|
|
1.IntroductionColocalization analysis is an invaluable and increasingly used tool to gain information about interactions between biomolecules, cellular uptake, and intracellular trafficking.1–7 Using multichannel microscopy, based on simultaneous or successive detection of several fluorochromes in a multilabeled sample, the colocalization of molecules with different fluorescent labels can be analyzed. Most of these studies are based on qualitative colocalization analysis by visual inspection of overlaid two-dimensional (2-D) microscopic images. This method may be subjective and misleading, because of the limited ability of the eye to perceive and interpret colors. For a more rigorous analysis, three-dimensional (3-D) quantitative colocalization is required.8 The results from quantitative colocalization analysis are usually presented with plots and coefficients that compare the intensity distributions in two detector channels or calculate the number of voxels with co-occurring fluorescence.8,9 Colocalization analysis is highly sensitive to artifacts. Sample preparation, image acquisition, and pre-processing are critical steps to minimize errors.3,8,10–15 A colocalization event is defined when the emitted light of two fluorochromes is collected in the same image element. However, the reproduction of the dimensions and location of objects in microscopic images depends on the optical resolution of the system and can be subject to artifacts caused by optical and mechanical aberrations. Deconvolution is a powerful technique that partly restores degraded images by reassigning out-of-focus blur, which is caused by diffraction, back to its focal plane of origin, thereby reducing noise and increasing the resolution in 3-D confocal data sets.8,10,16–18 Colocalization studies can provide information about the efficiency of gene delivery, which involves cellular uptake of exogenous DNA and its vector, intracellular trafficking to the nucleus, and transcription of the transgene. Chitosan is a nonviral polycationic gene delivery vector that efficiently condenses plasmid DNA (pDNA), shields its charges, and protects it from degradation.19 The superior biocompatibility of chitosan compared with other polycations makes it attractive for biomedical applications.20,21 The ability of chitosan to condense pDNA and improve the gene transfection efficacy strongly depends on its structural variables.22–25 We have synthesized a new generation of self-branched glycosylated (trisaccharide-substituted) chitosan oligomers (SBTCOs) that demonstrate higher gene transfection efficacy than linear chitosan oligomers (LCOs) of the same chain length.21,26,27 The reasons for the increased transfection efficacy of polyplexes (cationic polymer–pDNA complexes) formed by SBTCOs remain unclear, but we hypothesize that differences in the cellular uptake mechanisms and intracellular trafficking of the polyplexes are involved. Polyplexes are internalized via endocytotic pathways,1,5,7,28–30 of which the best characterized is clathrin-mediated endocytosis involving clathrin-coated pits that invaginate and form intracellular clathrin-coated vesicles. Depolymerization of the clathrin coat leads to the formation of early endosomes (EEs), which may mature into late endosomes (LEs) and further fuse with lysosomes.30 The escape of polyplexes from these acidic compartments is required for transfection and relies on, among other things, the buffering capacity of the polycations.31 The best-characterized clathrin-independent pathway is caveolae-mediated endocytosis, which has been reported to play an important role in gene transfection.1,28,32,33 Caveolae are small invaginations in the plasma membrane, containing the membrane protein caveolin-1.34 Caveolae have low mobility, and their uptake kinetics is generally slower than that of clathrin-mediated endocytosis.35 The intracellular faith of cargo internalized by caveolae is not clear, but many studies suggest that the internalized structures fuse with EEs.34,36–38 It has also been claimed that caveolae-mediated uptake can result in delivery of the cargo to caveolin-1-containing, pH-neutral structures, termed caveosomes,39,40 but this hypothesis is controversial.34 Numerous reports exist on quantitative colocalization analysis based on biological 2-D4 and 3-D3,10,41–44 images, but, to our knowledge, studies have not yet been published on the intracellular trafficking of polyplexes using quantitative 3-D colocalization analysis. The aim of the present work is to perform a quantitative 3-D colocalization analysis using different colocalization parameters in order to determine which are more suitable to give information about cellular uptake mechanisms, intracellular trafficking, and the dissociation of pDNA complexed with LCOs or SBTCOs, which are chitosans oligomers of equal length but with significantly different transfection efficacies.21,26,27 In this study, the colocalization of pDNA and chitosan, as well as the localization of free or complexed pDNA and chitosan in EEs, are analyzed based on multichannel confocal laser scanning microscopy (CLSM) images of HeLa cells that are incubated with fluorescently double-labeled pDNA-chitosan polyplexes before immunostaining of EEs. Imaging and analysis are performed at different time points to study the kinetics. 2.Materials and Methods2.1.Cell CulturesHeLa cells (human cervical carcinoma cell line) were grown in DMEM (Invitrogen, Gibco) supplemented with 10% fetal bovine serum (Invitrogen, Gibco), 1 mM nonessential amino acids (Invitrogen, Gibco), and 1 mM L-glutamine (Sigma-Aldrich). The cells were cultivated at 37 °C in a humidified atmosphere with 5% . 2.2.Plasmid DNAStock solutions of 6.7 kb pDNA (Aldevron) were prepared at a concentration of in sterile Milli-Q grade water. 2.3.ChitosansThe LCOs were prepared by nitrous acid depolymerization of completely de-N-acetylated chitosan (fraction of acetylated units ) to a number-average degree of polymerization (DPn) of 42 as previously described.45 The SBTCOs were prepared from LCOs by simultaneous self-branching and substitution with the trimer 2-acetamido-2-deoxy--glucopyranosyl-β-(1-4)-2-acetamido-2-deoxy-glucopyranosyl-β-(1-4)-2,5-anhydro-mannofuranose (A-A-M).21 The chitosans were dissolved at in Milli-Q grade water, sterile-filtered (0.2 µm Millipore), and stored in aliquots at . The properties of the chitosans used in this study are summarized in Table 1. Table 1Characteristics of chitosans.a
2.4.Antibodies and Fluorescent LabelsEEs were immunostained with rabbit anti-Early Endosomal Antigen 1 (anti-EEA1) (N-terminal) antibody (Sigma-Aldrich). Alexa Fluor 555 goat anti-rabbit IgG (Invitrogen) was used as a secondary antibody. The more photostable Alexa Fluor 555 was preferred over Alexa Fluor 546 as a label for the secondary antibody, because the latter showed significant photobleaching in preliminary experiments (data not shown). To label chitosans, Alexa Fluor 488 carboxylic acid, succinimidyl ester (Invitrogen) was chosen, because of its amine-reactive properties, high extinction coefficient, and photostability. The theoretical labeling degree was previously optimized to approximately one molecule of dye per 200 GlcN residues. Unconjugated dye was removed by extensive dialysis (Spectropor cutoff of 12 kDa), and the labeled chitosans were freeze-dried. For labeling of pDNA, a Cy5 Nucleic Acid Labeling Kit (Mirus Bio Corp.) that covalently binds Cy5 to pDNA was used according to the protocol from the supplier, and the unbound Cy5 was removed using microspin columns provided in the kit. 2.5.Preparation of PolyplexesThe pDNA-chitosan polyplexes were formed by self-assembly to a final pDNA concentration of . The required amount of pDNA stock solution was diluted in Milli-Q grade water. The sterile-filtered chitosan stock solution was added to the pDNA solution at an amino/phosphate () ratio of 10 during intense stirring on a vortex mixer. The polyplexes were incubated for 30 to 40 min at room temperature. 2.6.Incubation of Cells with Polyplexes and ImmunostainingHeLa cells were seeded on Lab-Tek 8-well detachable chamber slides (VWR International) at a density of cells per well and allowed to adhere overnight. The following day, the growth medium was removed, and the cells were incubated with LCO or SBTCO polyplexes in hypertonic OptiMEM (Invitrogen, Gibco) for 15 min, 1 h, and 3 h at 37 °C and 5% . The cells were subsequently fixed with 4% paraformaldehyde for 15 min and permeabilized for 10 min with 0.5% Triton X-100 (Sigma-Aldrich) at room temperature. The nonspecific binding sites were blocked with 5% goat serum and 0.3% Triton X-100 for 1 h. The antibody stock solutions were centrifuged at 1500 rpm at 5 min, the supernatants were diluted to in 1% bovine serum albumin and 0.3% Triton X-100, and the cells were incubated with the primary and secondary antibody for 1 h and 30 min, respectively. The optimal working concentrations of the antibodies were determined in preliminary titration experiments. To minimize the background fluorescence, the cells were washed in phosphate buffered saline (PBS) overnight after primary antibody incubation and for 2 h after secondary antibody incubation. The samples were mounted with ProLong Gold antifading reagent (Invitrogen), and the cover glasses were sealed with nail polish. The samples were allowed to cure for a minimum of 24 h. 2.7.Confocal Laser Scanning MicroscopyThe cells were imaged using a Zeiss LSM 510 META (Carl Zeiss GmbH, Jena) equipped with an α-Plan-FLUAR oil-immersion objective. Alexa Fluor 488, Alexa Fluor 555, and Cy5 were excited by the 488-, 543-, and 633-nm laser lines, respectively, and the fluorescence was detected using the filters BP 500 to 550 nm, BP 565 to 616, and the META detector with a spectral range of 651 to 704 nm, respectively. Sequential imaging (12-bit) was performed to minimize cross-talk. Oversampling of image -stacks for deconvolution was performed at a voxel size of . Four image averages and a scan zoom of 2 were suitable to obtain the desired image resolution without photobleaching. Equal optical section thicknesses for the three channels were obtained by setting pinhole diameters to 300, 262, and 192 µm for the 488-, 543-, and 633-nm laser lines, respectively. The 633-nm channel was used as a reference with a pinhole diameter corresponding to 1 Airy unit to avoid low signal-to-noise ratios (SNR) caused by smaller pinhole sizes. The detector gain and offset values were adjusted to utilize the entire dynamic range of the detector and to avoid oversaturated voxels and voxels of zero intensity. To minimize autofluorescence and cross-talk, the maximum laser power and detector gain were determined based on control samples comprising untreated cells, cells stained with the secondary antibody or the primary and secondary antibodies, and cells incubated with unlabeled pDNA complexed with labeled chitosan or labeled pDNA complexed with unlabeled chitosan. 2.8.Image Pre-Processing and RestorationDeconvolution requires knowledge of the point spread functions (PSFs) to calculate the statistical probability of the exact origin of the emitted photons for each voxel in each acquired image in the Z-stacks. In the present study, the PSFs were determined for all three emission channels by imaging subresolution beads from the Microscope Point Spread Source Kit (Invitrogen) with a diameter of (Fig. 1) and averaging a minimum of three beads for each channel using the Amira (Visage Imaging) BeadExtract module. The image data sets of the cell samples were subsequently subjected to deconvolution by applying an iterative maximum-likelihood image restoration algorithm (20 iterations) in Amira. Fig. 1Maximum intensity projections (MIPs) and volumetric representations of the PSF for the excitation wavelengths of 488 (a), 543 (b), and 633 nm (c). The , , and MIPs are located to the left, right, and bottom for each PSF, respectively. Each PSF data set has a volume of . The PSFs were estimated by imaging and averaging three different subresolution fluorescent microspheres with the same image acquisition settings as for the cell samples. 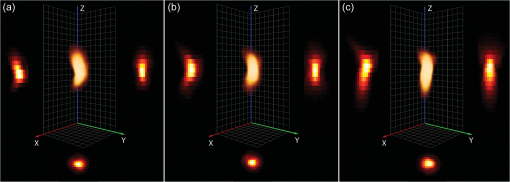 Chromatic shifts in the lateral and axial directions were measured from the deconvolved image data sets by determining the center of 1-µm FocalCheck™ Thin-Ring Fluorescent Microspheres Kit (Invitrogen) for the different channels using the ImageJ47 plug-in Sync Measure 3-D (written by Joachim Walter, http://rsbweb.nih.gov/ij/plugins/sync-windows.html). Five beads were averaged for each channel. Lateral [Figs. 2(a) and 2(c)] and axial [Figs. 2(b) and 2(c)] shifts rounded off to whole voxels were determined for the 543- and 633-nm channels relative to the 488-nm channel, which was used as a reference channel for the registration of the 3-D data sets. The 543-nm channel was shifted by 1 pixel in the X-axis, and the 633 nm channel was shifted by 1 voxel in all three (, , and ) axes, relative to the 488 nm channel. Both bead solutions were properly diluted, prepared on microscopy slides, mounted with ProLong Gold (Invitrogen), and captured using the same image acquisition parameters used for the cell samples. Fig. 2Representation of chromatic shifts in the lateral (a) and axial (b) axes for 1-µm fluorescent microspheres excited with the 488-, 543-, and 633-nm laser lines (using two types of beads with different emission spectra). The intensity profiles (c) adapted from (a,b) clearly demonstrate chromatic lateral (top) and axial (bottom) shifts and image registration (dotted line). The scale bar represents 1 µm. The asterisks indicate the intensity profiles of the registered data set. 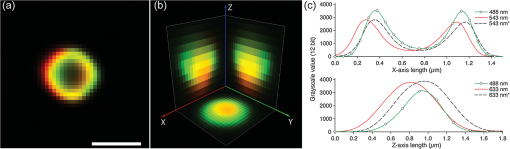 The restored and chromatic shift-corrected images were background-subtracted, converted to the 8-bit format, and thresholded before colocalization analysis. After several algorithms were tested, a manual threshold was considered the most suitable in this study, and a lower threshold cutoff of 13 grayscale values was sufficient to eliminate background and maintain the specific fluorescence signal for all three channels of the 8-bit pre-processed images. For the analysis of the raw image -stacks, higher threshold values were necessary, because of a higher background signal and noise level. The entire cells were defined as regions of interests (ROIs) for colocalization analysis, and the ROIs were drawn based on bright-field images [Fig. 3(a)]. The density of endosomes in each cell was calculated as the number of endosomes with respect to the volume of the cell’s 3-D ROI using the ImageJ47 3-D object counter plug-in (written by Fabrice Cordelires and Jonathan Jackson, http://rsbweb.nih.gov/ij/plugins/track/objects.html).8 Fig. 3Bright-field image (a) and raw (b) and pre-processed (c–f) fluorescence images of a HeLa cell that was incubated for 15 min with pDNA (red) complexed with SBTCO (blue) and immunostained with anti-EEA1 (green). The bright-field image was included to indicate the cell membrane and the location of the nucleus, whereas the nucleus in the fluorescence images is localized by the absence of EEs. The fluoresence signals from the labeled pDNA, chitosan, and EEs in the pre-processed image (c) are separated into three different channels in (d–f). Pre-processing included deconvolution, chromatic channel registration, and background subtraction. The scale bar (a) represents 4 µm. 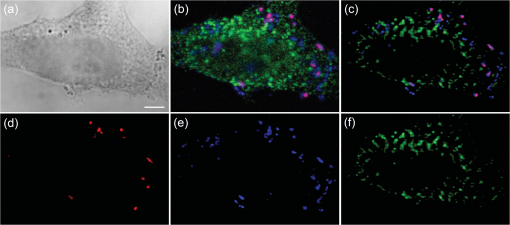 2.9.Quantitative ColocalizationTo compare the intracellular trafficking and dissociation of polyplexes formed by the two chitosans, quantitative colocalization analysis was based on the pre-processed, multichannel confocal -stacks of cells that had been incubated with the fluorescently labeled polyplexes and immunostained. Several parameters were used to quantify the 3-D colocalization of the objects in the multichannel images, in order to determine the optimal parameters to use and to achieve better reliability of the results. The colocalization between pDNA and chitosan was analyzed to indicate molecular binding and dissociation, and colocalization between the particles and EEs was analyzed to indicate particle transport via EEs. In the analysis of colocalization with EEs, pDNA and chitosan were included, regardless of whether they were free or bound in polyplexes. Scatter plots, Pearson’s correlation coefficient (PCC), Manders’ colocalization coefficients (MCCs), intensity correlation analysis (ICA) plots, and the intensity correlation quotient (ICQ) were obtained using the ImageJ44 plug-in Intensity correlation analysis (written by Tony Collins, http://www.uhnresearch.ca/facilities/wcif/software/Plugins/ICA.html). A brief description of each colocalization parameter follows. For convenience, we refer to the channel pair in a colocalization analysis as “channel A” and “channel B”. In a scatter plot, the intensity distributions of the two channels are plotted against one another, and a diagonal line indicates perfect proportional codistribution. The PCC quantifies the spread of the distribution with respect to this line by estimating the goodness of the linear approximation.8,14 The PCC is a parameter that is widely used to calculate the correlation between the voxel intensities in two channels and ranges from -1 (perfect negative linear relationship between the fluorescence intensities) to 1 (perfect positive linear relationship between the fluorescence intensities), although the interpretation of negative values and values close to zero are not straightforward. PCC reports the covariance and is independent of the signal offset (background) but sensitive to image noise and contributions from the PSF.10,48 Because it is sensitive to variations in fluorescence intensities,8,14 the PCC will fit a single line even to segregated groups of data points in samples with heterogeneous colocalization relationships. MCCs provide separate information for the two fluorescent molecules (i.e., indicates the fraction of the fluorescence signal in channel A colocalized with a signal in channel B, and vice versa for ) and range from 0 to 1.14 The coefficients strictly measure co-occurrence and are independent of the signal intensities. Thus, MCCs are a good choice if the intensities in the two channels are very different, but they are affected by the signal offset, noise, and contributions from the PSF.10,48 ICA describes the intensity synchrony between channels A and B. The product of the differences from the mean (PDM) is defined as , where A and are the voxel intensity and mean intensity, respectively, in Channel A, whereas B and are the voxel intensity and mean intensity, respectively, in Channel B. The ICA is based on the assumption that the sum of the PDM values approaches zero.9 If the fluorescence intensities in the two channels are dependent or segregated, then the PDM assumes a positive (colocalization) or negative (segregation) value, respectively. ICA results can be presented in two ICA plots where the voxel intensities in the two channels are plotted along the -axis as functions of the PDM values along the -axis. In ICA plots, colocalized and segregated voxels will be found on the right and the left side of the -axis, respectively. When such plots are difficult to interpret because of noise, background, or randomly distributed fluorescence, a more quantitative parameter that reflects the degree of dependency between the two channels, the ICQ, can be calculated. The ICQ is defined as the ratio of the number of positive PDM values to the total number of voxel pairs subtracted by 0.5, and it assumes values ranging from to 0.5. An ICQ of , 0, or 0.5 indicates totally segregated, random, or dependent staining, respectively. Similar to the MCCs, the ICA method can be applied in situations of heterogeneous staining. A high ICQ value does not necessarily imply a large number of colocalized voxels, but a high covariance (i.e., the voxel intensities of two fluorochromes vary together to a large extent). 2.10.StatisticsThe experimental groups ( to 35 cells in each group) consisted of two different polyplexes (LCO and SBTCO) and three different incubation times (15 min, 1 h, and 3 h). The average values of the quantitative colocalization parameters (PCC, MCCs, and ICQ) were calculated for each group and compared statistically using Student’s -test for normalized data or the Mann-Whitney test for sample populations with that did not follow a Gaussian distribution. A significance criterion of was used. 3.Results3.1.The Effect of Pre-Processing on Image Quality and Colocalization ResultsImage pre-processing [Figs. 3(c)–3(f)] reduced the noise and blur and provided better defined structures compared with raw confocal images. The bright-field image [Fig. 3(a)] of the cell is included to indicate the cell surface and nucleus. The raw images of the EEs were, in particular, subject to noise because of nonspecific binding. This noise was efficiently depressed by the deconvolution algorithm, which resulted in clearly defined EEs [Figs. 3(c) and 3(f)] in the pre-processed images. The effect of deconvolution, chromatic channel registration, and background subtraction of the images on scatter and ICA plots and quantitative colocalization parameters varied among different cells within a sample. The plots in Fig. 4 and the colocalization parameters in Table 2 present a comparison between colocalization analysis based on raw and pre-processed images of a cell. For this particular cell, image pre-processing reduced most of the colocalization parameters. The effect of pre-processing was different for the various quantitative parameters. Whereas the MCCs were, in general, highly influenced by image pre-processing, the effect on the ICQ and PCC was more random. Regarding , pre-processing reduced the fraction of pDNA located in EEs in most cells as well as the fraction of chitosan located in EEs in all of the analyzed cells. The effect was not as pronounced for the fraction of EEs colocalized with pDNA or chitosan (). Whereas the fraction of pDNA colocalized with chitosan was not consistently influenced by image pre-processing, the fraction of chitosan colocalized with pDNA () was higher in the raw images than in the pre-processed images for the majority of the cells. Fig. 4Resulting effect of image pre-processing (deconvolution, chromatic shift-correction, and background subtraction) on the intensity correlation analysis (ICA) represented by the scatter plots of the paired intensities of the two channels (left) and the individual fluorescence intensities of channel A (center) and channel B (right) plotted as functions of their respective PDM-values [(A–a)(B–b)]. The plots for pDNA and chitosan (a,b), pDNA and early endosomes (c,d), and chitosan and early endosomes (e,f) are based on raw (a,c,e) and pre-processed (b,d,f) confocal -stacks of one representative HeLa cell that was incubated with pDNA-LCO polyplexes for 1 h and immunostained with anti-EEA1. 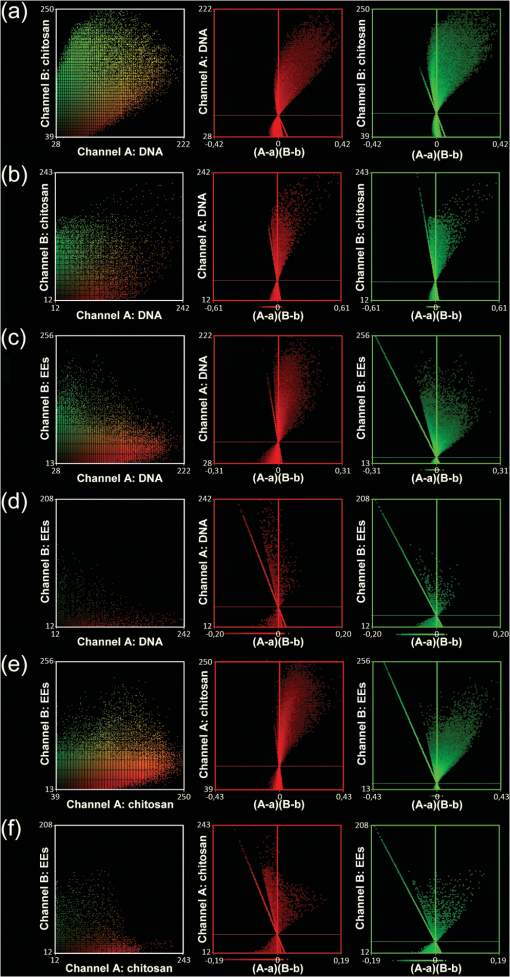 Table 2Quantitative colocalization parameters of raw and pre-processeda confocal Z-stacks. The degree of colocalization is estimated by Manders’ colocalization coefficients (i.e., MCCA is the fraction of objects in Channel A colocalized with objects in Channel B, and vice versa for MCCB), the intensity correlation quotient (ICQ), and Pearson’s coefficient (PCC).b The values correspond to the representative cell in Fig. 4.
3.2.Qualitative Interpretation of Uptake and Intracellular TraffickingClear differences in the cellular uptake and intracellular trafficking of polyplexes formed by the two chitosans were observable by the eye. More free or complexed pDNA overlapped with EEs or other well-defined intracellular structures for cells receiving pDNA-SBTCO compared with those receiving pDNA-LCO at all incubation times [Figs. 5(a)–5(c), 5(g)–5(i)]. Fig. 5HeLa cells that were incubated for 15 min (a,d,g,j), 1 h (b,e,h,k), and 3 h (c,f,i,l) with pDNA complexed with LCOs (a–f) and SBTCOs (g–l) and immunostained with anti-EEA1. (a–c,g–i): Confocal images of multilabeled samples (red, pDNA; dark blue, chitosan; and green, EEs). (d–f,j–l): Colocalized objects are indicated by voxels with positive PDM values after ICA analysis (yellow, pDNA colocalized with EEs; cyan, chitosan colocalized with EEs; and magenta, pDNA colocalized with chitosan). Arrows indicate colocalization between pDNA and EEs. Only cells containing pDNA and demonstrating colocalization are shown in the figure. In the cells incubated with pDNA-LCO for 15 min (a,d), uptake of pDNA was observed in only 57% of the cells. The deconvolved image stacks were surface-reconstructed in Amira (Visage Imaging). The scale bars represent 2 µm. 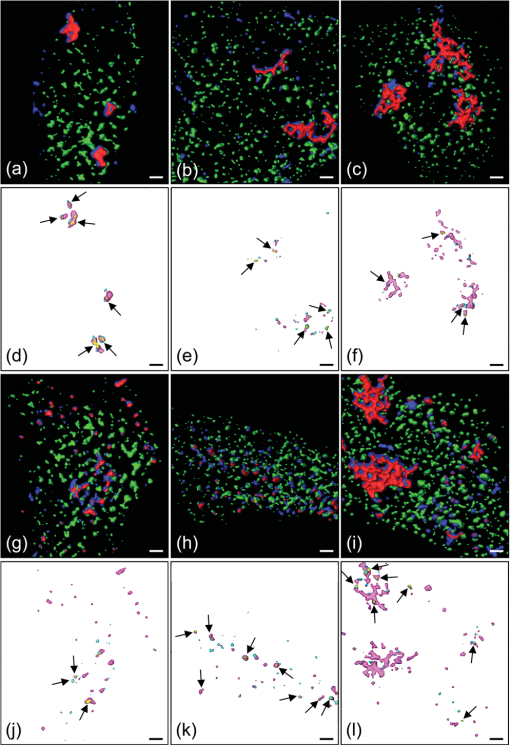 In cells incubated with pDNA-LCO for 15 min [Fig. 5(a)], free or complexed pDNA and LCOs were rarely observed in the EEs or other vesicles, and some polyplex aggregation was observed. More than half of the cells (57%) incubated for 15 min with LCO polyplexes did not contain any pDNA. All of the cells incubated with pDNA-SBTCO for 15 min [Fig. 5(g)] demonstrated polyplex uptake, and very little aggregation was observed. Free and complexed pDNA or SBTCO partly overlapped with EEs but mainly assembled in other small structures. After 1 and 3 h of incubation, polyplex aggregation, indicated by larger pDNA-chitosan structures, was observed in most of the cells incubated with pDNA-LCO [Figs. 5(b) and 5(c)], whereas pDNA-SBTCO aggregation mainly occurred after 3 h [Fig. 5(i)]. The large polyplex assemblies observed for both chitosans were apparently located intracellularly. However, confocal images may contain some contributions from out-of-focus light. Therefore, it could not be easily concluded whether these aggregates were internalized or partly attached to the cell surface. Although a few cells incubated with pDNA-LCO for 1 and 3 h showed atypical behavior with pDNA located in well-defined structures, free chitosan but less pDNA was generally observed in vesicles, compared with cells incubated with SBTCO polyplexes. 3.3.Quantitative Colocalization AnalysisQuantitative 3-D colocalization analysis was performed, and several colocalization parameters were determined (MCCs, ICQ, and PCC). 3-D reconstructions of images based on voxels with positive PDM values are presented to visualize colocalized objects [Figs. 5(d)–5(f), 5(j)–5(l)]. Quantitative results (Fig. 6) revealed a significant degree of colocalization between pDNA and chitosan for both LCOs and SBTCOs. Few polyplexes were located in EEs, but rather in other well-defined structures in the cytoplasma, and polyplexes formed by LCOs and SBTCOs showed different kinetics in intracellular trafficking. Fig. 6Quantitative colocalization analysis of HeLa cells that were incubated for 15 min, 1 h, or 3 h with pDNA complexed with LCOs or SBTCOs and immunostained with anti-EEA1. The fractions of objects in one channel colocalized with objects in another channel are presented by Manders’ colocalization coefficients (i.e., (a,e,h) and (b,f,i)). Colocalization was also quantified by the intensity correlation quotient (ICQ) (c,g,j) and by Pearson’s correlation coefficient (PCC) for pDNA and chitosan (d). Negative values of PCC were not considered to be valid and were excluded. The average values and standard deviations are based on to 35 cells. Colocalization after different incubation times were compared for polyplexes formed by the two chitosans; † indicates significant difference from 15 min, and ‡ indicates significant difference from 1 h. Significant differences between LCOs and SBTCOs for a respective incubation time are indicated by *. 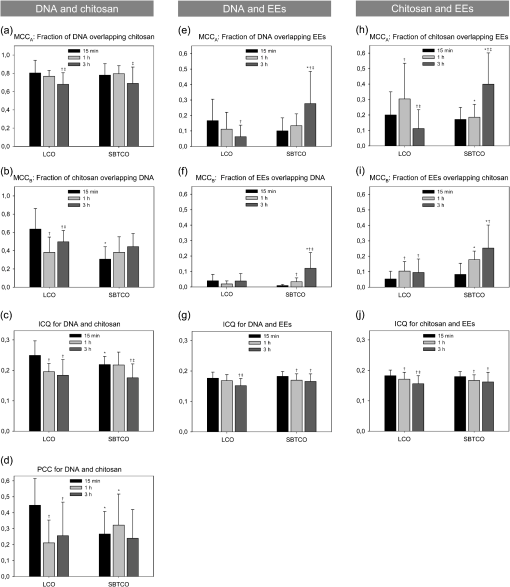 All of the parameters indicated a considerable degree of colocalization between pDNA and both chitosans at all of the incubation times, although most of the colocalization parameters decreased within 3 h of incubation. According to , the high fraction of pDNA colocalized with LCOs () and SBTCOs () after 15 min of incubation slightly decreased within 3 h ( and for LCOs and SBTCOs, respectively) [Fig. 6(a)], and no significant differences in colocalization () between pDNA and chitosan were observed for the two chitosans. For both polyplexes, the intracellular fractions of chitosans colocalized with pDNA () were lower than the fractions of pDNA colocalized with chitosans (), possibly due to an excess of free chitosan in the polyplex solution.49 The fraction of LCOs colocalized with pDNA after 15 min of incubation () was significantly higher than that of SBTCOs colocalized with pDNA () [Fig. 6(b)] but decreased and subsequently increased within 3 h. For cells incubated with polyplexes for 1 and 3 h, no significant differences were observed between the fractions of LCOs and SBTCOs colocalized with pDNA, and the latter remained unchanged for all of the incubation times. Furthermore, the ICQ for colocalization between pDNA and chitosan [Fig. 6(c)] was significantly higher at 15 min for LCOs () than for SBTCOs () and decreased faster for LCOs than for SBTCOs. Also, the PCC for pDNA and chitosan [Fig. 6(d)] was initially significantly higher for LCOs () than for SBTCOs (), but the situation was reversed after 1 h ( and for LCOs and SBTCOs, respectively). After 3 h of incubation, no significant differences between the pDNA complexed with LCOs and the pDNA complexed with SBTCOs were found for any of the colocalization parameters. Regarding the colocalization of pDNA and chitosan with EEs, the parameters do not take into account whether pDNA and chitosans were free or complexed. In general, the fractions of pDNA and chitosan colocalized with EEs were higher than the fractions of EEs colocalized with pDNA or chitosan, which indicates that most EEs did not contain pDNA or chitosans. indicated no significant difference between polyplexes formed by LCOs () and those formed by SBTCOs () with respect to the fractions of pDNA located in EEs after 15 min of incubation [Fig. 6(e)]. However, the kinetics for reaching the EEs in cells incubated with the two polyplexes were different. The fraction of pDNA located in EEs decreased with time for cells incubated with pDNA-LCO, whereas it reached, within 3 h, a more than four-fold increase compared with pDNA-LCO () for cells incubated with the SBTCO polyplexes (). The for the fraction of EEs colocalized with pDNA was initially low for both chitosans () and increased within 3 h to for cells incubated with pDNA-SBTCO [Fig. 6(f)], whereas it remained low for cells incubated with pDNA-LCO. A higher number of EEs was observed after 15 min and 1 h of incubation in cells incubated with pDNA-LCO compared with those incubated with pDNA-SBTCO, as estimated by the density of EEs (number of EEs per cell ROI volume). After 3 h of incubation, the number of EEs was equal (data not shown). Regarding the colocalization between free or complexed chitosan and EEs, indicated no significant differences after 15 min of incubation between the fractions of LCOs () and SBTCOs () colocalized with EEs [Fig. 6(h)]. However, after 3 h, the fraction of LCOs located with EEs had decreased to half its initial value, whereas the fraction of SBTCOs more than doubled its initial value. This trend is in accordance with the results for the fraction of pDNA in EEs for cells incubated with pDNA-SBTCO. The indicated that the fraction of EEs colocalized with chitosans was initially low and increased with time for both chitosans [Fig. 6(i)], reaching a value that was more than twice as high for SBTCOs () than for LCOs () within 3 h. The initial ICQ value of for the colocalization of EEs with pDNA or chitosan slightly decreased with time [Figs. 6(g) and 6(j)], and no significant differences were observed between the two chitosans at any time. Negative PCC values were reported for colocalization between EEs and pDNA or chitosans and were not considered valid. 4.DiscussionA quantitative 3-D colocalization analysis was used to study whether intracellular trafficking and the dissociation kinetics of pDNA-chitosan could explain the higher transfection efficacy of polyplexes formed by SBTCOs compared with those formed by LCOs.21,26,27 Most colocalization studies of the intracellular trafficking of polyplexes have been performed qualitatively by visual inspection of overlapping colors.1,2,5–7,50 This approach is possible if both channels display high fluorescence intensities. If the intensity in one of the channels is low, then the eye will conclude erroneously that the particles are not colocalized. Furthermore, colocalization has often been quantified by analyzing 2-D images without regard to image resolution or pre-processing, which both may have a great impact on quantitative results. Furthermore, 2-D lateral imaging gives incomplete information about the spatial organization of 3-D structures, which also have an axial dimension. Colocalization analysis of biological molecules and intracellular structures should therefore be performed in all three dimensions.8,9 In the present study, we performed quantitative 3-D colocalization analysis, and efforts were made to optimize the protocols for sample preparation, image acquisition, and pre-processing to minimize false colocalization events.10,11 4.1.Sample Preparation and Pre-Processing of ImagesThe results of the intracellular trafficking studies based on fluorescence imaging depend on cell type, cargo, sample preparation, choice of fluorescent dyes, imaging setup, and image pre-processing and analysis. Therefore, several considerations were taken when optimizing the experimental protocol of the present study. Although Cy5 as a DNA label may induce some aggregation of the polyplexes,49 its covalent attachment to DNA was preferred over intercalators. When immunostained samples are imaged, background fluorescence caused by nonspecific staining may reduce the SNR and, in turn, cause an overestimation of colocalization. To minimize the nonspecific staining, several procedures for sample preparation were preliminarily tested, and the cells were thoroughly washed during immunostaining to remove excess antibodies. Confocal microscopy is well suited for quantitative multichannel 3-D colocalization studies and has the advantage that the pinhole in front of the detector reduces contributions from out-of-focus light. However, the reduction of out-of-focus contributions may result in a loss of light and reduction in the SNR. Therefore, we chose pinhole sizes3 equal to and larger than 1 Airy unit when fulfilling the requirement of equal optical section thickness between the channels in quantitative multichannel 3-D colocalization analysis.51 Deconvolution and colocalization analyses require proper sampling of the image Z-stacks. According to the Nyquist sampling theorem, the image voxel size should be equal to the lateral or axial resolution limit divided by a minimum of two to avoid loss of spatial information (undersampling).8 Although oversampling reduces the signal intensity in each voxel to cause a dimmer image, it gives better image restoration when using 3-D deconvolution algorithms. Therefore, we sampled a minimum of three samples per resolvable element. Imaging artifacts, such as chromatic shifts between the channels, are not acceptable in colocalization analysis and were measured and corrected for, both laterally and axially. Chromatic shifts can be caused by several factors, such as chromatic aberrations, refraction index mismatch, irregularities and misalignments in the optical train, and/or mechanical shifts between filter cubes or dichroic mirrors.10,13,51 Image pre-processing influences quantitative colocalization results in several ways. Noise and blur in the raw images may induce randomly overlapping voxel intensities, in which there is no “real” colocalization. The reduction of noise and blur by background subtraction and image deconvolution may thus minimize false positive colocalization events.52 Chromatic shifts resulting from chromatic and mechanical aberrations also affect the overlap of voxels13,48 and increase or decrease the number of colocalizing voxels. In the present study, pre-processing had varying effects on the colocalization analysis. MCCs were highly influenced by image pre-processing and demonstrated, in general, a higher degree of colocalization in raw images than in pre-processed images. This effect was not unexpected, because MCCs are known to be highly affected by noise and image degradation.10,48 The effect from image pre-processing on the ICQ and PCC was more random and less pronounced. 4.2.Colocalization Analysis and Intracellular TraffickingTo obtain quantitative information, the colocalization was estimated by MCCs, ICQ, and PCC. Although the analysis is more robust when several parameters are used in combination, it is important to be aware of the slightly distinct interpretations of the colocalization parameters, and to remember that they may be suited for different cases. Before performing quantitative colocalization analysis, one should thus consider which parameter best provides information about the biological system under investigation. Whereas MCCs indicate colocalization even if the intensity is bright in one channel and dim in the other, the ICQ and PCC are based on the covariance of the voxel intensities. The ICQ and PCC may therefore be preferred when analyzing intermolecular binding and dissociation, such as interactions between pDNA and chitosan. When studying whether molecules are contained in vesicles or other organelles, a correlation of the intensity between the fluorescent labels of the cargo and those of the vesicle should not be expected,48 and we suggest that the MCCs are more reliable than the ICQ and PCC in such cases. This may be the reason the ICQ failed to identify the increasing colocalization with time between EEs and pDNA or SBTCOs, as reported by MCCs. MCCs may thus be more suitable to indicate the degree of colocalization between the particles and EEs. MCCs have the additional advantage of providing split information for the two channels, which means the amount of pDNA contained in the EEs can be studied independently of the amount of EEs involved in pDNA transport. This split information is in contrast to the ICQ and PCC, which indicate one combined value for the colocalization between the two channels. The PCC is the most commonly used quantitative estimate of colocalization for multichannel fluorescence imaging of biological samples,14,15 but it is only valid when a strong correlation of the voxel intensity occurs. PCC was only included for the correlation analysis between pDNA and chitosan, and it demonstrated partial colocalization in agreement with the other parameters. Effective gene delivery requires a balance between proper stability of the polyplexes to protect pDNA from degradation and pDNA release to activate transfection.22,53,54 Thus, studies of pDNA-chitosan dissociation are important.49 Colocalization between pDNA and chitosan in the cells was clearly indicated by MCCs, ICQ, and PCC. A reduction with time in these colocalization parameters indicates a transition from initially bound complexes to dissociation. Therefore, the faster decrease in ICQ and PCC for pDNA and LCOs compared with pDNA and SBTCOs indicates that the binding between pDNA and SBTCOs is more stable. The kinetics of polyplex dissociation may be crucial for efficient transfection. SBTCO polyplexes have demonstrated a slightly higher physical stability than pDNA-LCO, but also an earlier onset of transfection.21,49 This fine balance between polyplex stability and pDNA release may partly explain their higher transfection efficacy compared with pDNA-LCO.21,49 More pDNA was internalized by a higher number of cells when complexed with SBTCOs compared with LCOs, which has also recently been demonstrated.55,56 For both pDNA-LCO and pDNA-SBTCO, there was an excess of free chitosans in the cells. After polyplex preparation, the solution contains a substantial amount of unbound chitosans.49 The fraction of SBTCOs bound to pDNA in the polyplex solution has previously been reported to be lower than for LCOs.49 If the excess of free chitosan is taken up by the cells together with the polyplexes during incubation, it may explain why a higher fraction of free SBTCOs compared with free LCOs was observed intracellularly from the confocal images, and why the quantitative colocalization parameters demonstrated a higher initial uptake of free SBTCOs compared with LCOs. The higher excess of free SBTCO may contribute to an enhanced transfection efficacy due to chitosan’s buffering capacity.31 Particles internalized by clathrin-mediated endocytosis are known to be transported to EEs, and caveolae are also reported to fuse with EEs.34,36–38 Therefore, the internalization pathways of pDNA-chitosan polyplexes were studied by measuring colocalization with EEs. The colocalization analysis demonstrated that polyplexes formed by both chitosans were transported to EEs, but to a different extent and at different rates. In cells incubated with LCO polyplexes, the fraction of pDNA in EEs decreased with time during the 3 h of measurements, whereas the fraction of EEs colocalized with pDNA increased in cells incubated with SBTCO polyplexes. However, only a small fraction of the total pDNA and/or chitosan was contained in EEs, and the particles were located in the cytosol or in other well-defined, vesicular-like structures, particularly for samples incubated with pDNA-SBTCO. We have previously demonstrated that LCO polyplexes are taken up via both the clathrin- and caveolae-mediated pathways, whereas polyplexes formed by SBTCOs are primarily internalized by caveolae-mediated endocytosis.55,56 Most of the well-defined structures observed in the present study were therefore presumed to be a result of caveolae-mediated internalization. The difference in the kinetics of pDNA reaching EEs for the two types of polyplexes may occur because caveolae-mediated endocytosis is a slower internalization process and will deliver the cargo to the EEs later than clathrin-mediated endocytosis. Despite a higher uptake of pDNA-SBTCO compared with pDNA-LCO, a lower number of EEs was found in cells incubated with pDNA-SBTCO, which supports the idea that pDNA-SBTCO polyplexes are using a different endocytotic pathway. The decrease in the fractions of pDNA and LCOs in the EEs within 3 h may be attributed to particle transport from EEs to the cytosol or to LEs and lysosomes for degradation. The destination of the caveolae-mediated endocytosis is controversial, but most reports suggest that caveolae fuses with EEs, although some also suggest interaction with the Golgi apparatus or release of the cargo in the cytosol.34,36–38,57,58 Cargo internalized via this pathway may therefore escape lysosomal degradation, which may partly explain the higher transfection efficacy obtained for SBTCO polyplexes. The choice of endocytotic pathways and the degree of cellular uptake is partly determined by properties of the particle, such as charge and size.59,60 Although SBTCOs have a larger molecular weight (Table 1) and form polyplexes with a larger fraction of globular structures compared with LCOs,21,49 the observed aggregation of the particles may have a greater effect on the cellular uptake than small differences in the size and shape of single polyplexes. The observed higher tendency of pDNA-LCO, compared to pDNA-SBTCO, to aggregate and bind to the cell surface has also been reported previously.26,27,55,56 There are conflicting reports in the literature on how particle size affects the choice of uptake mechanisms.32,61 However, it has been suggested that the clathrin-mediated pathway is preferred by larger particles, whereas smaller particles are internalized by caveolin-coated pits.61 This is consistent with our results, considering that pDNA-LCO assembles into larger structures than pDNA-SBTCO. Altogether, the present colocalization study supports our previous results that polyplexes formed by LCOs and SBTCOs are internalized by different pathways. 5.ConclusionWe used a quantitative 3-D colocalization analysis to study dissociation and the intracellular trafficking of pDNA-chitosan polyplexes formed by LCOs and SBTCOs. Image deconvolution and correction for chromatic shifts in the confocal images were performed before colocalization analysis, and several colocalization parameters were determined. MCCs, which are independent of intensity variations between the two channels, were the more relevant parameter to study the localization of particles in intracellular compartments. ICQ and PCC, which are based on the covariance of the voxel intensities in the two channels, and the MCCs showed similar trends in molecular binding between pDNA and chitosan. The intracellular localization of the polyplexes, free pDNA, and free chitosans demonstrated differences between the two chitosans in the amount of cellular uptake, and in the intracellular pathways and rates of dissociation, which all may be important factors for effective transfection. Our results agree with previous conclusions that polyplexes formed by SBTCOs have properties that are more favorable for cellular uptake and intracellular trafficking compared with LCOs. This study demonstrates that quantitative 3-D colocalization analysis can yield valuable supportive and complementary information to other methods27,55,56 and help to explain the variations in the gene delivery efficiency of linear and branched pDNA-chitosan polyplexes. AcknowledgmentsWe gratefully acknowledge Kristin Sæterbø (Department of Physics, NTNU) for assistance with cell cultures and sample preparations and Michael Bruno Eric Marie (Department of Biomedicine, Molecular Imaging Center, University of Bergen) for advice about fixation procedures. ReferencesJ. RejmanA. BragonziM. Conese,
“Role of clathrin- and caveolae-mediated endocytosis in gene transfer mediated by lipo- and polyplexes,”
Mol. Ther., 12
(3), 468
–474
(2005). http://dx.doi.org/10.1016/j.ymthe.2005.03.038 1525-0016 Google Scholar
T. Lühmannet al.,
“Cellular uptake and intracellular pathways of PLL-g-PEG-DNA nanoparticles,”
Bioconjug. Chem., 19
(9), 1907
–1916
(2008). http://dx.doi.org/10.1021/bc800206r BCCHES 1043-1802 Google Scholar
S. V. Costeset al.,
“Automatic and quantitative measurement of protein-protein colocalization in live cells,”
Biophys. J., 86
(6), 3993
–4003
(2004). http://dx.doi.org/10.1529/biophysj.103.038422 BIOJAU 0006-3495 Google Scholar
M. Thibaultet al.,
“Intracellular trafficking and decondensation kinetics of chitosan-pDNA polyplexes,”
Mol. Ther., 18
(10), 1787
–1795
(2010). http://dx.doi.org/10.1038/mt.2010.143 1525-0016 Google Scholar
W. T. GodbeyK. K. WuA. G. Mikos,
“Tracking the intracellular path of poly(ethylenimine)/DNA complexes for gene delivery,”
Proc. Natl. Acad. Sci. U.S.A., 96
(9), 5177
–5181
(1999). http://dx.doi.org/10.1073/pnas.96.9.5177 PNASA6 0027-8424 Google Scholar
N. P. GabrielsonD. W. Pack,
“Efficient polyethylenimine-mediated gene delivery proceeds via a caveolar pathway in HeLa cells,”
J. Controlled Release, 136
(1), 54
–61
(2009). http://dx.doi.org/10.1016/j.jconrel.2009.02.003 JCREEC 0168-3659 Google Scholar
S. F. Penget al.,
“Mechanisms of cellular uptake and intracellular trafficking with chitosan/DNA/poly(gamma-glutamic acid) complexes as a gene delivery vector,”
Biomaterials, 32
(1), 239
–248
(2011). http://dx.doi.org/10.1016/j.biomaterials.2010.08.081 BIMADU 0142-9612 Google Scholar
S. BolteF. P. Cordelieres,
“A guided tour into subcellular colocalization analysis in light microscopy,”
J. Microsc., 224
(Pt. 3), 213
–232
(2006). http://dx.doi.org/10.1111/jmi.2006.224.issue-3 JMICAR 0022-2720 Google Scholar
Q. Liet al.,
“A syntaxin 1, Galpha(o), and N-type calcium channel complex at a presynaptic nerve terminal: analysis by quantitative immunocolocalization,”
J. Neurosci., 24
(16), 4070
–4081
(2004). http://dx.doi.org/10.1523/JNEUROSCI.0346-04.2004 JNRSDS 0270-6474 Google Scholar
T. AbrahamS. E. AllanM. K. Levings,
“Deconvolution and chromatic aberration corrections in quantifying colocalization of a transcription factor in three-dimensional cellular space,”
Micron, 41
(6), 633
–640
(2010). http://dx.doi.org/10.1016/j.micron.2010.03.005 MICNB2 0047-7206 Google Scholar
V. ZinchukO. Grossenbacher-Zinchuk,
“Recent advances in quantitative colocalization analysis: focus on neuroscience,”
Prog. Histochem. Cytochem., 44
(3), 125
–172
(2009). http://dx.doi.org/10.1016/j.proghi.2009.03.001 PHCCAS 0079-6336 Google Scholar
F. Sedaratet al.,
“Deconvolution of confocal images of dihydropyridine and ryanodine receptors in developing cardiomyocytes,”
J. Appl. Physiol., 97
(3), 1098
–1103
(2004). http://dx.doi.org/10.1152/japplphysiol.00089.2004 JAPYAA 0021-8987 Google Scholar
E. M. M. Manders,
“Chromatic shift in multicolour confocal microscopy,”
J. Microsc., 185
(Pt. 3), 321
–328
(1997). http://dx.doi.org/10.1046/j.1365-2818.1997.d01-625.x JMICAR 0022-2720 Google Scholar
E. M. M. MandersF. J. VerbeekJ. A. Aten,
“Measurement of colocalization of objects in dual-color confocal images,”
J. Microsc., 169
(Pt. 3), 375
–382
(1993). http://dx.doi.org/10.1111/jmi.1993.169.issue-3 JMICAR 0022-2720 Google Scholar
E. M. M. Manderset al.,
“Dynamics of 3-dimensional replication patterns during the S-phase, analyzed by double labeling of DNA and confocal microscopy,”
J. Cell Sci., 103
(Pt. 3), 857
–862
(1992). JNCSAI 0021-9533 Google Scholar
J. Boutet de MonvelS. Le CalvezM. Ulfendahl,
“Image restoration for confocal microscopy: improving the limits of deconvolution, with application to the visualization of the mammalian hearing organ,”
Biophys. J., 80
(5), 2455
–2470
(2001). http://dx.doi.org/10.1016/S0006-3495(01)76214-5 BIOJAU 0006-3495 Google Scholar
W. WallaceL. H. SchaeferJ. R. Swedlow,
“A working person’s guide to deconvolution in light microscopy,”
Biotechniques, 31
(5), 1076
–1097
(2001). BTNQDO 0736-6205 Google Scholar
J. R. Swedlow,
“Quantitative fluorescence microscopy and image deconvolution,”
Meth. Cell Biol., 81 447
–465
(2007). http://dx.doi.org/10.1016/S0091-679X(06)81021-6 MCBLAG 0091-679X Google Scholar
S. C. W. RichardsonH. V. J. KolbeR. Duncan,
“Potential of low molecular mass chitosan as a DNA delivery system: biocompatibility, body distribution and ability to complex and protect DNA,”
Int. J. Pharm., 178
(2), 231
–243
(1999). http://dx.doi.org/10.1016/S0378-5173(98)00378-0 IJPHDE 0378-5173 Google Scholar
M. Köping-Höggårdet al.,
“Chitosan as a nonviral gene delivery system. Structure-property relationships and characteristics compared with polyethylenimine in vitro and after lung administration in vivo,”
Gene Ther., 8
(14), 1108
–1121
(2001). http://dx.doi.org/10.1038/sj.gt.3301492 GETHEC 0969-7128 Google Scholar
S. P. Strandet al.,
“Tailoring of chitosans for gene delivery: novel self-branched glycosylated chitosan oligomers with improved functional properties,”
Biomacromolecules, 9
(11), 3268
–3276
(2008). http://dx.doi.org/10.1021/bm800832u BOMAF6 1525-7797 Google Scholar
M. Köping-Höggårdet al.,
“Improved chitosan-mediated gene delivery based on easily dissociated chitosan polyplexes of highly defined chitosan oligomers,”
Gene Ther., 11
(19), 1441
–1452
(2004). http://dx.doi.org/10.1038/sj.gt.3302312 GETHEC 0969-7128 Google Scholar
M. Lavertuet al.,
“High efficiency gene transfer using chitosan/DNA nanoparticles with specific combinations of molecular weight and degree of deacetylation,”
Biomaterials, 27
(27), 4815
–4824
(2006). http://dx.doi.org/10.1016/j.biomaterials.2006.04.029 BIMADU 0142-9612 Google Scholar
S. DanielsenK. M. VårumB. T. Stokke,
“Structural analysis of chitosan mediated DNA condensation by AFM: influence of chitosan molecular parameters,”
Biomacromolecules, 5
(3), 928
–936
(2004). http://dx.doi.org/10.1021/bm034502r BOMAF6 1525-7797 Google Scholar
S. P. Strandet al.,
“Influence of chitosan structure on the formation and stability of DNA-chitosan polyelectrolyte complexes,”
Biomacromolecules, 6
(6), 3357
–3366
(2005). http://dx.doi.org/10.1021/bm0503726 BOMAF6 1525-7797 Google Scholar
M. M. Issaet al.,
“Targeted gene delivery with trisaccharide-substituted chitosan oligomers in vitro and after lung administration in vivo,”
J. Controlled Release, 115
(1), 103
–112
(2006). http://dx.doi.org/10.1016/j.jconrel.2006.06.029 JCREEC 0168-3659 Google Scholar
J. MalmoK. M. VårumS. P. Strand,
“Effect of chitosan chain architecture on gene delivery: comparison of self-branched and linear chitosans,”
Biomacromolecules, 12
(3), 721
–729
(2011). http://dx.doi.org/10.1021/bm1013525 BOMAF6 1525-7797 Google Scholar
K. von Gersdorffet al.,
“The internalization route resulting in successful gene expression depends on both cell line and polyethylenimine polyplex type,”
Mol. Ther., 14
(5), 745
–753
(2006). http://dx.doi.org/10.1016/j.ymthe.2006.07.006 1525-0016 Google Scholar
C. PichonL. BillietP. Midoux,
“Chemical vectors for gene delivery: uptake and intracellular trafficking,”
Curr. Opin. Biotechnol., 21
(5), 640
–645
(2010). http://dx.doi.org/10.1016/j.copbio.2010.07.003 CUOBE3 0958-1669 Google Scholar
I. A. Khalilet al.,
“Uptake pathways and subsequent intracellular trafficking in nonviral gene delivery,”
Pharmacol. Rev., 58
(1), 32
–45
(2006). http://dx.doi.org/10.1124/pr.58.1.8 PAREAQ 0031-6997 Google Scholar
N. D. SonawaneF. C. SzokaA. S. Verkman,
“Chloride accumulation and swelling in endosomes enhances DNA transfer by polyamine-DNA polyplexes,”
J. Biol. Chem., 278
(45), 44826
–44831
(2003). http://dx.doi.org/10.1074/jbc.M308643200 JBCHA3 0021-9258 Google Scholar
J. Rejmanet al.,
“Size-dependent internalization of particles via the pathways of clathrin- and caveolae-mediated endocytosis,”
Biochem. J., 377
(Pt. 1), 159
–169
(2004). http://dx.doi.org/10.1042/BJ20031253 BIJOAK 0264-6021 Google Scholar
M. A. van der Aaet al.,
“Cellular uptake of cationic polymer-DNA complexes via caveolae plays a pivotal role in gene transfection in COS-7 cells,”
Pharm. Res., 24
(8), 1590
–1598
(2007). http://dx.doi.org/10.1007/s11095-007-9287-3 PHREEB 0724-8741 Google Scholar
A. Hayeret al.,
“Caveolin-1 is ubiquitinated and targeted to intralumenal vesicles in endolysosomes for degradation,”
J. Cell Biol., 191
(3), 615
–629
(2010). http://dx.doi.org/10.1083/jcb.201003086 JCLBA3 0021-9525 Google Scholar
B. Nichols,
“Caveosomes and endocytosis of lipid rafts,”
J. Cell Sci., 116
(23), 4707
–4714
(2003). http://dx.doi.org/10.1242/jcs.00840 JNCSAI 0021-9533 Google Scholar
L. Pelkmanset al.,
“Caveolin-stabilized membrane domains as multifunctional transport and sorting devices in endocytic membrane traffic,”
Cell, 118
(6), 767
–780
(2004). http://dx.doi.org/10.1016/j.cell.2004.09.003 CELLB5 0092-8674 Google Scholar
R. G. PartonK. Simons,
“The multiple faces of caveolae,”
Nat. Rev. Mol. Cell. Biol., 8
(3), 185
–194
(2007). http://dx.doi.org/10.1038/nrm2122 NRMCBP 1471-0072 Google Scholar
A. W. WongS. J. ScalesD. E. Reilly,
“DNA internalized via caveolae requires microtubule-dependent, Rab7-independent transport to the late endocytic pathway for delivery to the nucleus,”
J. Biol. Chem., 282
(31), 22953
–22963
(2007). http://dx.doi.org/10.1074/jbc.M611015200 JBCHA3 0021-9258 Google Scholar
L. PelkmansJ. KartenbeckA. Helenius,
“Caveolar endocytosis of simian virus 40 reveals a new two-step vesicular-transport pathway to the ER,”
Nat. Cell. Biol., 3
(5), 473
–483
(2001). http://dx.doi.org/10.1038/35074539 NCBIFN 1465-7392 Google Scholar
L. PelkmansA. Helenius,
“Endocytosis via caveolae,”
Traffic, 3
(5), 311
–320
(2002). http://dx.doi.org/10.1034/j.1600-0854.2002.30501.x 1398-9219 Google Scholar
R. Heilbronnet al.,
“ssDNA-dependent colocalization of adeno-associated virus Rep and herpes simplex virus ICP8 in nuclear replication domains,”
Nucleic Acids Res., 31
(21), 6206
–6213
(2003). http://dx.doi.org/10.1093/nar/gkg827 NARHAD 0305-1048 Google Scholar
P. Marbetet al.,
“Quantitative microscopy reveals 3D organization and kinetics of endocytosis in rat hepatocytes,”
Microsc. Res. Technol., 69
(9), 693
–707
(2006). http://dx.doi.org/10.1002/(ISSN)1097-0029 MRTEEO 1059-910X Google Scholar
D. E. Ferraraet al.,
“Quantitative 3D fluorescence technique for the analysis of en face preparations of arterial walls using quantum dot nanocrystals and two-photon excitation laser scanning microscopy,”
Am. J. Physiol. Regul. Integr. Comp. Physiol., 290
(1), R114
–R123
(2006). http://dx.doi.org/10.1152/ajpregu.00449.2005 0363-6119 Google Scholar
S. Wörzet al.,
“3D geometry-based quantification of colocalizations in multichannel 3D microscopy images of human soft tissue tumors,”
IEEE Trans. Med. Imaging, 29
(8), 1474
–1484
(2010). http://dx.doi.org/10.1109/TMI.2010.2049857 ITMID4 0278-0062 Google Scholar
K. Tømmeraaset al.,
“Preparation and characterisation of oligosaccharides produced by nitrous acid depolymerisation of chitosans,”
Carbohydr. Res., 333
(2), 137
–144
(2001). http://dx.doi.org/10.1016/S0008-6215(01)00130-6 CRBRAT 0008-6215 Google Scholar
K. Tømmeraaset al.,
“Preparation and characterisation of chitosans with oligosaccharide branches,”
Carbohydr. Res., 337
(24), 2455
–2462
(2002). http://dx.doi.org/10.1016/S0008-6215(02)00334-8 CRBRAT 0008-6215 Google Scholar
W. S. Rasband,
“ImageJ, National U. S. Institutes of Health,”
(1997) http://imagej.nih.gov/ij/ Google Scholar
K. W. DunnM. M. KamockaJ. H. McDonald,
“A practical guide to evaluating colocalization in biological microscopy,”
Am. J. Physiol. Cell. Physiol., 300
(4), C723
–C742
(2011). http://dx.doi.org/10.1152/ajpcell.00462.2010 0363-6143 Google Scholar
N. K. Reitanet al.,
“Characterizing DNA condensation by structurally different chitosans of variable gene transfer efficacy,”
Biomacromolecules, 10
(6), 1508
–1515
(2009). http://dx.doi.org/10.1021/bm900102d BOMAF6 1525-7797 Google Scholar
P. M. McLendonK. M. FichterT. M. Reineke,
“Poly(glycoamidoamine) vehicles promote pDNA uptake through multiple routes and efficient gene expression via caveolae-mediated endocytosis,”
Mol. Pharm., 7
(3), 738
–750
(2010). http://dx.doi.org/10.1021/mp900282e MOPMA3 0026-895X Google Scholar
A. J. North,
“Seeing is believing? A beginners’ guide to practical pitfalls in image acquisition,”
J. Cell Biol., 172
(1), 9
–18
(2006). http://dx.doi.org/10.1083/jcb.200507103 JCLBA3 0021-9525 Google Scholar
L. Landmann,
“Deconvolution improves colocalization analysis of multiple fluorochromes in 3D confocal data sets more than filtering techniques,”
J. Microsc., 208
(Pt. 2), 134
–147
(2002). http://dx.doi.org/10.1046/j.1365-2818.2002.01068.x JMICAR 0022-2720 Google Scholar
H. H. Chenet al.,
“Quantitative comparison of intracellular unpacking kinetics of polyplexes by a model constructed from quantum Dot-FRET,”
Mol. Ther., 16
(2), 324
–332
(2008). http://dx.doi.org/10.1038/sj.mt.6300392 1525-0016 Google Scholar
D. V. Schafferet al.,
“Vector unpacking as a potential barrier for receptor-mediated polyplex gene delivery,”
Biotechnol. Bioeng., 67
(5), 598
–606
(2000). http://dx.doi.org/10.1002/(ISSN)1097-0290 BIBIAU 0006-3592 Google Scholar
Z. Garaiovaet al.,
“Cellular uptake and photochemical internalization of DNA-chitosan nanoparticles: the role of clathrin- and caveolae-mediated pathways,”
Biomacromolecules, BOMAF6 1525-7797 Google Scholar
C. de L. Davieset al.,
“DNA-chitosan nanoparticles in gene delivery: endocytotic pathways and intracellular trafficking,”
in Proceedings #5404, 102th Annual Meeting American Assoc. Cancer Res.,
(2011). Google Scholar
J. S. ShinS. N. Abraham,
“Cell biology. Caveolae—not just craters in the cellular landscape,”
Science, 293
(5534), 1447
–1448
(2001). http://dx.doi.org/10.1126/science.1061079 SCIEAS 0036-8075 Google Scholar
R. G. PartonA. A. Richards,
“Lipid rafts and caveolae as portals for endocytosis: new insights and common mechanisms,”
Traffic, 4
(11), 724
–738
(2003). http://dx.doi.org/10.1034/j.1600-0854.2003.00128.x 1398-9219 Google Scholar
T. -G. IversenT. SkotlandK. Sandvig,
“Endocytosis and intracellular transport of nanoparticles: present knowledge and need for future studies,”
Nano Today, 6
(2), 176
–185
(2011). http://dx.doi.org/10.1016/j.nantod.2011.02.003 1748-0132 Google Scholar
J. AdlerI. Parmryd,
“Quantifying colocalization by correlation: the Pearson correlation coefficient is superior to the Manders’ overlap coefficient,”
Cytometry A, 77
(8), 733
–742
(2010). http://dx.doi.org/10.1002/cyto.a.20896 1552-4922 Google Scholar
S. Grosseet al.,
“Potocytosis and cellular exit of complexes as cellular pathways for gene delivery by polycations,”
J. Gene Med., 7
(10), 1275
–1286
(2005). http://dx.doi.org/10.1002/(ISSN)1521-2254 1099-498X Google Scholar
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||

