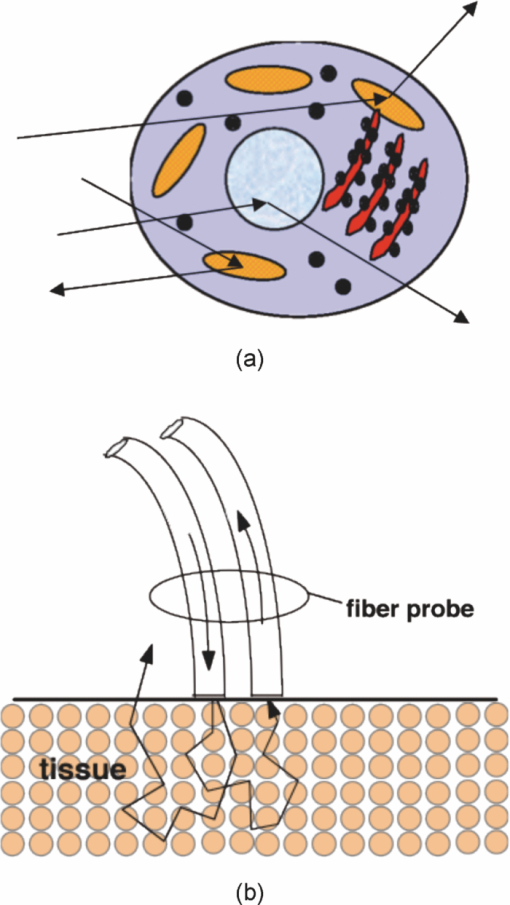 Ideally, when using elastic-scattering spectroscopy, some form of spectral analysis is performed on the collected measurements of the examined tissue. ESS was first applied in vivo in the urinary bladder by Mourant,38 where correlation of spectral features, determined “by inspection,” for the detection of malignant tissue in a retrospective analysis from a small sample size (110 biopsy sites from 10 patients) showed the feasibility of ESS as a diagnostic tool. Bigio 5 used artificial neural networks (ANN) to classify ESS spectra from breast tissue and sentinel nodes. Analysis resulted in sensitivities of 69 and 58%, and specificities of 85 and 93%, for breast tissue and sentinel nodes, respectively. In another study by Ge,2 light-scattering spectra of neoplastic and non-neoplastic colonic polyps could be distinguished using multiple linear regression analysis, linear discriminant analysis (LDA), and ANN. Sensitivities and specificities of 91% and 78%, 91% and 74%, and 79% and 91% were obtained, respectively, for each of the three methods. Dhar 1 used LDA and leave-one-out cross-validation to classify several colonic lesions, among them adenomatous versus hyperplastic polyps, with a sensitivity of 84% and specificity of 84%, adenocarcinoma versus normal colonic mucosa, with a sensitivity of 80% and specificity of 86%, and adenocarcinoma versus adenomatous polyps, with a sensitivity of 80% and specificity of 75%. In addition to these statistical pattern-recognition approaches, model-based classifications have been proposed, primarily based on extracting nuclear size distribution from the scattering spectra. These have yielded performances comparable to statistical approaches.4, 45, 46 The advantages of using ensemble classifiers and misclassification-rejection, described here, are intended to provide diagnostic information of higher reliability, with the benefit of providing an assessment of the risk of misclassification. The resulting improved method is tested prospectively on a naïve data set. 2.Materials and Methods2.1.InstrumentationThe ESS system and integrated forceps have been previously described.47 Briefly, when using ESS, or any point spectroscopic measurement, it is essential to coregister precisely the optical reading and the physical biopsy. With this in mind, an endoscopic tool that integrates an ESS probe with a biopsy forceps was designed. In order to incorporate an ESS probe into the biopsy forceps, it was necessary to reduce the diameter of the optical fibers. In previous studies, the illuminating fiber was a 400-μm core optical fiber and the backscatter detector a 200-μm fiber. For the integrated ESS optical biopsy forceps, two 200-μm core fibers (source and detector) were used, each with a numerical aperture of 0.22 in air. The center-to-center separation between the fibers was ∼250 μm. Biopsy forceps were built with a hollow central channel that extended to the space between the jaws (SpectraScience, San Diego, California) capable of accommodating the 0.470-mm diameter of the hypotube encasing the probe (Fig. 2). Fig. 2(a) Two-dimensional diagram of the forcep tip is depicted. The optical forcep is a modified traditional endoscopic jaw-type biopsy forcep (left) with a central channel through which fiberoptic probes can be introduced for tissue measurements (right). (b) A photograph of a clinically-usable unit, standard biopsy forceps (left), ESS integrated optical forceps (right). 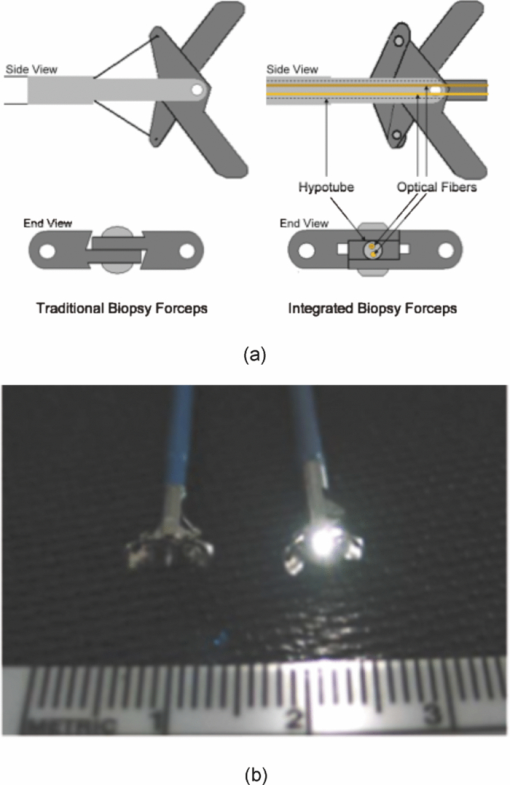 Similar to previously described ESS designs, the forceps connect to the ESS system, which consists of a pulsed Xenon-arc lamp (LS-1130-3, Perkin Elmer, Waltham, Massachusetts) broadband light source, a built-in computer with custom ESS software, and built-in spectrometer (S2000, Ocean Optics, Inc., Dunedin, Florida), microcontroller board and power supplies (Fig. 3). Before each procedure, the ESS forceps and spectrometer were calibrated for system response by measuring the reflectance from a spatially flat diffuse-reflector Spectralon (Labsphere Inc., North Sutton, New Hampshire) to standardize for variations in the light source, spectrometer, fiber transmission, and fiber coupling. It should be noted that the endoscope light source (Olympus 100 series with Evis Exera I and II systems) does not interfere with ESS readings, because the background light is measured separately and subtracted from the ESS measurement. 2.2.Clinical MeasurementsData collection was performed under an ongoing IRB-approved clinical study at the Veterans Affairs Medical Center, Boston, Massachusetts. Subjects were recruited from among individuals scheduled for a medically indicated screening or surveillance colonoscopy. When a polyp or suspicious growth was encountered by the endoscopist during the procedure, the forceps jaws were opened such that the central optical probe was placed in gentle contact with the identified polyp and five ESS measurements were taken in rapid succession. Once the optical readings were obtained, the forceps jaws were closed to obtain the physical biopsy. By using forceps with integrated ESS optics, precise coregistration of optical readings and physical biopsies is assured. Three specialist gastrointestinal pathologists, using predefined standard histopathological criteria, reviewed each endoscopic pinch biopsy independently. The “optical biopsies” were then correlated to the majority classification by histopathology. 2.3.Data Processing and Analysis2.3.1.PreprocessingThe spectra from the ESS measurements consist of ∼800 pixels in the wavelength range of 300–800 nm. All spectra were preprocessed before being analyzed. The five measurements taken at each site were averaged, smoothed, and then cropped. Smoothing is done by first using a moving average with a sliding window of a size of ten points (detector pixels), and then by averaging blocks of five points (corresponding to a spectral band of ∼3 nm). The spectra are then cropped from 330 to 760 nm, resulting in a spectrum of 126 points. Finally, each spectrum was normalized to the intensity at 650 nm, because we are interested in the spectral shape and not the relative intensity. 2.3.2.Principal component analysisGiven the high-dimensional nature of ESS data, classifiers are designed on lower-dimensional features extracted with principal component analysis (PCA). On the basis of the Karhunen–Loeve transform, PCA reduces dimensionality by restricting attention to those directions along which the variance of the data is greatest.13 These directions are obtained by selecting the eigenvectors of the pooled data covariance matrix that correspond to the largest eigenvalues. Reduction in dimensionality is then achieved by applying the linear transformation of the form Eq. 1[TeX:] \documentclass[12pt]{minimal}\begin{document}\begin{equation} \tilde {\bf X} = {\bf V}^{T} ({\bf X} - \mu _{\bf X}), \end{equation}\end{document}2.3.3.Support vector machinesFeatures obtained using PCA are used as inputs for our different classifiers based on support vector machines (SVMs).49, 50, 51 Support vector machines are, in their simplest form, binary linear classifiers, where the decision boundary obtained is a hyperplane that has the maximum separating margin between the classes. This allows this classifier to exhibit good generalization performance (i.e., good performance in the presence of unseen data).49 Training support vector machines involves solving the following quadratic optimization problem: Eq. 3[TeX:] \documentclass[12pt]{minimal}\begin{document}\begin{equation} \begin{array}{l} \mathop {\min }\limits_{{\bf w},b,\xi _i } \displaystyle\frac{1}{2}{\bf w}^T {\bf w} + C\sum\limits_i {\xi _i } \\[5pt] {\rm s}{\rm.t}{\rm.}\,\,{\rm y}_i ({{\bf w}^T {\bf x}_i + b}) \ge 1 - \xi _i \\[5pt] \quad \xi _i \ge 0, \\[5pt] \end{array} \end{equation}\end{document}2.3.4.Ensemble classifiersWhen developing an ensemble classifier for classification of ESS spectra, we intended to take advantage of the high dimensional nature of the data. For this purpose, a feature-level approach, as discussed earlier, is used. In this approach, the ensemble is created by designing each base classifier on a subset of the sample pattern's features. An important aspect of ensemble classifiers is diversity among the base classifiers (i.e., trying not to commit errors on the same samples).9, 11 With this in mind, we propose using the ensemble classifier architecture illustrated in Fig. 4. The system consists of a number of parallel classifiers, each trained on a region of the ESS spectrum. The design of each of the base classifiers involves a feature extraction (PCA) step followed by the training of a linear SVM classifier with those particular extracted features. Diversity is introduced by training the base classifiers on different subspaces generated by the extracted features. The final classification, or diagnosis, is obtained by combining the outputs of the base classifiers. Because we are working with SVM, whose output is a class label, combination rules that fuse label classifier outputs will be used. Two combination rules, majority-voting and naïve Bayes combiner, were considered. The majority-voting rule assigns a sample the class label that 50% + 1 or more of the base classifiers agree on. In the case of a tie when using even numbers of base classifiers, the member of the ensemble with the highest classification accuracy breaks the tie. The naïve Bayes combination rule is stated as follows:11 let x be a sample vector belonging to one of the possible ωk, k = 1, …, c classes. Also let L be the number of base classifiers in the ensemble. The ensemble classifies a sample x as belonging to class ωk if σk(x) is maximum, where Eq. 5[TeX:] \documentclass[12pt]{minimal}\begin{document}\begin{equation} \sigma _k ({\bf x})\propto \,P\left({\omega _k } \right)\prod\limits_{i = 1}^L P \left({y_i |\omega _k } \right), \end{equation}\end{document}Eq. 6[TeX:] \documentclass[12pt]{minimal}\begin{document}\begin{equation} \begin{array}{l} P\left({y_i = - 1|\omega _{neoplastic} } \right) = {\rm Sensitivity}, \\[5pt] P\left({y_i = 1|\omega _{neoplastic} } \right) = 1 - {\rm Sensitivity}, \\[5pt] P\left({y_i = 1|\omega _{non - neoplastic} } \right) = {\rm Sensitivity}, \\[5pt] P\left({y_i = - 1|\omega _{non - neoplastic} } \right) = 1 - {\rm Sensitivity}, \\[5pt] \end{array} \end{equation}\end{document}P(ωk) are the prior probabilities and can be estimated from the training data. Thus, this combination rule uses the performance of each base classifier, in the form of the sensitivity and specificity, to weight their individual decision when making the final classification. 2.3.5.Misclassification-rejectionSeveral methodologies that incorporate a misclassification-rejection option in SVM classifiers have been presented by others.23, 24, 25, 26, 27 Many of them accomplish this by rejecting samples that lie at a certain threshold distance from the decision boundary or by estimating probabilistic outputs from the SVM classifier and then applying Chow's rule.18, 19, 20 An alternative method of incorporating misclassification-rejection in SVMs is to obtain both the orientation of the hyperplanes and the width of the rejection region in the training phase.28, 29, 30 The decision region in this SVM with embedded MR (SVMMR), is defined as two parallel hyperplanes, where samples lying in-between them would be rejected. To accomplish this, the SVM training problem is reformulated by introducing a term in the minimization problem cost function in order to approximate the empirical error with rejections. Training is accomplished by solving the following problem:28, 29, 30 Eq. 7[TeX:] \documentclass[12pt]{minimal}\begin{document}\begin{equation} \begin{array}{l} \mathop {\min }\limits_{{\bf w},b,\xi _i,\varepsilon } \displaystyle\frac{1}{2}{\bf w}^T {\bf w} + C\sum\limits_i {h\left({\xi _i,\varepsilon } \right)} \\[5pt] {\rm s}{\rm.t}{\rm.}\,\,{\rm y}_i \left({{\bf w}^T {\bf x}_i + b} \right) \ge 1 - \xi _i \\[5pt] \quad \xi _i \ge 0 \\[5pt] \quad 0 \le \varepsilon \le 1. \\[5pt] \end{array} \end{equation}\end{document}This training problem is similar to the one for standard SVMs with the addition of ɛ, where 2ɛ/‖w‖ denotes the width between the separating hyperplanes defining the rejection region, and h(ξi,ɛ) is the modified cost function reflecting the trade-off between rejecting and misclassifying training samples. The decision function with the rejection option then becomes Eq. 8[TeX:] \documentclass[12pt]{minimal}\begin{document}\begin{equation} f({\bf x}) = \left\{ \begin{array}{l} + 1,\,\,\,\,\,\,\,\,\,\,\,\,{\rm if}\,{\bf w}^T {\bf x} - b \ge \varepsilon \\[5pt] - 1,\,\,\,\,\,\,\,\,\,\,{\rm if}\,{\bf w}^T {\bf x} - b \le - \varepsilon \\[5pt] 0,\,\,\,\,\,\,\,\,\,\,\,{\rm if} - \varepsilon < {\bf w}^T {\bf x} - b < \varepsilon \\[5pt] \end{array} \right.. \end{equation}\end{document}3.Results3.1.Data SetThe data set consists of 494 elastic-scattering spectroscopy (ESS) measurements from 297 polyps from 134 patients. Because our work will concentrate on binary classifiers, the samples were grouped into two clinically relevant classes, non-neoplastic samples and neoplastic samples. Of the 297 polyps measured 199 correspond to non-neoplastic tissue including hyperplastic polyps, histologically normal growths, and inflammatory polyps, while 98 correspond to neoplastic polyps and adenocarcinomas. A total of 325 spectra were collected from the non-neoplastic sites and 169 from the neoplastic sites. The data set was divided into separate training and testing sets for design and testing of the presented classifiers. Using the same training and testing sets will also permit the comparison and evaluation of the proposed classifiers in a standardized manner. The data was partitioned by randomly assigning 80 patients (60%) to the training set, from which 193 spectra from 111 non-neoplastic polyps and 92 spectra from 54 neoplastic polyps were acquired and the remaining 54 patients (40%) to the testing set, from which 132 spectra from 88 non-neoplastic polyps and 77 spectra from 44 neoplastic polyps were acquired. Table 1 summarizes the number of cases per pathology for the training and testing sets. Figure 5 illustrates the representative spectra for each class, as well as their standard deviation, for the training and testing set. Fig. 8Performance (sensitivity and specificity) as a function of rejection rate obtained using the error-rejection classification paradigm. 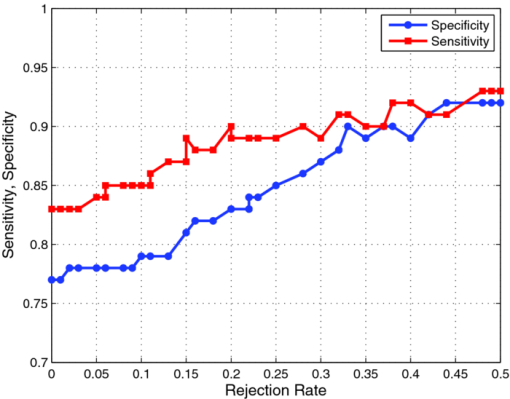 Table 1Data-set breakdown.
3.2.Spectral ClassificationTo establish a performance baseline for classifying neoplastic and non-neoplastic polyps using ESS, we used the standard classification paradigm of dimensionality reduction followed by classification, as this approach has been commonly used for spectral analysis. 1, 2, 5, 7, 16, 17, 52, 53, 54, 55, 56, 57 The training data was used to compute the transformation matrix V for PCA and to determine the number of PCs used to train the classifier, in this case an SVM with linear kernel. The number of PCs was determined by performing cross-validation on the training set while increasing the number of PCs used, and selecting the number after which no increase in performance was observed. SVM parameters were chosen from a finite set of values and by selecting those that showed the best performance on the training set. The full spectral range, 330–760 nm, was used and the first 15 principal components were used for classification. Results from the testing set reveal that this classification scheme can differentiate ESS spectra of neoplastic from spectra of non-neoplastic lesions with a sensitivity of 0.83 and specificity of 0.79 with an overall accuracy of 0.80. Figure 6 shows the averages and standard deviations of the reconstructed dataset, using Eq. 2, with the selected principal components. By using a single principal component, or a subset of components, to do this transformation, the reconstructed data will express the variance along those specific directions, revealing areas of the spectrum found to be informative by PCA. In Fig. 6b, it can be seen that the variance in the data is predominant at ∼330–600 nm when using the first PC. The variance is more contained around 330–380 nm, 410–440 nm, 480–625 nm, and 650–760 nm when using PC 2 to PC 15 [Fig. 6c]. In PC 3 to PC 15, we see that it is localized around 330–360 nm, 460–500 nm, and near 420 nm and 575 nm [Fig. 6d], with this trend remaining in subsequent PCs [Figs. 6e, 6f]. Fig. 6(a)Training set representative spectrum for each class, (b) reconstructed data using the first PC, (c) reconstructed data using PC2 to PC15, (d) reconstructed data using PC3 to PC15, (e) reconstructed data using PC4 to PC15, and (f) reconstructed data using PC5 to PC15. 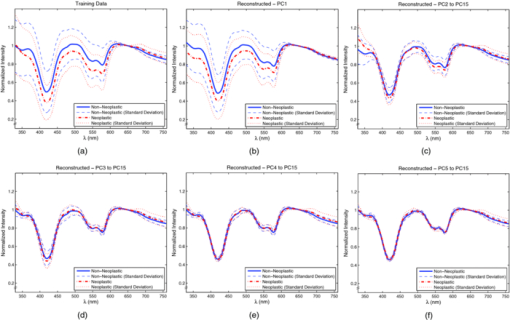 3.2.1.Ensemble classifiersTo construct the ensemble classifier proposed in Sec. 2.3.4 (Fig. 4) a series of classifiers were trained on specific regions of the ESS spectrum, using the standard classification paradigm, with PCA used for feature extraction and a linear SVM for classification. This set, or subsets, of classifiers will be used as the base classifiers in our scheme. A total of 12 regions were chosen empirically, taking into consideration the observations from principal component analysis discussed earlier. Regions were selected based on the spectral areas that exhibited the largest variance when the data were reconstructed with specific PCs (Fig. 6). These spectral areas are considered the most informative based on the PCA criterion. In addition, we made sure that spectral regions containing the hemoglobin Soret (∼400–440 nm) and Q (∼540–580 nm) bands were included, as well as the ranges of ∼330–370 nm and ∼380–500 nm since these have been found to be informative for distinguishing pathologies.3, 38 Table 2 summarizes the performance of the individual base classifiers. The best performance, accuracy of 0.80 was obtained using the full spectrum (i.e., 330–760 nm). The worst performance, accuracy of 0.67, was obtained in the region of 600–700 nm. Regions in the shorter wavelengths (<460 nm) exhibited better overall performance, with the performance of 330–400 nm, Se = 0.84, Sp = 0.73, on par with the performance when using the entire spectrum, Se = 0.83, Sp = 0.79. Table 3 shows, for each pair of regions, the fraction of the total misclassifications where both regions misclassified the same sample. It is desirable for this metric to have a small value as ideally the base classifiers in the ensemble commit errors on different samples, allowing for correction of these errors as the ensemble is taken as a whole. Although the regions were not completely independent and uncorrelated, because some overlap among them was allowed, it is observed that the resulting classifiers are committing errors on different samples. Even between regions with significant overlap (e.g., 330–760 nm and 330–600 nm, 360–460 nm, and 360–600 nm, 500–530 nm, and 500–570 nm), the fraction of total misclassifications where both regions misclassified the same sample was, at worst, around 0.70 (Table 3). This outcome is meaningful in the context of ensemble classifiers because this means that diversity is being promoted by this approach, including cases when regions overlap. Table 2Performance of classifiers trained on each region.
Table 3Fraction of total misclassification where region pairs misclassified the same sample.
Tables 4, 5 show the performance obtained for different ensembles, with a varying number of base classifiers, for the majority voting and naïve Bayes combination rules, respectively. For each number of base classifiers, the combination of regions with the best performance on the training set, using L base classifiers [i.e., evaluating all ( [TeX:] ${12}\atop{L} $ ) possible combinations]. Using majority voting, several ensembles, (using 7, 8, 9, and 10 base classifiers), yielded a Se = 0.86, Sp = 0.80 and accuracy of 0.82 in classifying neoplastic and non-neoplastic lesions. Using naïve Bayes combiner, a Se = 0.84, Sp = 0.83 and accuracy of 0.83 was obtained with ensembles composed of 9, 11, and 12 base classifiers. The use of this scheme showed improve results over the initial results of Se = 0.83, Sp = 0.79, accuracy of 0.80. Table 4Majority voting.
Table 5Naïve Bayes combiner.
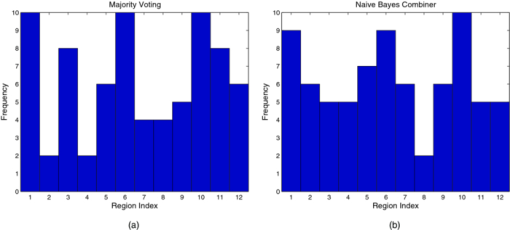
Figure 7 shows histograms of the chosen regions of the best ensembles for the different number of base classifiers. The most used regions were 330–760 nm, 500–530 nm, and 600 –700 nm, followed by 360–600 nm, 500–570 nm, and 330–400 nm. Although individually some of these regions were not the best in terms of performance, their combined performance as part of an ensemble results in improvement over any single classifier. Also of note is the fact that the fraction of total misclassifications where a pair of regions misclassified the same sample among these regions was at the highest 0.67, between 500–530 nm and 500–570 nm, and at the lowest 0.24, between 360–600 nm and 600–700 nm. This allows the ensembles to correct some of the misclassifications made by the individual base classifiers. 3.3.Spectral Classification with Misclassification-RejectionWe used the misclassification-rejection paradigm to evaluate the gain in performance as a function of the percentage of samples withheld from classification using this scheme. The SVM formulation with embedded misclassification-rejection, SVMMR, discussed earlier was used to design the classifier. Training was done using 15 principal components obtained from the full spectral range of 330–760 nm as was done in Sec. 3.2 above. Figure 8 shows the obtained sensitivity and specificity as a function of the rejection rate using this classification scheme. A trend of increasing performance can be observed as the rejection rate increases, with an initial sensitivity and specificity of 0.83 and 0.79 increasing to 0.89 and 0.85 with a 0.25 rejection rate, 0.91 and 0.90 with a 0.33 rejection rate and 0.93 and 0.92 with a 0.48 rejection rate. From Fig. 8, it can also be observed that this trend levels off at around a rejection rate of 0.33; after this point, increasing the rejection rate results in a small gain in performance, as opposed to the performance gain obtained below the rejection rate of 0.33. 3.3.1.Ensemble classifiers with misclassification-rejectionTo design ensemble classifiers using the misclassification-rejection paradigm, we used the architecture shown in Fig. 4, with the base classifiers designed using SVMMR. The same 12 spectral regions used in Sec. 3.2.1, as well as the number of features extracted with PCA, were used to train the base classifiers that comprised the ensembles. Because the different classifier designs lead to different rejection rates and, thus performance, we trained each of the base classifiers to have a rejection rate of around 0.33. At this rejection rate, the better performance-rejection trade-off was observed. Table 6 summarizes the performance of the individual classifiers using SVMMR. Using SVMMR resulted in an increase in overall accuracy of ∼10%, when compared to the results without misclassification-rejection (Table 6). In addition, sensitivities and/or specificities reach close to 0.90 at this rejection level for some spectral regions. We wish to point out that it is unlikely that better accuracy could be obtained by any method, because these values are on par with the “gold standard” of histopathology, in that the typical agreement rate among expert pathologists for this discrimination in colorectal biopsies is ∼90%. Table 6Performance of classifiers trained on each region using SVMMR.
We used a two-step process for combining the decision made by the base classifiers trained under the misclassification-rejection paradigm, noting that in this case the output of each of these could be either class label (neoplastic or non-neoplastic) or “not classified.” First, it was determined whether the ensemble would classify or reject the sample. The ensemble rejects a sample if the number of base classifiers withholding from classifying the sample were greater than or equal to a threshold T defined as Eq. 9[TeX:] \documentclass[12pt]{minimal}\begin{document}\begin{equation} T = {\rm ceil}\left({\frac{L}{2}} \right) + k, \end{equation}\end{document}Table 7Majority voting, k = 0.
Table 8Majority voting, k = 1.
Table 9Naïve Bayes combiner, k = 0.
Table 10Naïve Bayes combiner, k = 1.
4.DiscussionIn this paper, we presented work focused on the development of a diagnostic algorithm to provide improved classification of neoplastic lesions from non-neoplastic lesions in the colon using elastic-scattering spectroscopy. We started with the commonly used classification paradigm of an initial feature extraction step, in this case PCA, followed by classification, which in this work was done with linear SVM. A sensitivity of 0.83, a specificity of 0.79 and an accuracy of 0.80 were obtained with this type of approach. We then presented two different classification frameworks seeking to improve this performance. The first was the use of an ensemble classifier scheme, where a number of classifiers were trained on specific regions of the ESS spectrum. The parallel decisions made by the individual base classifiers were then combined to obtain the final diagnosis of the measured site. In our approach, the regions of the ESS spectrum where the base classifiers were trained were chosen empirically and a priori. The use of this scheme yielded a sensitivity of 0.84, a specificity of 0.83 and accuracy of 0.83, an improvement over the sensitivity, specificity, and accuracy of 0.83, 0.79, and 0.80 previously achieved. The second framework presented was the misclassification-rejection scheme. In this classification paradigm, the classifier is trained to withhold classification when samples are deemed to be at high risk of misclassification. Thus, a binary classifier has three possible outcomes in this paradigm: classify a sample as belonging to either class or not classifying the sample at all, labeling it as rejected or undecided. The use of this framework resulted in a sensitivity of 0.91 and specificity of 0.90, an improvement over the sensitivity and specificity of 0.83 and 0.79 initially obtained, and also better than the 0.84 and 0.83 obtained with ensemble classifiers alone. This improved classification performance was obtained at a cost of a rejection rate of 0.33. Finally, we presented a classifier that makes use of both these frameworks. The classifier had an ensemble architecture, where each of the base classifiers was trained using misclassification-rejection. With this classifier, sensitivities and specificities of 0.89/0.94, 0.90/0.89, 0.89/0.90, and 0.88/0.86, were obtained for rejection rates of 0.26, 0.24, 0.21, and 0.15, respectively. Although these sensitivities and specificities were comparable to the sensitivity of 0.91 and specificity of 0.90 previously obtained using misclassification-rejection, there was a significant decrease in the rejection rates from the value of 0.33. Thus, with the classifier combining the ensemble classifier and misclassification-rejection frameworks, the overall classification performance is improved (i.e., we are able to maintain the same performance level, in one case improving it, while decreasing the number of rejected samples needed to achieve this). An issue not addressed yet is the generalization ability of the different classifiers presented throughout this work. Although not the main objective of the presented work, assessing the generalizability of the discussed classifiers can provide insight into how, in their current form, these classifiers would perform prospectively. We employed k-fold cross-validation,13 with three folds, to assess generalizability. In this scheme, the data is partitioned into three sets, with two of them used for training and the remaining one for testing. The sets are cycled until all of them have been used for testing. Table 11 shows the results obtained with this cross-validation and compares them to the previous results obtained with the single testing set. The performances obtained with cross-validation exhibit a significant amount of variance [e.g., in the case of the single classifier the sensitivity and specificity had a standard deviation (SD) of 0.06 and 0.04 respectively, whereas for ensemble classifiers with misclassification-rejection SD can go as high as 0.12 and 0.04, respectively]. Results obtained from a single testing set generally fall within the one standard deviation range of the cross-validated results. Yet, as the classifier complexity increases (i.e., transitions from a single classifier to ensemble classifiers and/or misclassification rejection frameworks), it is observed that either the results from a single testing set fall out of this range or the variance in performance increases or both. This exemplifies a shortcoming with the proposed frameworks: as the classifier complexity increases, more training data are required to properly learn the data patterns.58, 59 In general, and especially considering the significant amount of variance observed in the cross-validated results, exhaustive validation with more data would be required in order to design a classifier suitable for prospective testing in clinical settings. Yet, the results from cross-validation show improvements in performances with the ensemble classifier and misclassification rejection frameworks introduced for spectral classification, in line with those shown above using a single testing set. Table 11Results from k-fold cross-validation compared to a single testing set. K-fold results shows as the average ± standard deviation.
4.1.Significance for Clinical Application to CRC ScreeningIn the context of colorectal cancer screening, the ability to accurately identify suspicious colonic lesions would allow non-neoplastic polyps to be left in situ and small adenomas to be resected and discarded without the need for histopathology because they have a very small risk of harboring cancer.60 As seen from the presented results, ESS, mediated through integrated forceps, holds such promise if measurements of colonic lesions are accurately classified by the diagnostic algorithm presented in this work. Yet, the use of the misclassification-rejection paradigm, while useful in improving classification performance, does introduce an additional design parameter to consider: the rejection rate. Previously, concerns in designing diagnostic algorithms were focused on obtaining acceptable sensitivities and specificities. In the proposed framework, an “acceptable” rejection rate becomes an additional design parameter and handling of “rejected” samples must be addressed. We have not yet incorporated instant diagnostic response in our system. If such were implemented, then, in the case of a diagnostic rejection, the physician could take additional readings on different parts of the polyp, possibly gaining a more decisive result, and reducing the effective nonclassified rate. Absent, the prospective study needed to address that option; the direct approach for handling unclassified samples would be to resect them for examination by a histopathologist. From the results shown earlier, this approach would result in 21–33% of the samples examined by ESS being resected for histological assessment, but would still obviate histopathology of 66% of the samples that would otherwise be reviewed by histopathologists under current practice. We have also shown that, by using a combined ensemble classifier with a misclassification-rejection framework, we can potentially obtain further reductions of the rejection rates with the same, if not a higher level of diagnostic performance, possibly reducing the number of samples needed to be sent for histopathological examination. In addition, any further improvements in feature extraction and classification at the base-classifier level, resulting from continuing study and/or understanding of this classification problem, could potentially serve to further improve classification performance, be it by an increase in accuracy and/or a reduction of rejection rates. The classifier architecture presented here (Fig. 4) could also potentially allow the incorporation of additional information that might be of significance in detecting neoplasia, such as biomarkers extracted from the ESS spectrum by way of theoretical/empirical models of light transport in tissue61 and/or diagnostically relevant polyp features, such as pit pattern and size.62, 63, 64 For the health-care system, the cost savings realized could be significant, while minimizing the risk of biopsy-related complications, because fewer benign lesions would be resected. In addition, because ESS measurements are obtained in milliseconds, there is the potential to reduce procedure time while improving care, as real-time, in vivo classification enables rapid interrogation of subtle, suspicious, and flat lesions as well. 5.ConclusionsIn this paper, we presented two spectral classification frameworks, ensemble classifiers, and misclassification-rejection, and applied them to the clinical problem of classifying non-neoplastic and neoplastic colorectal lesions based on ESS measurements. The result is an improvement in classification performance when these are applied individually, and an even better performance when used together in the development of a diagnostic algorithm. The capability to accurately classify colonic lesions, as a result of this improved diagnostic algorithm, will enable real-time and in vivo application, while seamlessly integrating with current screening procedures. This can lead to reductions in procedure time, healthcare cost, as well as a patient risk, because current screening involves the excision and histopathological assessment of all polyps found during a colonoscopy procedure. AcknowledgmentsThe authors acknowledge the financial support of the Bernard M. Gordon Center for Subsurface Sensing and Imaging Systems, under the Engineering Research Centers Program of the National Science Foundation (Award No. EEC-9986821) and by the National Cancer Institute (NIH) under Grant No. U54-CA104677. ReferencesA. Dhar, K. S. Johnson, M. R. Novelli, S. G. Bown, I. J. Bigio, L. B. Lovat, and
S. L. Bloom,
“Elastic scattering spectroscopy for the diagnosis of colonic lesions: initial results of a novel optical biopsy technique,”
Gastrointest. Endosc., 63
(2), 257
–261
(2006). https://doi.org/10.1016/j.gie.2005.07.026 Google Scholar
Z. Ge, K. T. Schomacker, and
N. S. Nishioka,
“Identification of colonic dysplasia and neoplasia by diffuse reflectance spectroscopy and pattern recognition techniques,”
Appl. Spectrosc., 52
(6), 833
–839
(1998). https://doi.org/10.1366/0003702981944571 Google Scholar
J. R. Mourant, I. J. Bigio, J. Boyer, T. M. Johnson, J. A. Lacey, A. G. Bohorfoush, and
M. Mellow,
“Elastic scattering spectroscopy as a diagnostic tool for differentiating pathologies in the gastrointestinal tract: preliminary testing,”
J. Biomed. Opt., 1 192
–199
(1996). https://doi.org/10.1117/12.231372 Google Scholar
G. Zonios, L. T. Perelman, V. Backman, R. Manoharan, M. Fitzmaurice, J. Van Dam, and
M. S. Feld,
“Diffuse reflectance spectroscopy of human adenomatous colon polyps in vivo,”
Appl. Opt., 38 6628
–6637
(1999). https://doi.org/10.1364/AO.38.006628 Google Scholar
I. J. Bigio, S. G. Bown, G. Briggs, C. Kelley, S. Lakhani, D. Pickard, P. M. Ripley, I. G. Rose, and
C. Saunders,
“Diagnosis of breast cancer using elastic-scattering spectroscopy: preliminary clinical results,”
J. Biomed. Opt., 5
(2), 221
–228
(2000). https://doi.org/10.1117/1.429990 Google Scholar
K. S. Johnson, D. W. Chicken, D. C. Pickard, A. C. Lee, G. Briggs, M. Falzon, I. J. Bigio, M. R. Keshtgar, and
S. G. Bown,
“Elastic scattering spectroscopy for intraoperative determination of sentinel lymph node status in the breast,”
J. Biomed. Opt., 9
(6), 1122
–1128
(2004). https://doi.org/10.1117/1.1802191 Google Scholar
L. B. Lovat, K. Johnson, G. D. Mackenzie, B. R. Clark, M. R. Novelli, S. Davies, M. O’Donovan, C. Selvasekar, S. M. Thorpe, D. Pickard, R. Fitzgerald, T. Fearn, I. Bigio, and
S. G. Bown,
“Elastic scattering spectroscopy accurately detects high grade dysplasia and cancer in Barrett's oesophagus,”
Gut, 55
(8), 1078
–1083
(2006). https://doi.org/10.1136/gut.2005.081497 Google Scholar
D. K. Rex, C. Kahi, M. O’Brien, T. R. Levin, H. Pohl, A. Rastogi, L. Burgart, T. Imperiale, U. Ladabaum, J. Cohen, and
D. A. Lieberman,
“The American Society for Gastrointestinal Endoscopy PIVI (Preservation and Incorporation of Valuable Endoscopic Innovations) on real-time endoscopic assessment of the histology of diminutive colorectal polyps,”
Gastrointest. Endosc., 73
(3), 419
–422
(2011). https://doi.org/10.1016/j.gie.2011.01.023 Google Scholar
M. Aksela and
J. Laaksonen,
“Using diversity of errors for selecting members of a committee classifier,”
Pattern Recogn., 39
(4), 608
–623
(2006). https://doi.org/10.1016/j.patcog.2005.08.017 Google Scholar
R. P. Duin and
M. Skurichina,
“Combining feature subsets in feature selection,”
Multiple Classifier Systems, 165
–175 Springer, Berlin
(2005). Google Scholar
L. Kuncheva, Combining Pattern Classifiers: Methods and Algorithms, Wiley, Hoboken, NJ
(2004). Google Scholar
K. Tumer, N. Ramanujam, J. Ghosh, and
R. Richards-Kortum,
“Ensembles of radial basis function networks for spectroscopic detection of cervical precancer,”
IEEE Trans. Biomed. Eng., 45
(8), 953
–961
(1998). https://doi.org/10.1109/10.704864 Google Scholar
R. O. Duda, P. E. Hart, and
D. G. Stork, Pattern Classification, Wiley, Hoboken, NJ
(2001). Google Scholar
Y. Freund and
R. E. Schapire,
“A decision-theoretic generalization of online learning and an application to boosting,”
J. Comput. Syst. Sci., 55
(1), 119
–139
(1997). https://doi.org/10.1006/jcss.1997.1504 Google Scholar
R. E. Schapire,
“Theoretical views of boosting,”
(1999). Google Scholar
S. K. Majumder, N. Ghosh, and
P. K. Gupta,
“Relevance vector machine for optical diagnosis of cancer,”
Lasers Surg. Med., 36
(4), 323
–333
(2005). https://doi.org/10.1002/lsm.20160 Google Scholar
N. Ramanujam, M. F. Mitchell, A. Mahadevan, S. Thomsen, A. Malpica, T. Wright, N. Atkinson, and
R. Richards-Kortum,
“Development of a multivariate statistical algorithm to analyze human cervical tissue fluorescence spectra acquired in vivo,”
Lasers Surg. Med., 19
(1), 46
–62
(1996). https://doi.org/10.1002/(SICI)1096-9101(1996)19:1<46::AID-LSM7>3.0.CO;2-Q Google Scholar
C. Chow,
“On optimum recognition error and reject tradeoff,”
IEEE Trans. Inf. Theory, 16
(1), 41
–46
(1970). https://doi.org/10.1109/TIT.1970.1054406 Google Scholar
G. Fumera, F. Roli, and
G. Giacinto,
“Multiple reject thresholds for improving classification reliability,”
Advances in Pattern Recognition, 863
–871 Springer, Berlin
(2000). Google Scholar
L. K. Hansen, C. Liisberg, and
P. Salamon,
“The error-reject tradeoff,”
Open Syst. Inf. Dyn., 4
(2), 159
–184
(1997). https://doi.org/10.1023/A:1009643503022 Google Scholar
L. P. Cordella, C. De Stefano, F. Tortorella, and
M. Vento,
“A method for improving classification reliability of multilayer perceptrons,”
IEEE Trans. Neural Networks, 6
(5), 1140
–1147
(1995). https://doi.org/10.1109/72.410358 Google Scholar
C. De Stefano, C. Sansone, and
M. Vento,
“To reject or not to reject: that is the question-an answer in caseof neural classifiers,”
IEEE Trans. Syst., Man. Cybern., Part C, 30
(1), 84
–94
(2000). https://doi.org/10.1109/5326.827457 Google Scholar
A. David and
B. Lerner,
“Support vector machine-based image classification for genetic syndrome diagnosis,”
Pattern Recogn. Lett., 26
(8), 1029
–1038
(2005). https://doi.org/10.1016/j.patrec.2004.09.048 Google Scholar
S. Mukherjee, P. Tamayo, D. Slonim, A. Verri, T. Golub, J. Mesirov, and
T. Poggio,
“Support vector machine classification of microarray data,”
(1999) Google Scholar
J. Platt,
“Probabilistic outputs for support vector machines and comparisons to regularized likelihood methods,”
Advances in Large Margin Classifiers, 61
–74 MIT Press, Cambridge, MA
(1999). Google Scholar
F. Tortorella and
I. Cassino,
“A ROC-Based Reject Rule for Support Vector Machines,”
Machine Learning and Data Mining in Pattern Recognition, 106
–120 Springer, Berlin
(2003). Google Scholar
C. Yuan and
D. Casasent,
“A Novel Support Vector Classifier with Better Rejection Performance,”
419
–424
(2003). Google Scholar
G. Fumera and
F. Roli,
“Support vector machines with embedded reject option,”
68
–82
(2002). Google Scholar
G. Fumera and
F. Roli,
“Cost-sensitive learning in support vector machines,”
(2002). Google Scholar
E. Rodriguez-Diaz and
D. A. Castanon,
“Support vector machine classifiers for sequential decision problems,”
2558
–2563
(2009). Google Scholar
Y. Zhu, T. Fearn, D. Samuel, A. Dhar, O. Hameed, S. G. Bown, and
L. B. Lovat,
“Error removal by orthogonal subtraction (EROS): a customised pre-treatment for spectroscopic data,”
J. Chemomet., 22
(2), 130
–134
(2008). https://doi.org/10.1002/cem.1117 Google Scholar
Y. Zhu, T. Fearn, G. Mackenzie, B. Clark, J. M. Dunn, I. J. Bigio, S. G. Bown, and
L. B. Lovat,
“Elastic scattering spectroscopy for detection of cancer risk in Barrett's esophagus: experimental and clinical validation of error removal by orthogonal subtraction for increasing accuracy,”
J. Biomed. Opt., 14
(4), 044022
(2009). https://doi.org/10.1117/1.3194291 Google Scholar
A. Jemal, R. Siegel, E. Ward, Y. Hao, J. Xu, T. Murray, and
M. J. Thun,
“Cancer statistics, 2008,”
CA: Cancer J. Clinicians, 58
(2), 71
–96
(2008). https://doi.org/10.3322/CA.2007.0010 Google Scholar
R. A. Smith, V. Cokkinides, and
O. W. Brawley,
“Cancer screening in the United States, 2009: a review of current American Cancer Society guidelines and issues in cancer screening,”
CA: Cancer J. Clinicians, 59
(1), 27
–41
(2009). https://doi.org/10.3322/caac.20008 Google Scholar
D. K. Rex, J. L. Petrini, T. H. Baron, A. Chak, J. Cohen, S. E. Deal, B. Hoffman, B. C. Jacobson, K. Mergener, B. T. Petersen, M. A. Safdi, D. O. Faigel, and
I. M. Pike,
“Quality indicators for colonoscopy,”
Gastrointest. Endosc., 63
(4 Suppl), S16
–28
(2006). https://doi.org/10.1016/j.gie.2006.02.021 Google Scholar
G. Postic, D. Lewin, C. Bickerstaff, and
M. B. Wallace,
“Colonoscopic miss rates determined by direct comparison of colonoscopy with colon resection specimens,”
Am. J. Gastroenterol., 97
(12), 3182
–3185
(2002). https://doi.org/10.1111/j.1572-0241.2002.07128.x Google Scholar
D. K. Rex, C. S. Cutler, G. T. Lemmel, E. Y. Rahmani, D. W. Clark, D. J. Helper, G. A. Lehman, and
D. G. Mark,
“Colonoscopic miss rates of adenomas determined by back-to-back colonoscopies,”
Gastroenterology, 112
(1), 24
–28
(1997). https://doi.org/10.1016/S0016-5085(97)70214-2 Google Scholar
J. R. Mourant, I. J. Bigio, J. Boyer, R. L. Conn, T. Johnson, and
T. Shimada,
“Spectroscopic diagnosis of bladder cancer with elastic light scattering,”
Lasers Surg. Med., 17
(4), 350
–357
(1995). https://doi.org/10.1002/lsm.1900170403 Google Scholar
I. J. Bigio, S. G. Bown, C. Kelley, L. Lovat, D. Pickard, and
P. M. Ripley,
“Developments in endoscopic technology for oesophageal cancer,”
J. R. Coll. Surg. Edinburgh, 45
(4), 267
–268
(2000). Google Scholar
L. Lovat and
S. Bown,
“Elastic scattering spectroscopy for detection of dysplasia in Barrett's esophagus,”
Gastrointest. Endosc. Clin. N Am., 14
(3), 507
–517
(2004). https://doi.org/10.1016/j.giec.2004.03.006 Google Scholar
L. B. Lovat, K. Johnson, M. R. Novelli, M. O’Donovan, S. Davies, C. R. Selvasekar, S. Thorpe, I. J. Bigio, and
S. G. Bown,
“Optical biopsy using elastic scattering spectroscopy can detect high grade dysplasia and cancer in Barrett's esophagus,”
Gastroenterology, 126
(4), A22
–A39
(2004). https://doi.org/10.1016/S0016-5107(04)01125-3 Google Scholar
J. M. Dunn, Y. Jiao, M. Austwick, C. A. Mosse, D. Oukrif, S. Thorpe, M. Novelli, M. R. Banks, S. G. Bown, and
L. Lovat,
“Elastic scattering spectroscopy is highly accurate for the detection of high grade dysplasia and DNA ploidy arising in Barrett's esophagus using a red enhanced spectrometer,”
Gastroenterology, 138
(5), S
–664
(2010). https://doi.org/10.1016/S0016-5085(10)63053-3 Google Scholar
J. P. Baak, F. J. ten Kate, G. J. Offerhaus, J. J. van Lanschot, and
G. A. Meijer,
“Routine morphometrical analysis can improve reproducibility of dysplasia grade in Barrett's oesophagus surveillance biopsies,”
J. Clin. Pathol., 55
(12), 910
–916
(2002). https://doi.org/10.1136/jcp.55.12.910 Google Scholar
H. Fang, M. Ollero, E. Vitkin, L. M. Kimerer, P. B. Cipolloni, M. M. Zaman, S. D. Freedman, I. J. Bigio, I. Itzkan, E. B. Hanlon, and
L. T. Perelman,
“Noninvasive sizing of subcellular organelles with light scattering spectroscopy,”
IEEE J. Sel. Top. Quantum Electron., 9
(2), 267
–276
(2003). https://doi.org/10.1109/JSTQE.2003.812515 Google Scholar
I. Georgakoudi, B. C. Jacobson, J. Van Dam, V. Backman, M. B. Wallace, M. G. Müller, Q. Zhang, K. Badizadegan, D. Sun, G. A. Thomas, L. T. Perelman, and
M. S. Feld,
“Fluorescence, reflectance, and light-scattering spectroscopy for evaluating dysplasia in patients with Barrett's esophagus,”
Gastroenterology, 120
(7), 1620
–1629
(2001). https://doi.org/10.1053/gast.2001.24842 Google Scholar
M. B. Wallace, L. T. Perelman, V. Backman, J. M. Crawford, M. Fizmaurice, M. Seiler, K. Badizadegan, I. Itzkan, R. R. Dasari, J. Van Dam, and
M. S. Feld,
“Endoscopic detection of dysplasia in patients with Barrett's esophagus using light-scattering spectroscopy,”
Gastroenterology, 119
(3), 677
–682
(2000). https://doi.org/10.1053/gast.2000.16511 Google Scholar
E. Rodriguez-Diaz, I. J. Bigio, and
S. K. Singh,
“Integrated Optical Tools For Minimally Invasive Diagnosis and Treatment at Gastrointestinal Endoscopy,”
Robot. Comput. Integr. Manuf., 27
(2), 249
–256
(2011). https://doi.org/10.1016/j.rcim.2010.06.006 Google Scholar
G. H. Golub, and
C. F. V. Loan, Matrix Computations, 694 3rd ed.Johns Hopkins University Press, Baltimore
(1996). Google Scholar
C. J. C. Burges,
“A tutorial on support vector machines for pattern recognition,”
Data Mining Knowl. Discov., 2
(2), 121
–167
(1998). https://doi.org/10.1023/A:1009715923555 Google Scholar
C. Cortes and
V. Vapnik,
“Support-vector networks,”
Mach. Learning, 20
(3), 273
–297
(1995). https://doi.org/10.1007/BF00994018 Google Scholar
V. N. Vapnik, Statistical Learning Theory, Wiley, Hoboken, NJ
(1998). Google Scholar
S. K. Chang, Y. N. Mirabal, E. N. Atkinson, D. Cox, A. Malpica, M. Follen, and
R. Richards-Kortum,
“Combined reflectance and fluorescence spectroscopy for in vivo detection of cervical pre-cancer,”
J. Biomed. Opt., 10
(2), 024031
(2005). https://doi.org/10.1117/1.1899686 Google Scholar
D. C. de Veld, M. Skurichina, M. J. Witjes, R. P. Duin, H. J. Sterenborg, and
J. L. Roodenburg,
“Autofluorescence and diffuse reflectance spectroscopy for oral oncology,”
Lasers Surg. Med., 36
(5), 356
–364
(2005). https://doi.org/10.1002/lsm.20122 Google Scholar
W. Lin, X. Yuan, P. Yuen, W. I. Wei, J. Sham, P. Shi, and
J. Qu,
“Classification of in vivo autofluorescence spectra using support vector machines,”
J. Biomed. Opt., 9
(1), 180
–186
(2004). https://doi.org/10.1117/1.1628244 Google Scholar
S. K. Majumder, N. Ghosh, and
P. K. Gupta,
“Support vector machine for optical diagnosis of cancer,”
J. Biomed. Opt., 10
(2), 024034
(2005). https://doi.org/10.1117/1.1897396 Google Scholar
G. M. Palmer, C. Zhu, T. M. Breslin, F. Xu, K. W. Gilchrist, and
N. Ramanujam,
“Comparison of multiexcitation fluorescence and diffuse reflectance spectroscopy for the diagnosis of breast cancer (March 2003),”
IEEE Trans. Biomed. Eng., 50
(11), 1233
–1242
(2003). https://doi.org/10.1109/TBME.2003.818488 Google Scholar
N. Ramanujam, M. F. Mitchell, A. Mahadevan-Jansen, S. L. Thomsen, G. Staerkel, A. Malpica, T. Wright, N. Atkinson, and
R. Richards-Kortum,
“Cervical precancer detection using a multivariate statistical algorithm based on laser-induced fluorescence spectra at multiple excitation wavelengths,”
Photochem. Photobiol., 64
(4), 720
–735
(1996). https://doi.org/10.1111/j.1751-1097.1996.tb03130.x Google Scholar
K. Fukunaga, Introduction to Statistical Pattern Recognition, Academic Press, New York
(1990). Google Scholar
C. Lee and
D. A. Landgrebe,
“Analyzing high-dimensional multispectral data,”
IEEE Trans. Geosci. Remote Sens., 31
(4), 792
–800
(1993). https://doi.org/10.1109/36.239901 Google Scholar
A. Ignjatovic, J. E. East, N. Suzuki, M. Vance, T. Guenther, and
B. P. Saunders,
“Optical diagnosis of small colorectal polyps at routine colonoscopy (Detect InSpect ChAracterise Resect and Discard; DISCARD trial): a prospective cohort study,”
Lancet Oncol., 10
(12), 1171
–1178
(2009). https://doi.org/10.1016/S1470-2045(09)70329-8 Google Scholar
R. Reif, O. A’Amar, and
I. J. Bigio,
“Analytical model of light reflectance for extraction of the optical properties in small volumes of turbid media,”
Appl. Opt., 46
(29), 7317
–7328
(2007). https://doi.org/10.1364/AO.46.007317 Google Scholar
A. Rastogi, J. Keighley, V. Singh, P. Callahan, A. Bansal, S. Wani, and
P. Sharma,
“High accuracy of narrow band imaging without magnification for the real-time characterization of polyp histology and its comparison with high-definition white light colonoscopy: a prospective study,”
Am. J. Gastroenterol., 104
(10), 2422
–2430
(2009). https://doi.org/10.1038/ajg.2009.403 Google Scholar
D. K. Rex,
“Narrow-band imaging without optical magnification for histologic analysis of colorectal polyps,”
Gastroenterology, 136
(4), 1174
–1181
(2009). https://doi.org/10.1053/j.gastro.2008.12.009 Google Scholar
S. Y. Tung, C. S. Wu, and
M. Y. Su,
“Magnifying colonoscopy in differentiating neoplastic from nonneoplastic colorectal lesions,”
Am. J. Gastroenterol., 96
(9), 2628
–2632
(2001). https://doi.org/10.1111/j.1572-0241.2001.04120.x Google Scholar
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||