|
|
1.IntroductionAlthough chromosome analysis is routinely used in diagnosing disease, predicting its prognosis and deciding the optimal treatment plan, the conventional chromosome banding analysis (karyotyping) has limitations due to its relatively lower resolution or poor contrast of the chromosome G- or Q-banging patterns. As a result, during the last two decades other new detection technologies have been developed and assessed to improve the accuracy of disease diagnosis using more effective genetic biomarkers. Among these new methods, fluorescent in situ hybridization (FISH) has been approved as a relatively simple, reliable, and robust method that has attracted great research interests in clinical applications. Studies have demonstrated that FISH enables researchers to discover cryptic abnormalities and identify structural and numerical abnormalities that may be missed by conventional cytogenetic studies.1 For example, aiming to improve accuracy of leukemia diagnosis, a number of prospective clinical trials were conducted in the last several years to assess the clinical utility of FISH technology.2–6 These studies confirmed that FISH supplemented standard the karyotyping method by detecting abnormalities in interphase nuclei and clarifying cryptic or complex abnormalities. As a result, the current USA National Cancer Institute guidelines recommended the incorporation of interphase FISH test to the diagnostic work-up of leukemia patients, in particular for all patients diagnosed and confirmed with chronic myeloid leukemia (CLL).7–10 Despite a number of advantages of FISH technology, detecting FISH-probed signals using current imaging methods may also introduce errors in some applications (i.e., detecting FISH signals related to the chromosome translocation). The occurrence of chromosomal translocation where a section of one chromosome attaches to the trail of another chromosome is commonly found in various cancers, such as acute lymphoblastic leukemia t(12;21) or Ewing sarcoma t(11;22)(q24;q12). By utilizing FISH-labeled deoxyribonucleic acid (DNA) probes, the translocation can be typically observed microscopically by the breaking-apart of the paired probes. Cytogenetic analysis for the purpose of disease diagnosis and its prognosis assessment relies on finding the related chromosome translocation from the acquired two-dimensional (2-D) FISH images. Although the level of residual cancer cells are significantly correlated with the risk of cancer relapse,11 the number of chromosomes with associated translocation are often be rare during remission. For example, in acute myeloid leukemia and acute lymphoblastic leukemia cases the abnormal interphase cells with chromosome translocation are less than 5%.12,13 Thus, accurately detecting all chromosomes with translocation can be important in achieving high diagnostic accuracy. The currently dominating microscopic imaging technology in cytogenetic laboratories1 may occasionally be unable to detect the trail of a chromosome. This is because viewing the 2-D projection image of a three-dimensional (3-D) object is not sufficient to capture the chromosomal translocation (two FISH-probed signal spots) when the chromosome stands along the depth direction that is perpendicular to the 2-D image plane (as shown in Fig. 1). Given that the cytogeneticists usually examine no more than 200 cells from a large pool of analyzable cells depicted on one specimen slide, a few false negatives will likely make an impact on the overall sensitivity. In this study, we report an observation of 3-D images of an interphase cell in which one pair of EWSR1 gene probes apparently break apart in one view perspective and fuse together in another. Fig. 1Illustration of an interphase cell processed with a fluorescence-hybridized LSI EWSR1 dual-color break-apart FISH probe, which shows the translocation happened in the depth direction resulting in the two break apart probes overlapped on the 2-D projection image (a) and using free rotation capability provided by a 3-D visualization approach to detect the separation of two FISH targeted DNA probes indicating, in this case, the chromosomal translocation (b). 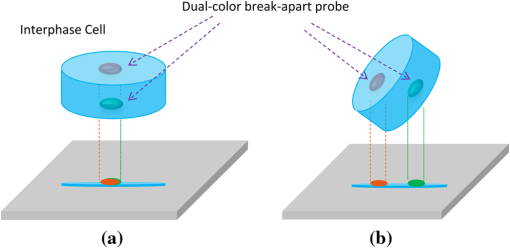 2.Materials and MethodsThe tumor tissue of interest was removed from a patient and prepared at the University of Oklahoma Health Sciences Center for FISH analysis, utilizing a LSI EWSR1 (22q12) dual color break-apart rearrangement probe, which is a whole painting probe-targeted chromosomes 21 and 22.14 The LSI EWSR1 dual color break-apart probe consists of two FISH DNA probes. One 500-kb painting probe is labeled orange. The term kb, or kilo-base pair, is a unit of measurement of DNA/RNA length, equal to 1,000 nucleotides.15 The other 1100 kb painting probe is labeled green. There is a 7-kb gap in between the two probes.14 The specimen slide was imaged under a Nikon A1 confocal microscope with oil objective lens. The imaged field of view (FOV) is with . The mechanical precision of the stage in direction is up to 0.1 μm. A stack of 27 digital image slices was acquired with 0.5-μm step interval, which is close to the theoretical field depth of the objective lens used in the microscope. Thus, these images fully cover the 3-D volume of the specimen, making the resolution of the image slice sufficient for detecting two FISH-probed signal spots indicating the associated chromosome translocation in the depth () direction. The acquired image slices are saved in red/green/blue (RGB) format. The 3-D interphase cell was assembled from the volume of interest of the RGB image stack using the 3-D viewer plug-in of the ImageJ program. 3.ResultsIn generic FISH analysis, FISH signals scattered throughout multiple focal planes may be captured to form a single 2-D projection image for examination. Whereas large component translocations in the lateral directions are visually recognizable, those in the depth direction are overlapped in the 2-D projection images and are thus undistinguishable, as illustrated in Fig. 2(a). When using the 3-D visualization approach, a stack of 27 image slices was reconstructed to avoid signal overlap in the depth direction. As a result, freely rotating the 3-D object in the 3-D visualization approach [Fig. 2(b)] allows for easy detection of the translocation, namely the separation of two (green and red) FISH signal spots in the depth direction. These would be concealed if only the 2-D image were examined. Fig. 2Demonstration of potential concealed translocation. The probes that manifest breaking apart are encircled in green dash lines. Observing a 3-D interphase cell from two perspective viewing directions in which (a) the cell is detected as normal in one view as the two pairs of separated FISH signal probes appear being fused together and (b) the cell is detected as abnormal from another perspective view due to distinguishing two separated FISH signal probes, which indicates the cell is positive for the disease of Ewing sarcoma. 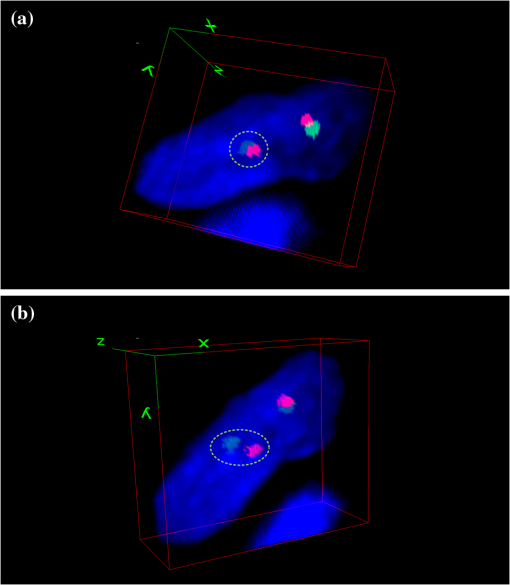 4.DiscussionsDespite the advantages and encouraging results of using FISH technology, we demonstrated the limitation of the existing FISH signal detection and analysis methods that are based on the conventional 2-D microscopic images. The 2-D projection of 3-D objects is incapable of revealing the separation of the FISH probe pair in the depth direction due to the signal spot overlapping, thus generating detection error. Although for the majority of disease cases, this kind of missed detection may not significantly impact the final diagnostic results due to the rarity of such interphase cells depicting a translocation chromosome standing vertically along the depth direction of the 2-D image plane. However, it may potentially impact detection and diagnosis of a fraction of early and subtle (e.g., heterogeneous) cases where the ratio between abnormal and normal interphase chromosome cells is relatively small. In addition, missing abnormal cells with two FISH translocation signal spots overlapped in the 2-D projection images may also reduce the accuracy in detecting residual disease and evaluating treatment efficacy. For example, to detect an early leukemia case and/or evaluate treatment efficacy of a leukemia patient, a blood specimen is typically taken from the patient and analyzed. Due to either the early status of the disease or the relatively effective treatment, the number of abnormal cells is either quite small or has been vastly diminished during treatment. Also, unlike the solid tumor, the abnormal cells are difficult to localize in the extracted blood specimen due to blood circulation.14 As a result, one can expect that the ratio of abnormal to normal cells in these cases to be quite small (i.e., one abnormal cell in every 100 or 1,000 normal cells). Hence, in the clinical practice physicians are required to examine approximately more than 2,000 cells to make a “confident” conclusion in the disease diagnosis and/or assessment of treatment efficacy. It is therefore vital to detect every abnormal cell with the FISH-probe-targeted chromosome translocation in order to discover an early relapse of the disease. The finding in this case report suggests that the 3-D visualization approach could be a useful accessory to leverage the accuracy of detecting early and/or residual disease. However, a number of limitations also need to be considered. First, to cover the entire depth and to accommodate the optical -resolution of the confocal microscope, it is often necessary to acquire many images along directions at each lateral position (e.g., 27 in this experiment). Consequently, fluorescent exposure will be extended and the lifespan of the slide will be reduced. Second, in order to obtain high-quality 3-D images, sophisticated multi-position image acquisition schemes and 3-D rendering algorithms are used. These processes are time consuming.16 Depending on the specific imaging equipment used, the image acquisition at one lateral position can easily take a few minutes. The accumulated workload, which is composed of scanning through hundreds of lateral positions for a whole slide, is therefore high. The two obstacles render the 3-D visualization a much less efficient alternative to traditional 2-D approaches, and making it less accessible in the clinical practice. There is a need for further investigation to make the 3-D visualization approaches more efficient and cost-effective for becoming viable to clinical practices and the general public. AcknowledgmentsThis research is supported in part by grant number RO1 CA136700 and RO1 CA115320 from the National Cancer Institute, National Institutes of Health. The authors would like to acknowledge the support of the Charles and Jean Smith Chair endowment fund at the University of Oklahoma. ReferencesX. Wanget al.,
“Automated detection and analysis of fluorescent in situ hybridization spots depicted in digital microscopic images of Pap-smear specimens,”
J. Biomed. Opt., 14
(2), 021002
(2009). http://dx.doi.org/10.1117/1.3081545 JBOPFO 1083-3668 Google Scholar
D. CampanaC.-H. Pui,
“Detection of minimal residual disease in acute leukemia: methodologic advances and clinical significance,”
Blood, 85
(6), 1416
–1434
(1995). BLOOAW 0006-4971 Google Scholar
M. C. Coxet al.,
“Comparison between conventional banding analysis and FISH screening with an AML specific set of probes in 260 patients,”
Hematol. J., 4
(4), 263
–270
(2003). http://dx.doi.org/10.1038/sj.thj.6200262 HJEOBZ 1466-4860 Google Scholar
G. H. Vanceet al.,
“Utility of interphase FISH to stratify patients into cytogenetic risk categories at diagnosis of AML in an Eastern Cooperative Oncology Group (ECOG) clinical trial (E1900),”
Leuk. Res., 31
(5), 605
–609
(2007). http://dx.doi.org/10.1016/j.leukres.2006.07.026 LEREDD 0145-2126 Google Scholar
K. E. Grimmet al.,
“Necessity of bilateral bone marrow biopsies for ancillary cytogenetic studies in the pediatric population,”
Am. J. Clin. Pathol., 134
(6), 982
–986
(2010). http://dx.doi.org/10.1309/AJCPHR1M1EERGEOK AJCPAI 0002-9173 Google Scholar
J. F. Colemanet al.,
“Diagnostic yield of bone marrow and peripheral blood FISH panel testing in clinically suspected myelodysplastic syndromes and/or acute myeloid leukemia: a prospective analysis of 433 cases,”
Am. J. Clin. Pathol., 135
(6), 915
–920
(2011). http://dx.doi.org/10.1309/AJCPW10YBRMWSWYE AJCPAI 0002-9173 Google Scholar
I. Chenet al.,
“Relationship of CRLF2 expression and outcome in pediatric B-cell precursor acute lymphoblastic leukemia (BCP-ALL): a report from the Children’s Oncology Group,”
J. Clin. Oncol., 29
(Suppl. 15), A-9505
(2011). JCONDN 0732-183X Google Scholar
M. Halleket al.,
“Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia (IWCLL) updating the National Cancer Institute-Working Group (NCI-WG) 1996 guidelines,”
Blood, 111
(12), 5446
–5456
(2008). http://dx.doi.org/10.1182/blood-2007-06-093906 BLOOAW 0006-4971 Google Scholar
E. Kuchinskayaet al.,
“Interphase fluorescent in situ hybridization deletion analysis of the 9p21 region and prognosis in childhood acute lymphoblastic leukemia (ALL): results from a prospective analysis of 519 Nordic patients treated according to the NOPHO-ALL 2000 protocol,”
Br. J. Haematol., 152
(5), 615
–622
(2011). http://dx.doi.org/10.1111/j.1365-2141.2010.08532.x BJHEAL 0007-1048 Google Scholar
J. Leeet al.,
“Cytogenetic and molecular cytogenetic studies of a variant of t(21;22), ins(22;21)(q12;q21q22), with a deletion of the 3’ EWSR1 gene in a patient with Ewing sarcoma,”
Cancer Genet. Cytogenet., 159
(2), 177
–180
(2005). http://dx.doi.org/10.1016/j.cancergencyto.2004.11.003 CGCYDF 0165-4608 Google Scholar
J. Ewing,
“Diffuse endothelioma of bone,”
CA: A Cancer J. Clin., 22
(2), 95
–98
(1921). Google Scholar
H. Cavéet al.,
“Clinical significance of minimal residual disease in childhood acute lymphoblastic leukemia,”
New Engl. J. Med., 339
(9), 591
–598
(1998). NEJMBH Google Scholar
M. R. O’Donnellet al.,
“Acute myeloid leukemia clinical practice guidelines in oncology,”
J. Natl. Compr. Canc. Netw., 4
(1), 16
–36
(2006). 1540-1405 Google Scholar
Abbott Company, “LSI EWSR1 Probe 20 Tests,”
http://www.intermedico.com/productpage.php?id=32-190059&p=1&pg=2 Google Scholar
M. P. Scottet al., Molecular Cell Biology, 5th ed.W.H. Freeman, San Francisco, CA
(2004). Google Scholar
Q. WuF. MerchantK. R. Castleman,
“Three-dimensional imaging,”
Microscope Image Processing, 329
–400 Academic Press, Waltham, MA
(2008). Google Scholar
|

