|
|
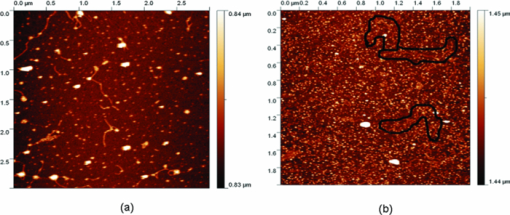
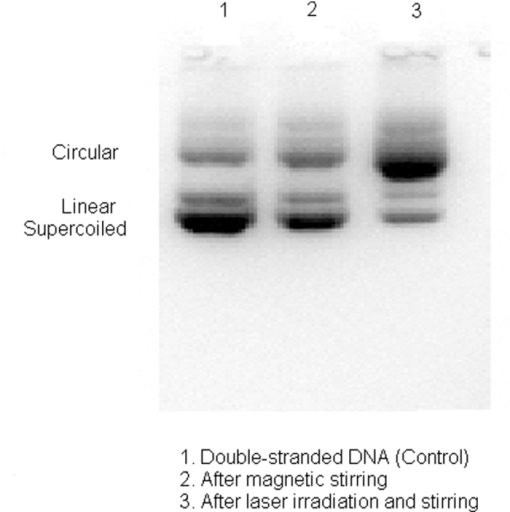
1.IntroductionRecently, a variety of viral systems, including M13 bacteriophage, tobacco mosaic virus, human papillomavirus, and human immunodeficiency virus have been shown to be inactivated by the irradiation of a visible femtosecond laser or a near-infrared subpicosecond fiber laser. 1, 2, 3, 4, 5, 6, 7, 8 Advantages of such a new laser technology over the presently prevailed disinfection methods are: 1. it is a noninvasive disinfection technology, because no foreign materials are added in the disinfection process; 2. it is an environmentally friendly disinfection method since no chemicals are used in the pathogen inactivation process; 3. it is a general method for selective disinfection of pathogens with potentially minimal side effects. These experimental results suggest that the inactivation of viruses by an ultrashort pulsed laser might involve disruption of their protein coat through laser-induced excitation of large-amplitude acoustic vibrations. Encephalomyocarditis virus (EMCV), which is a nonenveloped virus with a single-stranded RNA, is a virus commonly found in the blood supply. Salmonella typhimurium, which is a gram-negative bacterium, usually is found in the contaminated food and can cause severe human disease, particularly for those who have compromised immune systems. In this work, we report experimental results on the inactivation of EMCV, M13 bacteriophage, and Salmonella typhimurium by using a visible femtosecond laser derived from the second harmonic generation of a continuous-wave (CW) mode-locked Ti-sapphire laser system, in an attempt to gain some insight into the possible inactivation mechanisms. We demonstrate that inactivation of viruses by a visible femtosecond laser involves the breaking of hydrogen/hydrophobic bonds or the separation of the weak protein links in the protein shell of a viral particle. On the other hand, inactivation of bacteria is related to the disruption of their metabolism by the DNA relaxation process due to irradiation of a visible femtosecond laser. 2.Samples and MethodologyThe excitation source employed in this work was a diode-pumped CW mode-locked Ti-sapphire laser. The laser produced a continuous train of 60 fs pulses at a repetition rate of 80 MHz.1, 2, 3, 4, 5 As shown in Fig. 1, the output of the second harmonic generation system of the Ti-sapphire laser was used to irradiate the sample. The excitation laser was chosen to operate at a wavelength of λ = 425 nm and with an average power of about 50 mW. It has a pulse width of full-width at half maximum (FWHM) ≅100 fs. An achromatic lens was used to focus the laser beam into about 100 μm in diameter in the sample area. In order to facilitate the interaction of laser with the pathogens which were placed inside a Pyrex cuvette and diluted in 0.1 ml buffer solution, a magnetic stirrer was set up so that pathogens would enter the laser-focused volume as described above and interact with the photons. The typical exposure time of the sample to laser irradiation was about 1 h, unless otherwise indicated. All the experimental results reported here are obtained at T = 22°C and with the single laser beam excitation. Fig. 1Experimental set up for the inactivation of viral particles and bacteria with a visible femtosecond laser. M.O.: focusing lens; M: mirror; S: vial containing viruses/bacteria in buffer solutions. 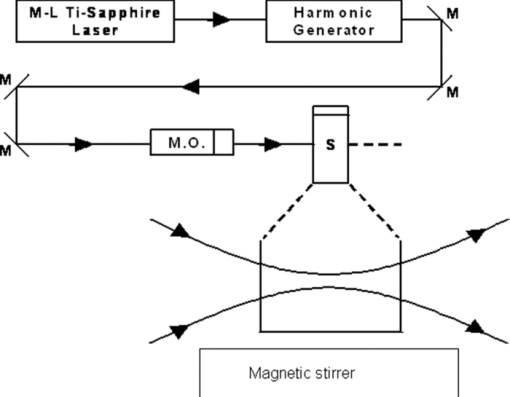 EMCV samples (from Washington University, St. Louis, Missouri) were diluted in the Dulbecco's Modified Eagle Medium (DMEM) buffer solution. M13 bacteriophage samples (from Stratagene, La Jolla, California) were diluted in the phosphate buffered saline (PBS) buffer solution. Salmonella typhimurium samples (from ASU, Tempe, Arizona) were diluted in the PBS buffer solution. The EMCV, M13 bacteriophage, and Salmonella typhimurium samples, which were prepared in 0.1 ml buffer solution, were loaded into small Pyrex vials (of about 1 cm in diameter and 3 cm in height) for laser irradiation. A typical number of particles per vial for EMCV, M13 bacteriophage, and Salmonella typhimurium was about 2×103, 1×107, and 1×106, respectively. Atomic force microscope imaging of M13 bacteriophages, optical properties of bovine serum albumin (BSA), gel electrophoresis experiments of single-stranded DNA of M13 bacteriophage, and double-stranded DNA from plasmid were carried out to clarify the possible mechanisms underlying the inactivation of EMCV, M13 bactriophage, and Salmonella typhimurium. The infectivity of EMCV was determined by plaque counts or plaque forming units (pfu) on plates made up of cells and cellular nutrients; the infectivity of M13 bacteriophage was measured by plague forming units on plates made up of bacteria and bacterial nutrients; whereas the activity of Salmonella typhimurium was measured by colony forming units (cfu) on soft agar plates containing bacterial culture. 2.1.Procedure for Atomic Force Microscopy ImagingBiomolecules such as protein, DNA, and RNA can be easily captured on the aminopropyltriethoxysilane (APTES) modified surface using glutaradehyde. The laser-irradiated viral particles were immobilized on the APTES mica and imaged under atomic force microscopy (AFM).9, 10, 11, 12 2.2.APTES Mica PreparationFresh cleaved mica was placed in a desiccator with 30 μl APTES (99%, Sigma-Aldrich, St. Louis, Missouri) and 10 μl N,N-diisopropylethylamine (99%, distilled, Sigma-Aldrich) in the bottom. The desiccator was then purged with argon for 3 min, and the mica was allowed to remain in the APTES vapor for 1 h to achieve good modification. 2.3.Sample ImmobilizationOne hundred microliters of 2 μm glutaradehyde (grade I, Sigma-Aldrich) was deposited onto the APTES mica surface for 10 min, and the surface was then washed gently with distilled water. After that, 200 μl of virus samples with concentration ranging from 1×106 to 1×108 particles per millimeter were pipetted onto the glutaradehyde-treated mica surface and allowed to incubate for 40 to 80 min. The mica surface, now containing the immobilized sample, was then gently rinsed with distilled water and dried with nitrogen. 2.4.ImagingImaging were carried out with a PicoPlus 2500+ AFM [molecular imaging; now 5500 AFM (N9410S) from Agilent] equipped with a Si3N4 cantilever (AppNano SPM) with a spring constant range from 25 to 75 N/m and the resonance frequency around 300 kHz. 2.5.Procedure for Protein Structure EvaluationBSA (≥99% purity) was purchased from Sigma-Aldrich (St. Louis, Missouri). BSA solutions (34 and 340 μm) were prepared in 10 mm phosphate buffer (pH = 6.0). Concentrations were determined by UV absorbance at 280 nm (ɛ = 43,824 m−1 cm−1) using a Cary50 UV-Vis spectrophotometer (Varian). After laser irradiation, aliquots from the two samples were diluted to a final concentration between 1.4 and 2 μm to avoid signal saturation in subsequent measurements. Two identical samples that were not laser irradiated, but otherwise underwent identical treatment, were also diluted to a final concentration between 1.4 and 2 μm. The circular dichroism and fluorescence of all four samples were measured. 2.6.Fluorescence Spectroscopy of BSAFluorescence spectra were measured in a 1 cm quartz cuvette (Hellma QC) using a QuantaMaster 40 spectrofluorometer (Photon Technology International) with a 0.5 mm bandpass. Excitation and emission spectra were measured from 250 to 325 nm (λem = 336 nm) and 283 to 450 nm (λex = 281 nm), respectively, and buffer subtracted. 2.7.Circular Dichroism Spectroscopy of BSACircular dichroism (CD) spectra13, 14 of the samples were measured in a 1 mm quartz cuvette (Starna Cells) using a Jasco J-710 spectropolarimeter (Jasco Company) with a 1 nm bandwidth. For each sample, four spectra taken between 250 and 190 nm with a 0.2 nm pitch at a 50 nm/min scan speed were averaged and then buffer subtracted. Spectra were rescaled by the samples’ respective concentrations, determined by the UV absorbance values, before data were analyzed. 2.8.Procedure for Gel Electrophoresis ExperimentsSingle-stranded DNA from M13 bacteriophages was purchased from New England Biolaboratory (Ipswich, Massachusetts). The double-stranded DNA was from an ASU-homemade plasmid which contains mouse mammary tumor virus (MMTV). All the DNA samples with 10× nondenaturing dye were loaded to the gel wells of a 0.8× agrose gel that was prepared in 1× tris-borate-ethylenediamine tetra-acetic acid buffer. The gel was run at room temperature at constant voltage at 80 V for 1.5 h.15 3.Experimental Results3.1.Inactivation of EMCV, M13 Bacteriophage, and Salmonella Typhimurium by a Visible Femtosecond LaserFigures 2, 3 show the pfu on a logarithmic scale as a function of fluence on a logrithmic scale for EMCV and M13 bacteriophage samples, respectively. Log-kill factors of 3 and 5 have been found for EMCV and M13 bacteriophage, respectively, with 1 h of laser exposure time. Fig. 2Log-kill factor as a function of the log laser fluence for the femtosecond laser-irradiated, CW laser-irradiated EMCV, as indicated. 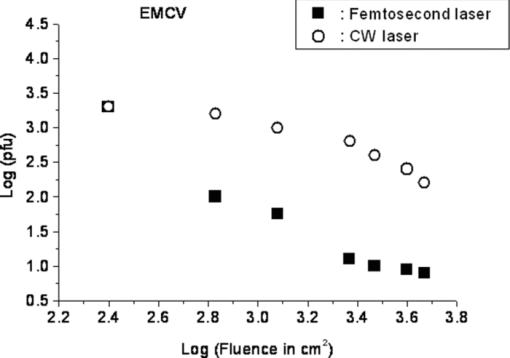 Fig. 3Log-kill factor as a function of the log laser fluence for the femtosecond laser-irradiated, CW laser-irradiated M13 bacteriophages, as indicated. 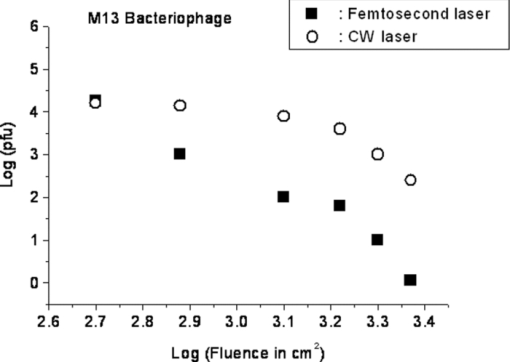 Figure 4 shows the cfu on a logarithmic scale as a function of fluence on a logarithmic scale for Salmonella typhimurium. A log-kill factor of about 6 was observed with 1 h of laser exposure time, suggesting that visible femtosecond laser irradiation can very effectively inactivate the bacteria. Fig. 4Log-kill factor as a function of the log laser fluence for the femtosecond laser-irradiated, CW laser-irradiated Salmonella typhimurium, as indicated. 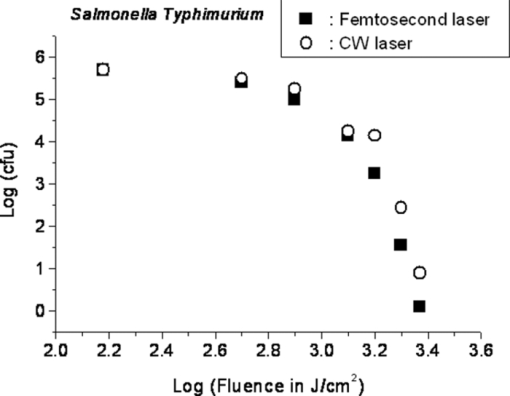 We have found that log (pfu)/log (cfu) versus log (fluence) in Figs. 2, 3, 4 are nonlinear. 3.2.Atomic Force Microscope Images of M13 BacteriophagesFigure 5 shows atomic force microscope pictures of M13 bacterioaphage without laser irradiation [Fig. 5a] and with laser irradiation [Fig. 5b] by a visible femtosecond laser. The relatively smooth worm-like features having a diameter of about 6 nm and about 850 nm in length in Fig. 5a revealed the presence of M13 bacteriophages in the control. Figure 5b showed, in contrast to Fig. 5a, appearance of a lot of small structures which were about 6 nm in diameter after laser irradiation. As discussed later, these small structures were consistent in size with the size of individual α − helix protein unit of which the protein shell of the M13 bacteriophage is composed. As a result, these small structures are most likely individual α − helix protein units of the M13 bacteriophage. In addition, some zigzagged worm-like features (encircled by artificially drawn black curves) were observed. The facts that its length was about 850 nm and it was in zigzagged structure indicated that these zigzagged structures were bare DNAs from M13 bacteriophages. The observation of the bare DNAs in the laser-irradiated M13 bacteriophage sample indicated that irradiation of the visible femtosecond laser severely alters the structural integrity of the protein shell of the M13 bacteriophages. 3.3.Effects of Irradiation of a Visible Femtosecond Laser on Proteins and DNAsTo elucidate the possible inactivation mechanisms for viruses and bacteria by a visible femtosecond laser, we have tested the effects of irradiation of a visible femtosecond laser on the structure of BSA proteins, double-stranded DNA from homemade plasmid, and single-stranded DNA from M13 bacteriophages. The luminescence, excitation spectra, and CD from amino acids of proteins are very sensitive to the structural changes of proteins. Therefore, these optical characterization methods were employed to detect the primary and secondary structural changes of the proteins before and after the laser irradiation. Figures 6a, 6b, 6c show our preliminary results for BSA proteins in buffer solution with and without irradiation with a visible femtosecond laser. In Fig. 6a, the excitation spectrum corresponded to the broad structure centered around 280 nm. The luminescence spectrum represented the broad peak around 340 nm. Each spectrum contained 4 curves in which two of them were control and two were laser-irradiated samples, as indicated. The two control samples and two laser-irradiated samples had 60 and, 300 μm of BSA proteins, respectively. For clarity, the spectra shown were normalized to the concentration of BSA proteins. In Fig. 6b, the far ultraviolet CD contained four curves, in which two of them were control and two were laser-irradiated samples. The two control samples and two laser-irradiated samples had 60 and 300 μm of BSA proteins, respectively. For clarity, the spectra shown were normalized to the concentration of BSA proteins. In Fig. 6c, the near ultraviolet CD included four curves in which two of them were control and two were laser-irradiated samples. The two control samples and two laser-irradiated samples had 60 and 300 μm of BSA proteins, respectively. For clarity, the spectra shown were normalized to the concentration of BSA proteins. The experimental results indicated that, within experimental uncertainty, the luminescence, excitation spectra, and circular dichroism of BSA proteins remained practically the same before and after the laser irradiation, indicative of minimal structural change of BSA proteins by the irradiation of a visible femtosecond laser. Figure 7 shows the effect of visible femtosecond laser irradiation on the single-stranded DNA from M13 bacteriophages, as detected by the agarose gel electrophoresis method. The control sample revealed the presence of a dark band from the single-stranded DNA of M13 bacteriophages. The laser-irradiated sample showed a single dark band similar in width to, and located in the same position, as that of the control sample. Figure 8 shows the agarose gel electrophoresis results on the single-stranded DNAs from the M13 bacteriophages (control) and the laser-irradiated M13 bacteriophage sample. The dark band, interpreted to be from the single protein units of the laser irradiated M13 bacteriophage sample, is out of scale and is, therefore, not shown here. Again, the laser-irradiated M13 bacteriophage sample showed a single dark band similar in width to, and located at, the same position as that of the control sample. Therefore, these experimental results indicated that, within the experimental uncertainty, irradiation of a visible femtosecond laser caused no denaturing or severe structural change of the single-stranded DNAs of M13 bacteriophages. Fig. 7Gel electrophoresis experiments on single-stranded DNA of M13 bacteriophages before and after the visible femtosecond laser irradiation, as indicated, for 1 h. 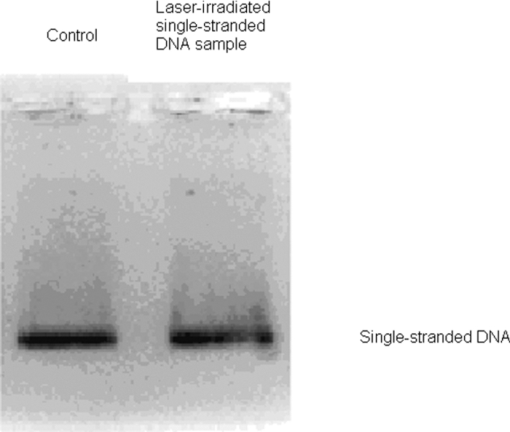 Fig. 8Gel electrophoresis experiments on single-stranded DNAs of M13 bacteriophages (control) and the laser-irradiated M13 bacteriopahges after the visible femtosecond laser irradiation for 1 h. For clarity, on the laser irradiated sample, an additional band resulting from the α − helix protein units of M13 bacteriophages, which appears on a different scale, is not shown. 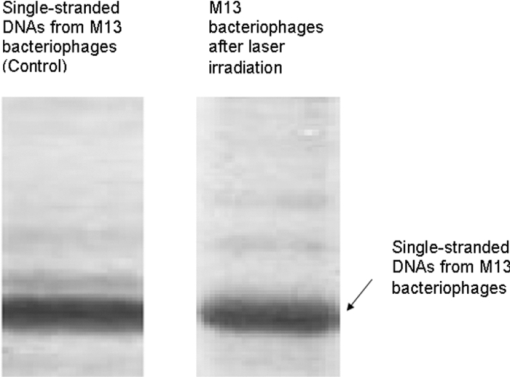 Figure 9 demonstrates our preliminary results for double-stranded DNA in buffer solution before and after irradiation by a visible femtosecond laser, as detected by the agarose gel electrophoresis method. The control sample (labeled no. 1) revealed the presence of three dark bands corresponding to circular, linear, and super-coiled double-stranded DNA, respectively. Sample no. 2 showed that stirring the sample slightly changed the relative darkness of the bands. On the other hand, the laser-irradiated sample (labeled no. 3) showed that the relative darkness of the three bands was altered greatly. These data suggest that the effects of visible femtosecond laser irradiation primarily relaxed the super-coiled double-stranded DNA or linear double-stranded DNA to the circular double-stranded DNA. 4.Discussions4.1.Insight into the Inactivation of a Virus by a Visible Femtosecond LaserThe gel electrophoresis results of Figs. 7, 8 on the single-stranded DNAs of M13 bacteriophages, indicate that irradiation of a visible femtosecond laser does not alter the structure of single-stranded DNA. In fact, this interpretation is consistent with the observation of the bare, undamaged DNAs in our AFM images of Fig. 5b for the laser-irradiated M13 bacteriophage sample. The experimental results on the optical characterization of BSA proteins (Fig. 6) suggest that there is practically no structural change of BSA proteins upon laser irradiation. Because the capsid of a M13 bacteriophage is mostly composed of α − helix protein units, these results suggest that visible femtosecond laser irradiation under our experimental conditions most likely will not damage the individual protein unit of which the protein shell of M13 bacteriophage is made up. Fig. 6(a) Excitation and luminescence spectra of BSA proteins. (b) Far UV circular dichroism spectra of BSA proteins. (c) Near UV circular dichroism spectra of BSA proteins. 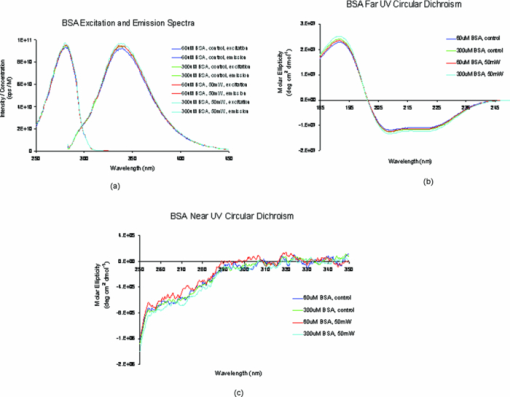 By taking into account the size of small structures about 6 nm in diameter in Fig. 5b, the resolution of the tip of AFM used in the imaging, and the actual size of the α − helix protein unit which forms the capsid of M13 bacteriophage, we have found that the small structures observed in Fig. 5b are consistent in size with those of the α − helix protein units of the capsid of M13 bacteriophages. This estimate further supports our conclusion that visible femtosecond laser irradiation under our experimental conditions does not damage individual protein units in M13 bacteriophages. Therefore, the AFM images of Fig. 5, together with the DNA gel electrophoresis results of Figs. 7, 8 and optical results of BSA proteins of Fig. 6, demonstrate that irradiation of a visible femtosecond laser alters the structural integrity of the protein shells of M13 bacteriophages by breaking their weak links without damaging either the single-stranded DNAs or the basic protein units of M13 bacteriophages. Deposition of laser energy onto the capsid of M13 bacteriophages can cause the breaking of weak links such as hydrophobic contacts and hydrogen bonds of the capsids. As a matter of fact, it has been estimated16 that simultaneous (or within a 100 fs time interval) deposition of a sufficient number of photons (of the order of 100) on the capsid of cowpea chlorotic mottle virus (CCMV) could break the hydrogen/hydrophobic bonds on the capsid of CCMV, leading to the subsequent disintegration of the capsid of CCMV into trimers. Based on the above arguments, the inactivation of M13 bacteriophages by a visible femtosecond laser is most likely due to the deposition of laser energy onto the capsids of M13 bacteriophages. Possible mechanisms that can account for our experimental observations for the inactivation of M13 bacteriophages are discussed in Secs. 4.1.1, 4.1.2, 4.1.3. 4.1.1.Linear absorption processThe linear absorption process can deposit laser energy to the capsid of M13 bacteriophages. Possible chromophores on the protein shells of M13 viruses, which absorb 425 nm light, can lead to the breaking of hydrophobic contacts and hydrogen bonds of the capsid of M13 bacteriophages. 4.1.2.Two-photon Raman scattering processIrradiation of an intense ultrashort pulsed laser, such as a femtosecond laser, can deposit laser energy onto the protein shell of a viral particle by the excitation of low-frequency acoustic vibrations on the capsid of the virus. This so-called impulsive stimulated Raman scattering process has been used to deposit laser energy to the solid state systems as well as to the biological molecules. 17, 18, 19, 20, 21, 22, 23, 24, 25, 26 In this two-photon Raman scattering process, the deposited laser energy on the protein shell of a viral particle can be shown17 to be proportional to the square of the laser intensity and to the Raman scattering cross section. If the deposited laser energy is large enough to break the weak links (for example, hydrogen bonds or hydrophobic contacts) between the proteins, damage to the capsid of the virus occurs, leading to viral inactivation. We have performed absorption experiments on the M13 bacteriophages, which measures the total deposited laser energy from both the linear absorption process and two-photon Raman scattering process described above. We have found that, on average, at a laser wavelength of 425 nm, the deposition of laser energy per M13 bacteriophage is about 1.2×10−17 J/pulse. We have also performed absorption experiments on the bare single-stranded DNAs of the M13 bacteriophages. We have found that, under our experimental conditions, there was negligible laser energy deposited for the bare single-stranded DNA of the M13 bacteriophage, indicating that it is the protein shells and not the single-stranded DNAs of M13 bacteriphages into which the laser energy is deposited. We note that both the linear absorption process and two-photon Raman scattering process described above can only deposit a very small amount of laser energy to the M13 bacteriophage per laser pulse; however, the damage to the capsid caused by this energy deposition, which leads to the breaking of hydrophobic contacts and hydrogen bonds, can be irreversible and accumulated; as a result, a long enough laser exposure time or a sufficient amount of laser dose is able to alter the structural integrity of the capsid of M13 bacteriophage, as observed in the AFM image of Fig. 5b. 4.1.3.Possibility of shock wave generation by the visible femtosecond laserThe other likely mechanism is the generation of a shock wave by the ultrashort pulsed laser. An intense, ultrashort pulsed laser has been known27 to produce a shock wave when interacting with materials including biological molecules, which can lead to damage of the capsid of M13 bacteriophages. In general, the energy relaxation process in the biological system is shorter than 1 ns; because the time interval between two subsequent laser pulses is about 13 ns for the femtosecond visible laser used in our experiments, it is the energy per laser pulse that is important to consider. From the absorption of the visible femtosecond laser passing through the sample, we estimate that, under our experimental conditions, the average absorption laser energy for a M13 bacteriophage is about1.2×10−17 J/pulse or 2.3×10−8 J/cm2. However, in order to generate a shock wave of sufficient amplitude in materials to cause damage, the laser energy absorbed has to exceed 1 J/cm2.28 Therefore, the shock wave generation mechanism seems to be unlikely. 4.2.Insight into the Inactivation of Bacteria by a Visible Femtosecond LaserTo obtain insight into the inactivation mechanisms, we have performed inactivation of a mutant Salmonella typhimurium by a visible femtosecond laser. The mutant is deficient in RECA proteins which are responsible for the repair of damaged DNA. In other words, the mutant is very sensitive/vulnerable to the damage of DNA. Figure 10 shows the inactivation of both the wild and mutant Salmonella typhimurium by a visible femtosecond laser as a function of the laser fluence. In general, the log–load reduction factor at a given laser dose is higher for the mutant than for the wild strain. In particular, our experimental results indicate that by using the visible femtosecond laser, with laser dose of about 800 J/cm2, a log–load reduction factor of about 5 for mutant Salmonella typhimurium was observed; however, by employing the same laser parameter, a log-kill factor of only 0.5 for the wild Salmonella typhimurium was found. Because the only difference between these two stains of Salmonella typhimurium is the RECA proteins which are in charge of the repair of damaged DNA, these experimental results indicate that irradiation of a visible femtosecond laser causes DNA damage and subsequent inactivation of the Salmonella typhimurium. Fig. 10Log-kill factor as a function of laser fluence for the wild, mutant Salmonella typhimurium, as indicated. 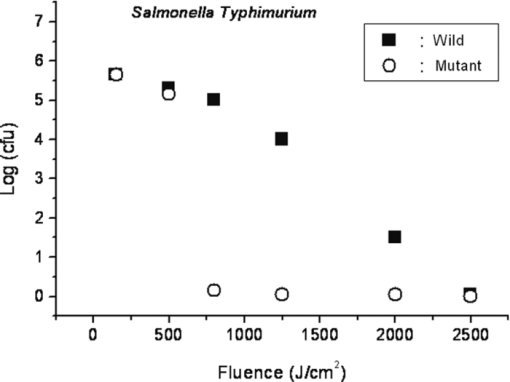 It has been established that29, 30, 31, 32, 33, 34 photostimulation of endogenous intracellular porphyrin molecules in the bacteria by continuous wave visible light irradiation results in the production of reactive oxygen species, predominantly singlet oxygen, and consequently, damage to the DNA and the death of bacteria. We believe that this is the dominant mechanism for the inactivation of Salmonella typhimurium by a visible femtosecond laser. We have also carried out absorption experiments on Salmonella typhimurium. Our results indicated that, on average, deposited laser energy for a Salmonella typhimurium is about 1.1×10−16 J/pulse or 1.1×10−9 J/cm2, indicating that the shock wave generation mechanism28 is not the inactivation mechanism for Salmonella typhimurium. 4.3.Inactivation of Viruses and Bacteria by a Continuous-Wave Laser at a Wavelength of 425 nmWe have also performed inactivation of M13 bacteriophages, EMCV, and Salmonella typhimurium as a function of laser fluence with an InGaN CW laser operated at 425 nm. The results are shown in Figs. 2, 3, 4, respectively. Because a CW laser is expected to inactivate viruses and bacteria dominantly through linear absorption process, comparison of inactivation between a CW visible laser and the visible femtosecond laser operated at the same wavelength provides valuable information on the inactivation mechanisms. For example, the substantial differences of inactivation between a CW laser and a femtosecond laser in Figs. 2, 3 reflect significant inactivation of M13 bacteriophages and EMCV due to a nonlinear two-photon Raman scattering process. On the other hand, Fig. 4 shows that inactivation between a CW laser and a femtosecond laser is very similar for Salmonella typhimurium, indicating that the linear absorption process is the primary mechanism for the inactivation of Salmonella typhimurium by a visible femtosecond laser. 4.4.Two-Photon Absorption ProcessBased on the assumption that the two-photon absorption cross section is 10−50 cm4 s photon−1 for a typical molecule, the excitation laser power density is about 100 MW/cm2, and for the laser parameters used in our experiments, we estimate that the number of ultraviolet (UV) photons at wavelength λ = 2.12.5 nm produced in the sample by the excitation laser through two-photon absorption process is about 15. This number of generated UV photons is too small to contribute to the inactivation of M13 bacteriophages, EMCV, and salmonella. Therefore, we believe the two-photon absorption process cannot account for the inactivation of M13 bacteriophages, EMCV, or salmonella bacteria observed in our experiments. 5.ConclusionWe have studied the inactivation of EMCV, M13 bacteriophage, and Salmonella typhimurium by a visible femtosecond laser. Very efficient inactivation was observed for Salmonella typhimurium. Our experimental results suggest that the inactivation of Salmonella typhimurium is related to the damage of their DNA. On the other hand, inactivation of viral particles, such as M13 bacteriophages, by a visible femtosecond laser involves the breaking of hydrogen/hydrophobic bonds or the separation of the weak protein links in the protein shell of a viral particle. We have also discussed possible mechanisms for the inactivation of viruses and bacteria by a visible femtosecond laser. AcknowledgmentsThis work was supported by the National Science Foundation. The opinions or assertions contained herein are the private views of the authors and are not to be construed as official or reflecting the views of the Armed Forces Radiobiology Research Institute, Uniformed Services University of the Health Sciences, or the U.S. Department of Defense. ReferencesK. T. Tsen, S.-W. D. Tsen, C.-L. Chang, C.-F. Hung, T. C. Wu, and
J. G. Kiang,
“Inactivation of viruses by coherent excitations with a low power visible femtosecond laser,”
Virology J., 4 50
(2007). https://doi.org/10.1186/1743-422X-4-50 Google Scholar
K. T. Tsen, S-W. D. Tsen, C.-L. Chang, C.-F. Hung, T. C. Wu, and
J. G. Kiang,
“Inactivation of viruses with a very low power visible femtosecond laser,”
J. Phys.: Condens. Matter, 19 322102
(2007). https://doi.org/10.1088/0953-8984/19/32/322102 Google Scholar
K. T. Tsen, S.-W. D. Tsen, O. F. Sankey, and
J. G. Kiang,
“Selective inactivation of microorganisms with near-infrared femtosecond laser pulses,”
J. Phys.: Condens. Matter, 19 472201
(2007). https://doi.org/10.1088/0953-8984/19/47/472201 Google Scholar
K. T. Tsen, S.-W. D. Tsen, C.-L. Chang, C.-F. Hung, T. C. Wu, and
J. G. Kiang,
“Inactivation of viruses by laser-driven coherent excitations via impulsive stimulated Raman scattering process,”
J. Biomed. Opt., 12 064030
(2007). https://doi.org/10.1117/1.2821713 Google Scholar
K. T. Tsen, S.-W. D. Tsen, C.-L. Chang, C.-F. Hung, T. C. Wu, B. Ramakrishna, K. Mossman, and
J. G. Kiang,
“Inactivation of viruses with a femtosecond laser via impulsive stimulated Raman scattering,”
Proc. SPIE, 6854 68540N
(2008). https://doi.org/10.1117/12.762324 Google Scholar
K. T. Tsen, S.-W. D. Tsen, Q. Fu, S. M. Lindsay, K. Kibler, B. Jacobs, T.-C. Wu, B. Karanam, S. Jagu, R. Roden, C.-F. Hung, O. Sankey, B. Ramakrishna, and
J. G. Kiang,
“Photonic approach to the selective inactivation of viruses with a near-infrared subpicosecond fiber laser,”
J. Biom. Opt., 14 064042
(2009). https://doi.org/10.1117/1.3275477 Google Scholar
S.-W. D. Tsen, Y.-S. D. Tsen, K. T. Tsen, and
T. C. Wu,
“Selective inactivation of viruses with femtosecond laser pulses and its potential use for in vitro therapy,”
J. Healthcare Engineering, 1
(2), 185
–196
(2010). https://doi.org/10.1260/2040-2295.1.2.185 Google Scholar
R. Won,
“Selective disinfection,”
Nature Photonics, 4 136
(2010). https://doi.org/10.1038/nphoton.2010.10 Google Scholar
H. D. Wang, R. Bash, J. G. Yodh, G. L. Hager, D. Lohr, and
S. M. Lindsay,
“Glutaraldehyde modified mica: A new surface for atomic force microscopy of chromatin,”
Biophys. J., 83 3619
–3625
(2002). https://doi.org/10.1016/S0006-3495(02)75362-9 Google Scholar
X. Ji, J. Oh, A. K. Dunker, and
K. W. Hipps,
“Effects of relative humidity and applied force on atomic force microscopy images of the filamentous phage fd,”
Ultramicroscopy, 72 165
–176
(1998). https://doi.org/10.1016/S0304-3991(97)00169-1 Google Scholar
K.-T. Nam, B. R. Peelle, S.-W. Lee, and
A. M. Belcher,
“Genetically driven assembly of nanorings based on the M13 virus,”
Nano Lett., 4 23
–27
(2004). https://doi.org/10.1021/nl0347536 Google Scholar
D. Anselmetti, R. Luthi, E. Meyer, T. Richmond, M. Dreier, J. E. Frommer, and
H. J. Guntherodt,
“Attractive-mode imaging of biological materials with dynamic force microscopy,”
Nanotechnology, 5 87
–94
(1994). https://doi.org/10.1088/0957-4484/5/2/004 Google Scholar
S. Y. Venyaminov and
J. T. Yang,
“Determination of protein secondary structure,”
Theory of Circular Dichroism and the Conformational Analysis of Biomolecules, 109
–157 Plenum Press, New York
(1996). Google Scholar
R. W. Woody and
A. K. Dunker,
“Aromatic and cysteine side-chain circular dichroism in proteins,”
Theory of Circular Dichroism and the Conformational Analysis of Biomolecules, 109
–157 Plenum Press, New York
(1996). Google Scholar
R. C. Ogden and
D. A. Adams,
“Electrophoresis in agarose and acrylamide,”
Gels. Meth. Enzymol., 152 61
–87
(1987). https://doi.org/10.1016/0076-6879(87)52011-0 Google Scholar
M. F. Thorpe, Google Scholar
Y.-X. Yan, K. A. Nelson,
“Impulsive stimulated scattering: General importance in femtosecond laser pulse interactions with matter, and spectroscopic applications,”
J. Chem. Phys., 83 5391
–5399
(1985). https://doi.org/10.1063/1.449708 Google Scholar
K. A. Nelson, R. J. D. Miller, D. R. Lutz, and
M. D. Fayer,
“Optical generation of tunable ultrasonic waves,”
J. Appl. Phys., 53 1144
–1149
(1982). https://doi.org/10.1063/1.329864 Google Scholar
S. De Silvestri, J. G. Fujimoto, E. P. Ippen, E. B. Gamble Jr., L. R. Williams, and K. A. Nelson,
“Femtosecond time-resolved measurements of optic phonon dephasing by impulsive stimulated Raman scattering in α-perylene crystal from 20 to 300 K,”
Chem. Phys. Lett., 116 146
–152
(1985). https://doi.org/10.1016/0009-2614(85)80143-3 Google Scholar
K. A. Nelson,
“Stimulated Brillouin scattering and optical excitation of coherent shear waves,”
J. Appl. Phys., 53 6060
–6063
(1982). https://doi.org/10.1063/1.331556 Google Scholar
G. C. Cho, W. Kutt, and
H. Kurz,
“Subpicosecond time-resolved coherent-phonon oscillations in GaAs,”
Phys. Rev. Lett., 65 764
–766
(1990). https://doi.org/10.1103/PhysRevLett.65.764 Google Scholar
T. K. Cheng, J. Vidal, H. J. Zeiger, G. Dresselhaus, M. S. Dresselhaus, and
E. P. Ippen,
“Mechanism for displacive excitation of coherent phonons in Sb, Bi, Te, and Ti2O3,”
Appl. Phys. Lett., 59 1923
–1925
(1991). https://doi.org/10.1063/1.106187 Google Scholar
J. M. Chwalek, C. Uher, J. F. Whittaker, and
G. A. Mourou,
“Subpicosecond time- resolved studies of coherent phonon oscillations in thin-film YBa2Cu3O6+x (x<0.4),”
Appl. Phys. Lett., 58 980
–982
(1991). https://doi.org/10.1063/1.104462 Google Scholar
R. Merlin,
“Generating coherent THz phonons with light pulses,”
Solid State Commun., 102 207
–220
(1997). https://doi.org/10.1016/S0038-1098(96)00721-1 Google Scholar
Y. R. Shen and
N. Bloembergen,
“Theory of simulated Brillouin and Raman scattering,”
Phys. Rev., 137 A1787
–A1805
(1965). https://doi.org/10.1103/PhysRev.137.A1787 Google Scholar
Y. R. Shen, The Principles of Nonlinear Optics, Wiley, New York
(1984). Google Scholar
M. Boustie, L. Berthe, T. de Resseguier, and
M. Arrigoni,
“Laser shock waves: fundamentals and applications,”
(2008). Google Scholar
H. Ashkenazi, Z. Malik, Y. Harth, and
Y. Nitzan Eradication,
“Propionibacterium acnes by its endogenic porphyrins after illumination with high intensity blue light,”
FEMS Immunol. Med. Microbiol., 35 17
–24
(2003). https://doi.org/10.1111/j.1574-695X.2003.tb00644.x Google Scholar
M. Elman, M. M. Slatkine, and
Y. Harth,
“The effective treatment of acne vulgaris by a high-intensity, narrow band 405–420 nm light source,”
J. Cosmet. Laser Ther., 5 111
–116
(2003). https://doi.org/10.1080/14764170310001276 Google Scholar
O. Feuerstein, N. Persman, and
E. I. Weiss,
“Phototoxic effect of visible light on Porphyromonas gingivalis and Fusobacterium nucleatum: an in vitro study,”
Photochem. Photobiol., 80 412
–415
(2004). Google Scholar
R. A. Ganz, J. Viveiros, A. Ahmad, A. Ahmadi, A. Khalil, M. J. Tolkoff, N. S. Nishioka, and
M. R. Hamblin,
“Helicobacter pylori in patients can be killed by visible light,”
Laser Surg. Med., 36 60
–265
(2005). https://doi.org/10.1002/lsm.20161 Google Scholar
N. S. Soukos, S. Som, A. D. Abernethy, K. Ruggiero, J. Dunham, C. Lee, A. G. Doukas, and
J. M. Goodson,
“Phototargeting oral blackpigmented Bacteria,”
Antimicrob Agent Chemother, 49 1391
–1396
(2005). https://doi.org/10.1128/AAC.49.4.1391-1396.2005 Google Scholar
M. Maclean, S. J. MacGregor, J. G. Anderson, and
G. Woolsey,
“High- intensity narrow-spectrum light inactivation and wavelength sensitivity of staphylococcus aureus,”
FEMS Microbiol. Lett., 285 227
–232
(2008). https://doi.org/10.1111/j.1574-6968.2008.01233.x Google Scholar
|