|
|
1.IntroductionLung cancer ranked first in terms of incidence and mortality, with a five-year survival rate of 15%.1 The routine diagnosing process includes a series of examinations such as physical examination, imaging tests, and biopsies, which are time-consuming and invasive.2 Raman spectroscopy provides molecular changes of tissues and biofluids in the human body, but the low sensitivity obstructs its application. Surface-enhanced Raman spectroscopy (SERS) can increase the intensity of normal Raman (NR) by an order of 4 to 15 owing to different substrates,3,4 and it has been widely used in the low-concentration biofluid detection that NR is hardly qualified to perform.5–7 Saliva has shown the ability to predict many diseases via several methods, including liquid chromatography, mass spectrometry,8 and enzyme-linked immunosorbent assay (ELISA).9 But most of them are time-consuming and require a large sample compared to SERS.10 Because of the mechanism of SERS, certain medications of nano-metal particles are usually used, such as biotinylated Ag powder11 and DNA aptamers capped Ag-nanoparticle.12 Nevertheless, some studies that apply naked SERS-active nanoparticles to the detection of body fluids have been performed. SERS of serum has exhibited differences between type II diabetes mellitus and diabetic complication groups at both the Raman shifts and the principal components.6 A concentration of 1 g/mL of 5-fluorouracil in saliva has been measured by the removal of thiocyanate and a pretreatment of SERS-active capillaries.13 A difference in saliva SERS between healthy people and oral-cancer patients at three wavenumbers has been detected using a gold nanoparticle assembly.14 In addition, SERS has been used in the differentiation of saliva for the AIDS virus by a support vector machine algorithm.15 Although it has not been studied very much, we have seen the potential of using SERS of saliva as a tool for the diagnosis of diseases. In this paper, the saliva SERS of 21 lung cancer patients and 20 healthy people was measured. The peak variations between the healthy and lung cancer groups were investigated for the diagnosis of lung cancer. Oven-heated silver colloids were used as the SERS substrate, using capillaries as the holder of the mixture of silver colloid and liquid-state saliva to reserve the bioactivity. Then, peaks were assigned in accordance with the possible components of saliva. Finally, multivariate analysis of principal component analysis (PCA) combined with linear discriminant analysis (LDA) were used on the spectroscopy to observe the diagnostic accuracy. 2.Materials and Methods2.1.Saliva SamplingWhole saliva was collected from 21 lung cancer patients and 20 healthy people. In each case, about 1 mL of saliva was collected between 9:00 a.m. and 11:00 a.m. to prevent the interference of food. Saliva providers were asked to rinse their mouths three times using pure water. A nonstimulated collection method was used: providers spit the saliva into a disposable cup. Then the saliva was transferred to a 1.5-mL centrifuge tube and was centrifuged at 14,000 rpm for 10 min to remove the oral mucous epithelial cells and food debris.8 The whole collection process was performed at a temperature of 4 °C. The average age of the lung cancer patients was 56, and that of the healthy volunteers was 60. Most of the patients were smokers (Table 1). For better comparison between the two groups, a control group with similar age and smoking history was also selected. Table 1Demographics of the study population.
2.2.SERS MeasurementSilver hydrosol was synthesized by the deoxidizing method, using nanoparticles and silver nitrate heated in a microwave oven, that is described in Liu’s article.16 This method can produce more uniform silver nanoparticles due to the heating mode of the microwave oven. A 2-μL sample of centrifuged saliva and 2 μL of silver colloid were mixed thoroughly by an oscillator for 10 min, and then the mixture was sucked into capillaries for later spectroscopy collection. Spectroscopy was collected from an inverted microscope (inVia, Renishaw plc., England) in the range of to , with a 632.8-nm He-Ne laser at a power maintained at 3.5 mW. The exposure time is 100 s. 2.3.Spectral AnalysisRaw spectra were processed by vector normalization, smoothing, and baseline correction to avoid errors or artificial interference during the sample preparation and spectral acquisition. Changes of Raman peaks between the two groups were compared, and the intensity change was defined as:17 Raman peaks were assigned in accordance with the specifications of Raman spectroscopy or SERS of tissues, body fluids, or bio-molecules. Peaks with wavelength difference within were seen as the same peak, considering different detection conditions and experimental errors. PCA-LDA algorithm was used to discriminate the saliva SERS of the lung cancer group from the control group. PCA is very effective in dimension reduction and can transform a large amount of data into fewer new variables. LDA is a linear method used to classify two or more groups, and it is often combined with PCA. PCA-LDA has been widely used in the classification of Raman spectroscopy for disease diagnosis, and high prediction results have been obtained.18,19 We used the Curve Fitting Toolbox and Statistical Toolbox in the Matlab software (The Mathworks Inc., Natick, MA) in our data processing. 3.Results and Discussion3.1.Raman SpectroscopyThe saliva SERS standard deviation (SD) of both the lung cancer group () and the control group () were compared (Fig. 1). There were 15 major peaks at the wavelengths of about 523, 622, 696, 735, 789, 822, 884, 909, 925, 1009, 1077, 1280, 1369, 1393, and . Fig. 1(a) Comparison of the average spectra for the lung cancer patients (red line, ) versus that of the normal group (green line, ). The broken lines represent the standard deviations of the averages. Also shown at the bottom (blue line) is the difference spectrum. (b) Plot showing the average intensity value and the corresponding standard deviations of the selected peaks. The peaks that are marked wavenumbers are those that have values lower than 0.05 of Student’s test.  Some differences between the two groups still existed, and the difference spectrum made them apparent. The major changes were at the nine wavenumbers of 822, 884, 909, 925, 1009, 1077, 1369, 1393, and . Most of the peaks decrease between the control group to the lung cancer group. At 822, 884, 925, 1009, 1280, 1369, and (Student’s test, ), the peak intensities were greater for the control group than for the lung cancer group, while the bands at 909, 1077, and were more intense in the saliva of the lung cancer patients (Fig. 1). Those Raman peaks were assigned to amino acids and nucleic acid bases (Table 2), and the decreases of peak intensities represented the corresponding decrease of those bio-molecules. Table 2Assignments of SERS peaks and the changes between healthy people and lung cancer patients. (Peaks were regarded as the same Raman peaks with a wavenumber difference of less than 10 cm−1.)
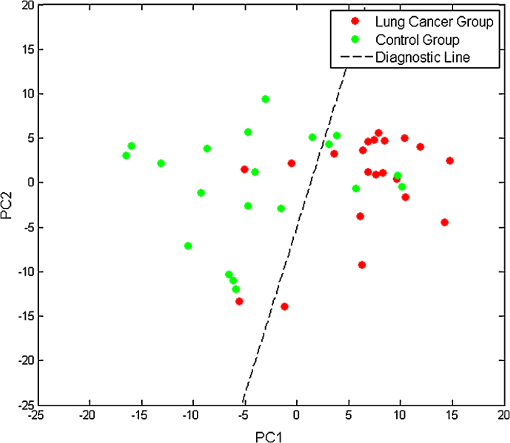
Nucleic acid bases: A (adenine), T (thymine), C (cytosine), U (uracil), G (guanine)Amino acids: Trp (tryptophan), Phe (phenylalanine), Lys (lysine), Pro (proline), Glu (glutamate), Val (valine), Gly (glycine), Hyd (hydroxyprolin), Met (methionine). 3.2.PCA-LDAPCA was used on our preprocessed SERS of saliva. The contribution rate to the total variation of spectra of the first two PCs (PC1 and PC2) was 83%; hence, the first two principal components can account for 83% of the total variability. Therefore, only the first two PCs were used for further analysis. The scatterplot of the two groups was obtained using the first two PCs, the spots representing different groups distributed separately, and they can be discriminated well. LDA was used on the first two PCs to examine the diagnostic ability of our method. The Fisher’s discriminant function (diagnostic line) of LDA is as follows: Most of the spots belonged to different groups distributed at the two sides of the diagnostic line (Fig. 2), which means that our SERS can be discriminated well using PCA-LDA. An accuracy of 86%, a sensitivity of 94%, and a specificity of 81% were obtained (Table 3). Table 3Diagnostic results of PCA-LDA on the SERS of saliva.
4.DiscussionAlthough some of the peaks were assigned to certain unit structures of proteins or nucleic acids (that is, certain amino acids or nucleic acid bases), the bonds existed in most of the units. For example, existed in all 20 common amino acids, all 5 kinds of nucleic acids have the structure of , and 3 kinds of amino acids in the aromatic group have aromatic rings. So the assignment may be expanded to those series of molecules. Such as the peak at , it was assigned to both phenylalanine20 and other proteins.21 Our results showed an overall decrease of protein and nucleic acids in the lung cancer group than in the control group. This decrease trend may have the same cause as the Raman spectroscopy of human lung epithelial tumor cells, which is induced by the breakdown of several kinds of molecular bonds.22 As blood is one of the means for disease diagnosis, and saliva is the ultra-filtration of blood, saliva can reflect the condition of tissues to a certain degree. Hence, the decrease of intensity of certain Raman bands of saliva might be a reflection of lung tissue. Further research is needed to determine if there is a distinct corresponding relationship. SERS is not the pure enhancement of NR. There are some wavelength shifts in SERS and new peaks will appear compared to NR.23 As the fingerprint property of Raman spectroscopy, the same bond will have different Raman shifts in different groups. For example, stretch vibration has a Raman shift as in lauric acid, but it moves to in myristic acid, and in palmitic acid.24 Therefore, certain errors in the peak assignments based on the literature might exist due to totally different measuring conditions. Some high-abundance proteins, like amylase, lysozyme, and mucin25 in saliva, have a very small diagnosis significance and might immerse the Raman peaks of low-abundance biomarkers. Removal of those substances can exclude the disturbance and improve the expression of those low-abundance biomarkers. Silver colloid in SERS is more uniform than that made by traditional water-heating, but the Ag particles are smaller; thus, there is a consequent decrease in the enhancement level. A highly ordered substrate can create a higher enhancement and more reproducible results. SERS effect is influenced by many factors, like laser power, temperature, solvent of samples, SERS substrate, the state of the analyte, and exposure time. The uniformity of SERS spectrometers and measuring conditions can eliminate the above fluctuations greatly and is the future orientation of our research. 5.ConclusionSaliva has shown the potential to diagnose many diseases, and recently, SERS has been used in the detection of saliva because of the high enhancement effect. The peak changes between the lung cancer group and the control group and an 80% diagnosis accuracy in our study showed the potential of saliva SERS for diagnosing lung cancer. SERS of saliva has several advantages, such as fast, noninvasive, and low-analyte requirements, and it has prospective clinic applications. High-abundance proteins will be depleted, and better-ordered SERS substrate will be introduced in our future experiments to eliminate the disturbance of those proteins and increase the enhancement and the reproducibility of SERS measurements. Possible saliva biomarkers will be selected and detected for more exact peak assignments. AcknowledgmentsWe thank The Fourth Affiliated Hospital of China Medical University for providing the saliva sample, and Dalian University of Technology for the technical support on SERS measurement. ReferencesD. M. Parkinet al.,
“Global Cancer Statistics, 2002,”
CA Cancer J. Clin., 55
(2), 74
–108
(2005). http://dx.doi.org/10.3322/canjclin.55.2.74 CAMCAM 0007-9235 Google Scholar
J. Langwith,
“Diagnosis and Staging,”
Lung Cancer, 71
–77 Greenhaven Press, New York
(2007). Google Scholar
X. M. Linet al.,
“Surface-enhanced Raman spectroscopy: substrate-related issues,”
Anal. Bioanal. Chem., 394
(7), 1729
–1745
(2009). http://dx.doi.org/10.1007/s00216-009-2761-5 ABCNBP 1618-2642 Google Scholar
S. NieS. R. Emory,
“Probing single molecules and single nanoparticles by surface-enhanced Raman scattering,”
Science, 275
(5303), 1102
–1106
(1997). http://dx.doi.org/10.1126/science.275.5303.1102 SCIEAS 0036-8075 Google Scholar
S. Fenget al.,
“Nasopharyngeal cancer detection based on blood plasma surface-enhanced Raman spectroscopy and multivariate analysis,”
Biosens. Bioelectron., 25
(11), 2414
–2419
(2010). http://dx.doi.org/10.1016/j.bios.2010.03.033 BBIOE4 0956-5663 Google Scholar
H. Hanet al.,
“Analysis of serum from type II diabetes mellitus and diabetic complication using surface-enhanced Raman spectra (SERS),”
Appl. Phys. B: Lasers Optics, 94
(4), 667
–672
(2009). Google Scholar
K. VirklerI. K. Lednev,
“Forensic body fluid identification: the Raman spectroscopic signature of saliva,”
Analyst, 135
(3), 512
–517
(2010). ANLYAG 0365-4885 Google Scholar
S. Huet al.,
“Salivary proteomics for oral cancer biomarker discovery,”
Clin. Cancer Res., 14
(19), 6246
–6252
(2008). http://dx.doi.org/10.1158/1078-0432.CCR-07-5037 CCREF4 1078-0432 Google Scholar
M. N. Brookset al.,
“Salivary protein factors are elevated in breast cancer patients,”
Mol. Med. Rep., 1
(3), 375
–378
(2008). MMROA5 1791-2997 Google Scholar
X. X. HanB. ZhaoY. Ozaki,
“Surface-enhanced Raman scattering for protein detection.,”
Anal. Bioanal. Chem., 394
(7), 1719
–1727
(2009). http://dx.doi.org/10.1007/s00216-009-2702-3 ABCNBP 1618-2642 Google Scholar
K. KimH. K. ParkN. H. Kim,
“Silver-particle-based surface-enhanced Raman scattering spectroscopy for biomolecular sensing and recognition,”
Langmuir, 22
(7), 3421
–3427
(2006). http://dx.doi.org/10.1021/la052912q LANGD5 0743-7463 Google Scholar
L. Fabriset al.,
“Aptatag-based multiplexed assay for protein detection by surface-enhanced Raman spectroscopy,”
Small, 6
(14), 1550
–1557
(2010). http://dx.doi.org/10.1002/smll.v6:14 1613-6829 Google Scholar
S. Farquharsonet al.,
“Detection of 5-fluorouracil in saliva using surface-enhanced Raman spectroscopy,”
Vibrational Spectrosc., 38
(1–2), 79
–84
(2005). http://dx.doi.org/10.1016/j.vibspec.2005.02.021 VISPEK 0924-2031 Google Scholar
K. W. Khoet al.,
“Surface-enhanced Raman spectroscopic (SERS) study of saliva in the early detection of oral cancer,”
Proc. SPIE, 5702 84
–91 2005). http://dx.doi.org/10.1117/12.590142 Google Scholar
W. Yanet al.,
“Preliminary study on the quick detection of Acquired Immune Deficiency Syndrome by saliva analysis using surface-enhanced Raman spectroscopic technique,”
in 31st Annual Int Conf IEEE EMBS,
885
–887
(2009). Google Scholar
K. Liuet al.,
“Surface-enhanced Raman spectrum of rat serum in the novel silver colloid,”
Spectroscopy Spectral Anal., 28
(2), 339
–342
(2008). Google Scholar
Z. ZhangY. Xu,
“The molecular mechanism of the photosensitization of photofrin (YHPD),”
Sci. China Chem., 21
(6), 595
–601
(1991). SCCCCS 1869-1870 Google Scholar
A. T. Harriset al.,
“Potential for Raman spectroscopy to provide cancer screening using a peripheral blood sample,”
Head Neck Oncol., 1
(1), 34
(2009). HNOEAE 1758-3284 Google Scholar
S. K. Tehet al.,
“Diagnostic potential of near-infrared Raman spectroscopy in the stomach: differentiating dysplasia from normal tissue,”
Brit. J. Cancer, 98
(2), 457
–465
(2008). http://dx.doi.org/10.1038/sj.bjc.6604176 BJCAAI 0007-0920 Google Scholar
M. C. ChenR. C. LordR. Mendelsohn,
“Laser-excited Raman spectroscopy of biomolecules, V. conformational changes associated with the chemical denaturation of lysozyme,”
J. Am Chem. Soc, 96
(10), 3038
–3042
(1974). http://dx.doi.org/10.1021/ja00817a003 JACSAT 0002-7863 Google Scholar
J. L. Pichardo-Molinaet al.,
“Raman spectroscopy and multivariate analysis of serum samples from breast cancer patients,”
Lasers Med. Sci., 22
(4), 229
–236
(2007). Google Scholar
I. Notingheret al.,
“Spectroscopic study of human lung epithelial cells (A549) in culture: living cells versus dead cells,”
Biopolymers, 72
(4), 230
–240
(2003). http://dx.doi.org/10.1002/(ISSN)1097-0282 BIPMAA 0006-3525 Google Scholar
J. Guicheteauet al.,
“Raman and surface-enhanced Raman spectroscopy of amino acids and nucleotide bases for target bacterial vibrational mode identification,”
Proc. SPIE, 6218 62180O 2006). http://dx.doi.org/10.1117/12.670294 Google Scholar
J. De Gelderet al.,
“Reference database of Raman spectra of biological molecules,”
J. Raman Spec., 38
(9), 1133
–1147
(2007). http://dx.doi.org/10.1002/(ISSN)1097-4555 JRSPAF 0377-0486 Google Scholar
K. VirklerI. K. Lednev,
“Analysis of body fluids for forensic purposes: from laboratory testing to non-destructive rapid confirmatory identification at a crime scene,”
Forensic Sci. Int., 188
(1–3), 1
–17
(2009). http://dx.doi.org/10.1016/j.forsciint.2009.02.013 FSINDR 0379-0738 Google Scholar
N. Stoneet al.,
“Raman spectroscopy for identification of epithelial cancers,”
Faraday Discuss., 126 141
–157
(2004). http://dx.doi.org/10.1039/b304992b FDISE6 0301-7249 Google Scholar
U. Neugebaueret al.,
“Identification and differentiation of single cells from peripheral blood by Raman spectroscopic imaging,”
J. Biophoton., 3
(8–9), 579
–587
(2010). Google Scholar
C. Ottoet al.,
“Surface-enhanced Raman spectroscopy of DNA bases,”
J. Raman Spectrosc., 17
(3), 289
–298
(1986). http://dx.doi.org/10.1002/(ISSN)1097-4555 JRSPAF 0377-0486 Google Scholar
K. VirklerI. K. Lednev,
“Raman spectroscopic signature of blood and its potential application to forensic body fluid identification,”
Anal. Bioanal. Chem., 396
(1), 525
–534
(2010). http://dx.doi.org/10.1007/s00216-009-3207-9 ABCNBP 1618-2642 Google Scholar
A. Barhoumiet al.,
“Surface-enhanced Raman spectroscopy of DNA,”
J. Am Chem. Soc., 130
(16), 5523
–5529
(2008). http://dx.doi.org/10.1021/ja800023j JACSAT 0002-7863 Google Scholar
P. O. Andradeet al.,
“Study of normal colorectal tissue by FT-Raman spectroscopy,”
Anal. Bioanal. Chem., 387
(5), 1643
–1648
(2007). http://dx.doi.org/10.1007/s00216-006-0819-1 ABCNBP 1618-2642 Google Scholar
B. G. FrushourJ. L. Koenig,
“Raman scattering of collagen, gelatin, and elastin,”
Biopolymers, 14
(2), 379
–391
(1975). BIPMAA 0006-3525 Google Scholar
A. Synytsyaet al.,
“Raman spectroscopy of tissue samples irradiated by protons,”
Int. J. Radiat. Biol., 80
(8), 581
–591
(2004). http://dx.doi.org/10.1080/09553000412331283515 IJRBE7 0955-3002 Google Scholar
W. L. Peticolas,
“Application of Raman spectroscopy to biological macromolecules,”
Biochimie, 57
(4), 417
–428
(1975). http://dx.doi.org/10.1016/S0300-9084(75)80328-2 BICMBE 0300-9084 Google Scholar
C. Krafftet al.,
“Near infrared Raman spectra of human brain lipids,”
Spectrochim. Acta Part A: Mol. Biomol. Spectroscopy, 61
(7), 1529
–1535
(2005). Google Scholar
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||