|
|
1.IntroductionFilamentous fungi grow in the form of multicellular filaments called hyphae. A connected network of these tubular branching hyphae (called mycelium) has multiple, genetically identical nuclei and is considered a single organism. In vitro susceptibility testing of filamentous fungi is becoming increasingly important because of the frequency and diversity of infections caused by them.1 Additionally, these micro-organisms produce enzymes that can be used in various industrial and commercial applications. Some of them have the potential to alleviate environmental problems, as in the case of the white rot fungus, Phanerochaete chrysosporium.2 Some authors reported the mean of movement rates of fungal mycelia of P. chrysosporium measured as growth ranged from 10.9 to 12 mm after 2 and 5 days of incubation, respectively.3 Bacteria are a large group of single cells (prokaryote micro-organisms) with few micrometers of length. They have a wide range of shapes, ranging from spheres to rods and spirals. Bacterial populations can double by binary fission every 9.8 min. Motile bacteria move at an average speed of 10 to .4 Some bacteria with pathogenic and biodegradative properties such as Pseudomonas aeruginosa,5 have wide metabolic versatility and high intrinsic and acquired resistance to antimicrobials related to its motility and further biofilm formation. The correct identification of micro-organisms is of fundamental importance to microbial systematists as well as to scientists involved in many other areas of applied research and industry. The phenotypic approach is obviously also insufficient when studying bacteria and fungi grown on a substrate at early stage of development. The identification of filamentous fungi at early stage of development is time-consuming due to cultivation and microscopic examination. A rapid In vitro discrimination between motile bacteria and filamentous fungi in watery supports, as in a tissue, may help doctors to take quick decisions indicating the proper antibiotic or antifungal. On the other hand, an early identification of contamination in a biological reactor or assay would avoid non-useful results and reduce the time for micro-organism identification. A rapid and accurate diagnosis of fungal presence is still hampered by shortcomings in the currently available methodology. A more recent approach to the classification and identification of micro-organisms involves the comparison of genotypic or genetic characteristics. Several PCR (polymerase chain reaction) and enzyme-linked immunosorbent assay methods have recently been reported.6 Sometimes the lack of standardization and clinical validation has led to its exclusion from consensus criteria in fungi discrimination. The selectivity of an existing commercial SPC (solid phase cytometry) procedure for fungi, proposed by one manufacturer has to be substantially modified to reduce the interference from bacteria and to optimize the labeling of fungi.7 Also, FTIR (Fourier transform infrared spectroscopy) coupled with chemometric methods, is becoming an important tool in micro-organisms differentiation.8 These methodologies are expensive or time consuming. However, the isolation of one micro-organism on an agar plate followed by macroscopic, microscopic, biochemical, and/or molecular identification remains the backbone of the laboratory diagnosis in daily routine. This type of procedure may take up to five days. Therefore, it is of great interest to develop new methods to improve the detection in situ (without sample manipulation) and reduce the detection time. Optical techniques are non-invasive and have advantages that deserve exploring. We have recently reported on the use of dynamic laser speckle (or biospeckle) techniques for motility and chemotactic detection in bacteria.9 The random dynamic fluctuation of speckles obtained when an active sample is illuminated by laser light is characteristic of objects activity, typically biological tissues, and it carries useful information about their biological and physiological activity, for example, their time scales. The study of the temporal evolution of the speckle patterns provides an interesting tool to characterize the parameters involved in these processes. With the extended use of laser technology in the medical and biologic fields, the applications of biospeckle techniques have been increased in the last years. It has been applied to the measurement of blood flow, parasite motility, cerebral activity of rats, atherosclerotic plaque detection, and botanical specimens.10 In this paper, we present a dynamic laser speckle method to easily discriminate filamentous fungi from motile bacteria in soft surface, such as agar plate. This method is appropriate for a fast preliminary screening, previous to conventional (biochemical, molecular, and/or morphologic) tests which usually guide to determine the correct genus and species. The new procedure could be applied to hospital supply waters, dialysis fluids, endoscopic rinse waters, microbial effluent analysis, and infected tissues. Since biospeckle activity depends on microscopic movements, micro-organisms with different type of motility, such as chemotactic bacteria (Pseudomonas aeruginosa), non chemotactic mutant motile bacteria (Pseudomonas putida F1 cheA) and filamentous fungi (Panerochaete chrysosporium) were chosen to evaluate the proposed method. Although different micro-organisms differ very much in the speeds of their movements, fungi are one to three orders of magnitude slower than bacteria ( as compared with 10 to , respectively).4,11 This difference produces very smaller Doppler shifts in the scattered light thus leading to experimental observations of much lower activity. Therefore, biospeckle techniques are able to discriminate dynamic movements of micro-organisms with different speeds. 2.Experimental Procedures2.1.Culture of Phanerochaete ChrysosporiumPhanerochaete chrysosporium RP-78 was grown on Petri dishes with mineral salts (MS) liquid medium. MS liquid medium composition was (in grams per liter): , 11.2, , 4.8 (), 2, ·, 0.246, ·, , pH 7, supplemented with glucose (0.1% ) and peptone (0.1% ) as attractants, and high-purity agar agar (0.35% ). The fungi were cultured for 5 days at 25°C. Dilutions were made in motility buffer (MB) (10 mM potassium phosphate pH 7, and 0.1 mM EDTA), to a final turbidity of 1 (). Aliquots (0.003 mL) of that suspension were used to inoculate each soft agar plate in motility assays. 2.2.Culture of P. aeruginosa and Pseudomonas putidaIn the present work two strains were used: Pseudomonas aeruginosa isolated from soil12 and another strain of Pseudomonas putida F1 cheA. The last one is a motile strain but does not show chemotactic response due to a mutation in the cheA gen (kinase). They were suspended into MS medium supplemented with peptone at a final concentration of 1% (). After 24 to 40 h of incubation at 25°C in an orbital shaker at 120 rpm, the cells were harvested at mid log phase by centrifugation at 3500 rpm during 15 min. Then, they were washed (one time) with sterile motility buffer (MB) (10 mM potassium phosphate pH 7, and 0.1 mM EDTA) and centrifuged again. The resulting precipitated cells were re-suspended in MB, previously vortexed to achieve good aeration. After that, the cells were re-suspended in 2 mL of MB (without any source of carbon and energy), in order to achieve a desired cell density of cells., and placed again in the shaker at 120 rpm for 24 h. Aliquots (0.003 mL) of that suspension were used to inoculate each soft agar plate in the motility assays. 2.3.Motility AssaysThe swarm plates for bacterial assays contained MS and yeast extract (0.025% ) as attractant and agar-agar (0.35% ). For fungi assays, glucose (0.05% ) and peptone (0.05% ) were used as attractants, and high-purity soft agar-agar (0.35% ). A drop of buffer (MB) without bacteria or fungi was used as negative controls. The middle surface of the plate with agar was inoculated with 0.003 mL of cells re-suspended in motility buffer (either bacteria or fungi). Simultaneously, soft agar plates containing yeast (0.05% ) and glucose (0.05% ) as attractants, with agar-agar (0.35% ) as support, were prepared. Bacteria and fungi suspensions, as well as MB (negative control) were placed in the same soft agar plate as inocula. The chemoattractants uptake generates a concentration gradient necessary for the bacterial chemotactic response,13 which can be inferred from the expanding rings. All assays were done by triplicate and repeated at least 10 times. 2.4.Experimental SetupThe dynamic laser speckle technique was previously implemented for the analysis of bacteria motility and chemotaxis, as reported by Murialdo et al.9 The experimental set-up is shown in Fig. 1. An expanded and attenuated HeNe laser (632.8 nm wavelength and 30 mW) illuminated the plate under study from the bottom through a ground glass diffuser in order to increase the speckle contrast as was done in a previous work.9 A CCD camera connected to a frame grabber registered sequences of 100 images (8 bits and squared pixels of 8.3 µm) and stored them into the computer. A constant 25 Hz sampling frequency was used, and the camera integration (exposure) time was set in 40 ms. The camera lens was focused on the plate, and its pupil was empirically adjusted using the camera diaphragm to obtain the best spatial resolution with a speckle grain diameter comparable to the pixel size. The images were strongly demagnified, so that the individual micro-organisms were clearly not resolved by the optical system. Fig. 1The dynamic laser speckle technique employed to easily discriminate filamentous fungi from motile bacteria in soft surface, such as agar plate. (a) Scheme of the setup for speckle imaging (the ground glass diffuser was removed and the mirror was replaced by a circular fluorescent tube over a dark surface for the white light imaging), and (b) an example of a speckle image. 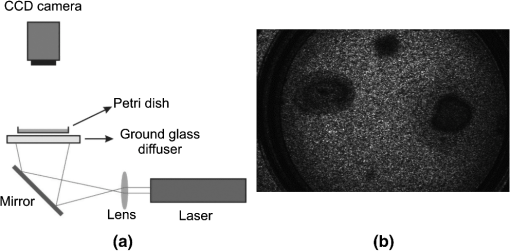 Soft agar assays for motility, chemotaxis, and aerotaxis are well described in detail in Parkinson.14 Briefly, in soft agar media, flagellated bacteria swim within the water filled tunnels in the agar matrix. The expanding colony typically extends from the bottom of the plate to the top surface of the plate with the majority of the cells embedded in the agar rather than at the air interface. These colonies can contain one or more rings of cells that are concentrated near the outer boundaries of nutrient, energy, or electron acceptor gradients generated through the cells’ metabolic activities. The size, thickness, relative position, and depth of each ring provide important information about a strain’s motility and tactic behavior. Direct light from above is ill-suited for viewing or photographing soft agar colonies. Cells on the surface of the agar reflect much of the incident light, whereas those within the agar receive and reflect much less light. Consequently, this type of illumination obscures much of the detailed structure of the colonies.14 In contrast, indirect light from behind the agar more evenly illuminates the colonies and brings out their fine structure features, such as internal rings or fuzzy edges. However, the light should not pass from the source directly upward through the plate, but rather should enter only the bottom half of the plate and at an oblique angle, approximating a dark field effect. The morphologies of colonies in soft agar media can provide a wealth of information about a strain’s locomotor and chemotactic abilities. Photographic images are often the simplest and most effective means of documenting these behavioral phenotypes. Uniform, indirect, transmitted illumination of the plates is essential for obtaining good colony images. Therefore, the conventional technique to detect motility, chemotaxis, and aerotaxis consists in observing how the plate evolves in time by illuminating it with white light from the bottom.14 In our case, the images were obtained with the CCD camera using also the experimental setup described in Fig. 1, where the diffuser was removed and the mirror was replaced by a circular fluorescent lamp placed over a dark surface. 2.5.Image ProcessingThe time-varying intensity at each pixel of the image sequence was considered as a stationary signal, in a wide sense. The latter is due to the fact that micro-organism population movement did not exhibit relevant changes during the acquisition lapse (4 s). Many descriptors have been previously addressed to characterize the speckle laser dynamics of the observed phenomena.10 But given that the aim of this process is to discriminate image areas, where different motilities are expected (fungi and bacteria), and that the mobility affects the beating frequency, the spectral bands energy descriptor was chosen because of its ability to describe these phenomena.15 As decomposition in spectral bands was explained in detail in a previous work,15 here we give only a brief description. The power density spectrum of the dynamic speckle spreads significantly till approximately 6 Hz, with sampling frequency of 25 Hz. Hence, a three band filter bank is designed to filter the dynamic speckle signals: a low pass from 0 to 0.25 Hz, a band pass filter from 0.25 to 5 Hz and a high pass with cut frequency higher at 5 Hz. The losses specification in each of this filters are the following: maximum tolerated at the pass band 1 dB and the minimum losses required at the reject band is the 40 dB. Instead of using Butterworth approximation like in previous work,15 elliptic (Cauer) filters were used because, in spite of their higher implementation complexity, they exhibit a more selective frequency response than the Butterworth solution. Then the Average Energy for each filtered signal (lowpass, bandpass and highpass) was computed as characteristic indices at each , image location: where is the filtered intensity of the pixel at , location in the image, and is the quantity of images in the sequence. In this way, three values of energy for each pixel were obtained, each of them belonging to one of the frequency bands. The digital filter bank was computed using Matlab® software routines.Hence three grayscale images were assembled with the energy of each filtered sequence of speckle patterns (low pass image, band pass image, and high pass image). Grayscale digital images are those in which the value of each pixel is a single number, that is, it carries only intensity information, within a range that goes from 0 (black), or minimum intensity, up to 255 (white), or maximum intensity, having many shades of gray in between. In order to improve the visualization of the differences among regions of filtered speckle images, we proposed to assemble pseudo-colored images using the Red Green Blue color model (RGB). In the RGB model, the color of each pixel is determined by the combination of the red, green, and blue intensities stored in each color plane at the pixel’s location. The aim of this experiment was to detect three different regions: the region where bacteria grow, the region of fungi colonies, and the background (agar support). Samples of these three areas where then collected by an expert, using a set of 30 images. These images were the result of the processing using the band pass filtering, as mentioned in the Motility assay section. Hence a total of thirty images were composed of energy filtered signals, captured under same experimental conditions where the location of each zone was well determined. A histogram equalization process was applied to the whole set of images to adjust their contrast, achieving a domain support that covered the whole intensity range (0 to 255). These images were then used as training examples to estimate a likelihood function for each class (bacteria, fungi, and agar support). This function is a continuous probability distribution with parameters obtained from the samples using the Maximum Likelihood Estimation (MLE) method.16 Consequently, every pixel of a descriptor image could be assigned with the probability level of each class, as a vector of three components. Each vector component was allocated to a different color plane. Therefore, each color plane was filled with the estimated probability values of one class. The red plane mapped a probability function of the existence of bacteria, the green plane showed the probability of existence of fungi, and the blue plane indicated the presence of agar support. The probability functions were estimated using a Gaussian kernel within the range of energy intensities of the samples. This procedure was applied to both the white light photographs and the processed speckle images [Fig. 2(a) and 2(b), respectively]. Fig. 2Biospeckle and white light images comparison. Pseudomonas aeruginosa (right side), Pseudomonas putida F1 cheA (left side), and P. chrysosporium (middle upper) were used as inocula in soft agar plates (see annotations). (a) White light photograph of a Petri dish inoculated with bacteria and fungi, (b) equalized energy of the filtered speckle images, (c) RGB discrimination probability of the white light photograph (a), and (d) RGB discrimination probability of the processed speckle image (b). Note about the color meaning (only in the speckle RGB image): , , and support. Both (c) and (d) images are masked to enhance the region of interest of the petri dish. The bright spot in the lower part in (a) is probably due to a contamination. 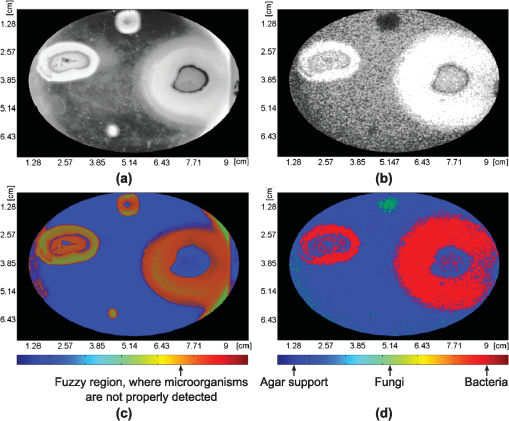 3.Results and DiscussionPseudomonas aeruginosa, Pseudomonas putida F1 cheA, and P. chrysosporium were used as inocula in soft agar plates. The white light photograph of the plate, after 72 h from inoculation [Fig. 2(a)], shows that it is very difficult to discriminate between bacteria and fungi. When the same plate is illuminated with a laser speckle pattern a sequence of images is registered and processed. After filtering the dynamic speckle signals using the filter bank, three images corresponding to the average energy of the filtered signals were assembled (lowpass, bandpass, and highpass). Comparing the images assembled with the energy of each one of the three filters, the image of the energy of the bandpass signal is the one that more efficiently detects and segments areas according with the speckle dynamic, which Fig. 2(b) shows in grayscale. The brightest areas of Fig. 2(b) (left middle and right middle) correspond to bacteria regions, while the darkest zone (middle top) matches with the fungi zone. The time-varying speckle signal of each image pixel is produced by microscopic movements (micro-organism movements) or other physical reasons such as variations in the refractive index. The latter can be neglected in these experiments.9 The intensity of the processed images is proportional to the energy of the filtered signal. In these experiments bacteria movements produce time-varying speckle (biospeckle) that result in high intensities [Fig. 2(b)]. On the other hand, the white light image [Fig. 2(a)] shows higher intensity not only for bacteria but also for fungi. This is due to the fact that the white light intensity is proportional to the density of both motile and non-motile micro-organisms, while the time varying speckle is produced only by motile ones. After comparison of the white light and speckle-processed images we found that both are complementary images. Although the white light conventional technique does not clearly distinguish bacteria from fungi [Fig. 2(a)], the speckle-processed image is only sensitive to the presence of motile bacteria [Fig. 2(b)]. On the other hand, control assays with chemotactic bacteria [Fig. 2(a) and 2(b), right side] and non chemotactic bacteria [Fig. 2(a) and (b), left side] were assessed with both white light [Fig. 2(a)] and dynamic laser speckle [Fig. 2(b)], in order to compare the difference in their migration pattern. The speckle-processed image showed that the pattern size of motility coincided with the pattern size of the optic density appreciated with white light. To improve the visualization of both bacteria and fungi, RGB images were assembled [Fig. 2(c) and 2(d)]. Previously to the building of the RGB planes, a mask that delimits the field of interest was applied to the descriptor images to avoid noise near the plate edges. The RGB image obtained with the filtered dynamic speckle signals using the bandpass filter is displayed in Fig. 2(d). In this figure, bacteria, fungi, and medium were clearly detected, practically no overlap among them is displayed, given that they only primary colors are observed. While in Fig. 2(c), where the RGB image is built from the white light information, the combination of red and green colors produces the brownish one, which represents the inability for discriminating between both micro-organisms. As already found before in Fig. 2(a), the white light image cannot distinguish between bacteria and fungi. On the other hand, the processed speckle image clearly distinguishes between both, showing no brownish colors. However, we propose the use of both techniques simultaneously in order not only to confirm results, but also to observe the effect of the optical density of micro-organisms. At the bottom of Fig. 2(a) (white light image) a small spot of bright intensity is found (due to a possible contamination). While in Fig. 2(c) (white light processed photo) it appears as a brownish change of intensity against the background that cannot be distinguished as fungi or bacteria, in the speckle pattern processed image, Fig. 2(b), it appears as a region of very low activity. Its analysis in the processed RGB speckle image (doing a zoom of the area) shows a region where fungi exhibit a higher probability (green spots) than the bacteria while with Fig. 2(c) no micro-organism differentiation is possible. 4.ConclusionsWe have shown a new method to macroscopically discriminate between bacteria and fungi in agar-agar assay over Petri dishes. Fast in situ preliminary discrimination between filamentous fungi and motile bacteria with dynamic laser speckle will guide to take faster decisions and achieve lower costs for further identifications. This initial method will require other subsequent reliable and confirmatory methods (by morphology, molecular, and biochemical tests) normally used in the laboratory for taxonomic identification. Besides, the combination of white light and processed-speckle images can be used to analyze density and motility of micro-organisms. Characteristics of each colony ring provide important information about a strain’s motility and tactic behavior.14 In this sense, the motility grades obtained from the speckle processed images may provide useful information in future investigations. AcknowledgmentsThis work was supported by a ANPCyT Grant PICT 2008-1430, Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), Comisión de Investigaciones Científicas de la Provincia de Buenos Aires (CIC), Secretaría de Estado de Ciencia y Técnica (SECYT), Universidad Nacional de Mar del Plata (UNMDP), and Universidad Nacional de La Plata (UNLP). Silvia Murialdo is member (Adjunct Researcher) of CIC. ReferencesJ. Meletiadiset al.,
“Analysis of growth characteristics of filamentous fungi in different nutrient media,”
J. Clin. Microbiol., 39
(2), 478
–484
(2001). http://dx.doi.org/10.1128/JCM.39.2.478-484.2001 JCMIDW 1070-633X Google Scholar
D. Morales-Fonsecaet al.,
“Desarrollo de un bioadsorbente laminar con Phanerochaete chrysosporium hipertolerante al cadmio, al níquel y al plomo para el tratamiento de aguas,”
Revista Iberoamericana de Micología, 27
(3), 111
–118
(2010). Google Scholar
S. K. O. NtwampeM. S. Sheldon,
“Quantifying growth kinetics of Phanerochaete chrysosporium immobilised on a vertically orientated polysulphone capillary membrane: biofilm development and substrate consumption,”
Biochem. Eng. J., 30
(2), 147
–151
(2006). http://dx.doi.org/10.1016/j.bej.2006.03.001 BEJOFV 1369-703X Google Scholar
L. Cisneroset al.,
“Reversal of bacterial locomotion at an obstacle,”
Phys. Rev. E, 73
(3), 030901
(2006). http://dx.doi.org/10.1103/PhysRevE.73.030901 PLEEE8 1063-651X Google Scholar
A. AlonsoF. RojoJ. L. Martinez,
“Environmental and clinical isolates of Pseudomonas aeruginosa show pathogenic and biodegradative properties irrespective of their origin,”
Environ. Microbiol., 1
(5), 421
–430
(1999). http://dx.doi.org/10.1046/j.1462-2920.1999.00052.x ENMIFM 1462-2912 Google Scholar
B. Posteraroet al.,
“Update on the laboratory diagnosis of invasive fungal infections,”
Mediterr. J. Hematol. Infect. Dis., 3
(1), e2011002
(2011). http://dx.doi.org/10.4084/MJHID.2011.002 Google Scholar
M. M. De VosH. J. Nelis,
“An improved method for the selective detection of fungi in hospital waters by solid phase cytometry,”
J. Microbiol. Meth., 67
(3), 557
–565
(2006). http://dx.doi.org/10.1016/j.mimet.2006.05.020 JMIMDQ 0167-7012 Google Scholar
L. Marieyet al.,
“Discrimination, classification, identification of microorganisms using FTIR spectroscopy and chemometrics,”
Vib. Spectrosc., 26
(2), 151
–159
(2001). http://dx.doi.org/10.1016/S0924-2031(01)00113-8 VISPEK 0924-2031 Google Scholar
S. E. Murialdoet al.,
“Analysis of bacterial chemotactic response using dynamic laser speckle,”
J. Biomed. Opt., 14
(6), 064015
(2009). http://dx.doi.org/10.1117/1.3262608 JBOPFO 1083-3668 Google Scholar
H. RabalR. Braga, Dynamic Laser Speckle and Applications, CRS Press, Taylor and Francis Publisher, Boca Ratón, FL, USA
(2009). Google Scholar
G. JimenezM. Pennincx,
“Nuevo proceso de biodegradación de colorantes, utilizando hongos de la podredumbre blanca de la madera,”
VITAE, 10
(2), 52
–59
(2003). 1317-987X Google Scholar
S. E. Murialdoet al.,
“Degradation of phenol and chlorophenols by mixed and pure cultures,”
Water SA, 29
(4), 457
–463
(2003). WASADV 0378-4738 Google Scholar
G. Pandeyet al.,
“Chemotaxis of a Ralstonia sp. SJ98 toward co-metabolizable nitroaromatic compounds,”
Biochem. Biophys. Res. Commun., 299
(3), 404
–409
(2002). http://dx.doi.org/10.1016/S0006-291X(02)02601-3 BBRCA9 0006-291X Google Scholar
J. S. Parkinson,
“A “bucket of light” for viewing bacterial colonies in soft agar,”
Methods Enzymol., 423 432
–435
(2007). http://dx.doi.org/10.1016/S0076-6879(07)23020-4 MENZAU 0076-6879 Google Scholar
G. H. Sendraet al.,
“Decomposition of biospeckle images in temporary spectral bands,”
Opt. Lett., 30
(13), 1641
–1643
(2005). http://dx.doi.org/10.1364/OL.30.001641 OPLEDP 0146-9592 Google Scholar
J. Aldrich,
“R.A. Fisher and the making of maximum likelihood 1912 to 1922,”
Stat. Sci., 12
(3), 162
–176
(1997). http://dx.doi.org/10.1214/ss/1030037906 STSCEP 0883-4237 Google Scholar
|

