|
|
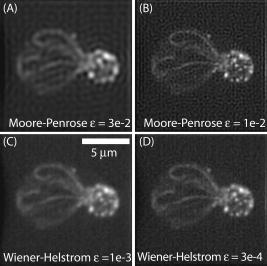
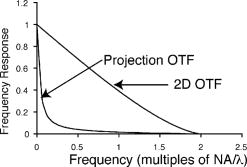
1.IntroductionThe rate of acquisition of fluorescent signals in 3-D using digital imaging-based microscopy is a critical factor in a number of applications, particularly in high-throughput microscopy. Because the depth of focus is relatively shallow, and becomes shallower as the resolving power of the objective lens improves (as the numerical aperture increases), it is necessary to acquire image samples at multiple levels of focus in the object of interest to avoid missing potentially important sources of signal. In applications requiring large sample sizes, the rate of acquisition sets an upper limit on possible throughput. In applications where the object of interest moves during acquisition, this rate correlates directly with the amount of blur in the acquired images. The method presented here is designed specifically to increase the rate of acquisition of signal originating throughout 3-D samples of interest. Current practice is to acquire individual 2-D image planes at discrete levels in the specimen by moving focus between each acquisition, then to combine these planes to produce 3-D stacks (or “volumes”), which are processed further as needed. Collection of multiple individual planes introduces delays because: 1. quickly changing focus introduces vibrations in the sample, which degrade the image, but waiting for the vibrations to dampen or slowly moving focus slows the process; and 2. images must be collected, digitized, and transmitted individually. In wide-field microscopy, delays from the second of these problems arise in part from time intervals required to open and close illumination and camera shutters. In confocal scanning microscopy (CSM) and multiphoton fluorescence excitation microscopy (MPFEM), long image-collection time is due to the need to scan the illumination over the specimen. Shorter confocal acquisition times may be possible with alternative confocal methods, but images made with each of the confocal methods include only a thin layer of the specimen, and it is likely that not all the structures of interest are contained in that thin layer. In this regard, image volumes acquired using wide-field microscopy benefit from detecting fluorescence further away from each plane of focus, while simultaneously suffering image degradation by the addition of out-of-focus blur. Deconvolution reduces the blur in 3-D image volumes, but is computationally burdensome. An extended depth of focus (EDOF) method, i.e., one that would allow obtaining 2-D images in which all portions of a thick specimen appear in focus, would greatly benefit high-throughput microscopy. An existing method for EDOF is wavefront coding. In this method, a phase mask is placed in the objective back focal plane (BFP) (or at a plane conjugate to the BFP) that distorts the imaging characteristics of the objective in such a way that, although they are severely degraded, they are practically independent of the depth at which the objective is focused.1 Wavefront coding, however, is not yet applicable to the high numerical aperture (NA) objectives necessary for high-resolution imaging. Here we present a method for EDOF that is applicable to any objective and, although slower than wavefront coding, is more rapid than the existing methods outlined before. The method is based on collecting a through-focus image as a single 2-D image and then deconvolving this image using existing algorithms for deconvolution. Part of this work is described by us in a United States patent.2 This work is organized as follows. Section 2 describes the mathematical foundations of the proposed method. Section 3 presents the results of applying the proposed method to fluorescent images. In Sec. 4 we describe research projects in which we have successfully applied the EDOF method we developed. Section 5 compares results using different deconvolution methods, in particular linear versus nonlinear. In Sec. 6 we analyze the sensitivity of our method to using a point spread function (PSF) that is not at a depth representative of the specimen, and show that the method is rather robust to a mismatch in the PSF depth. In Sec. 7 we present important earlier developments that relate to the proposed method, but that in one way or another fell short of the goal of the method we propose here of a fast and robust method for rendering 2-D images with extended depth of focus. Lastly, Sec. 8 summarizes our conclusions and describes directions for possible enhancements to the proposed method. 2.Description of the MethodUnder imaging conditions that often hold, fluorescent image formation can be approximated as the 3-D convolution where is the specimen function, i.e., the concentration of fluorescent dye at in object space ; is the image intensity, i.e., the recorded pixel value at in image space ; and is the point spread function (PSF) of the microscope, the intensity at a point in the image space due to a unit point source of light at in object space.In our method, the image is not collected as a 3-D stack, but as a 2-D image. More specifically, it is collected by opening the camera and illumination shutters and moving the microscope stage or objective through focus. This has the effect of accumulating in the charge-coupled device (CCD) chip the intensity at all the planes through which the microscope is focused. The resulting 2-D image is where is a 2-D point in image space and is the pixel value at of the resulting 2-D image. If the data collection is taken over a large enough depth such that the integrals along the axes ( and ) can be considered to be practically from minus to plus infinity, Eq. 2 can be written asThe innermost integral in Eq. 3,is independent of the -axes coordinates. We define the 2-D projection of the specimen function asThat is, is an image of in which all structures are in focus at the same time, regardless of their depth, i.e., an extended depth-of-focus image of . Substituting Eqs. 4, 5 into Eq. 3, we getEquation 6 is the 2-D convolution of and defined by Eqs. 3, 4. Therefore, to obtain the extended depth of focus (EDOF) projection of the specimen function in Eq. 5, one needs to invert (i.e., deconvolve) Eq. 6 using a 2-D PSF that is an integrated-intensity projection of the 3-D PSF. It is possible to obtain Eq. 6 using the central slice theorem of the Fourier transform. However, the previous derivation more clearly shows the relation between the image collected by moving the specimen through focus while the shutter is open and the 2-D projections of the specimen function and PSF.There are many approaches for deconvolving incoherent imagery. For the images presented here, we use a maximum-likelihood (ML) approach based on the expectation-maximization (EM) formalism of Dempster, Laird, and Rubin.3 The ML-EM algorithm that results can be found in Ref. 4. However, the algorithm is somewhat slow for routine application, and thus in our regular research we use faster constrained ML deconvolution algorithms that we have derived.5 3.Experimental Methods and ResultsWe tested the method in two ways. First, we collected a 3-D stack of images and then collapsed the stack into a 2-D integrated-intensity projection image that was deconvolved as described before. The original 3-D stack was also deconvolved with the same algorithm. This approach somewhat allows us to compare the resolution obtainable with the method presented here against that of the much slower method of 3-D data acquisition and deconvolution. The second test was done with images collected by leaving the camera and illumination shutters open while the microscope stage was moving through focus. For both cases, the 3-D PSF of the microscope was computed from the model of Gibson and Lanni6 using Gauss-Kronrod quadrature7, 8 to numerically evaluate the integrals. The 2-D PSF was calculated from the 3-D PSF using Euler’s method. We are currently investigating how the choice of more precise quadrature methods affects the PSF and, more importantly, the deconvolved image. 3.1.Three-Dimensional StackThe first test image is from fluorescently labeled actin filaments in yeast cells collected with a objective. The image has cubic pixels on each side (3-D stack courtesy of Karpova at the National Cancer Institute, National Institutes of Health, Bethesda, Maryland). Figure 1 shows the recorded and deconvolved EDOF images. Figure 1a is a 2-D projection of the 3-D stack obtained by adding all the 2-D optical slices into a single image. Figure 1b is the 2-D deconvolution of Fig. 1a using the 2-D PSF defined in Eq. 4 and 3000 iterations of the EM-M algorithm. Figures 1c and 1d are obtained by deconvolving the 3-D stack using a 3-D PSF and the EM-ML algorithm for 1000 and 3000 iterations, respectively. The time required by the 2-D deconvolution in Fig. 1b is about two orders of magnitude shorter than the time required for Fig. 1d that uses the same number of iterations. Despite this large time difference, the results obtained by the two methods are similar. 3.2.Two-Dimensional ProjectionFigure 2 shows images of 4',6-diamidino-2-phenylindole-stained nuclei from an approximately -thick section of mouse kidney. The images were collected with a oil-immersion objective and a Roper Scientific (Trenton, New Jersey) Quantix57 CCD camera driven by custom-written software. The pixels are squares. The through-focus excursion was (from approximately above to approximately below the specimen). For PSF computation, the fluorescent wavelength , and we assumed that the specimen was immediately below the coverslip. We obtained nearly identical results, assuming the specimen to be under the coverslip. Fig. 2Images of nuclei in mouse kidney. Top: (a) and (b) focused at the top and bottom of the volume, respectively. (c) 2-D projection collected by moving the specimen through focus with the camera and excitation shutters open. (d) 2-D deconvolution of (c) using the EM-ML algorithm. (Scale bar .) 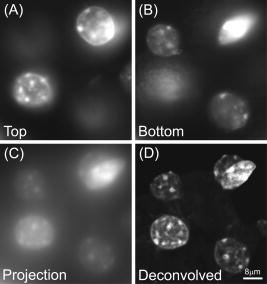 Two nuclei that appear bright and well focused when the focus is set at the top of the section [Fig. 2a] are blurred when the focus is set at the bottom of the section [Fig. 2b] and, in one case (in the upper right-hand corner), obscured by an in-focus nucleus. In the through-focus image [Fig. 2c], all the nuclei are visible but are severely blurred. The blur, however, is removed in the deconvolved image [Fig. 2d]. 4.Application to Our ResearchThe method presented here was developed in response to our need to obtain location information from thick specimens when the time to collect a 3-D stack is inadequately long. Here we list two examples in which the acquisition of full stacks are limiting: 1. time-lapse acquisition of fluorescent spots that rapidly move out of focus, and 2. identification of labeled cells within a relatively large subject. In both instances, our approach significantly increases throughput at the imaging stage. The reduced image acquisition time is because our method avoids multiple start-and-stop focusing movements and delays caused by the multiple shutter opening and closing, and by multiple data transfers from camera to computer to storage. Furthermore, deconvolution times and data storage requirements are considerably reduced. Deconvolution time is reduced not only because of the reduction from 3-D to 2-D, but also because for routine work, instead of the EM algorithm, we use the faster algorithms we developed for maximum-likelihood constrained deconvolution.5 In addition, although the result is a 2-D image, it is entirely adequate to address common research problems. In our time-lapse acquisition of fluorescent spots, the approach presented here allows us to track fluorescent protein-tagged chromosome regions as they move around the living Saccharomyces cerevisiae nucleus, imaging at a rate of two full frames of pixels per second. With our equipment, this is a significantly better temporal resolution than we can achieve with 7-slice stacks using only 1/8 of the frame, and thus greatly increases our efficiency in terms of imaging time per cell. In this experiment, the issues are to identify when rapid movements begin and stop, as assessed in large numbers of cells so that rare events are not missed, and to determine whether the rates of movement are different in different cell types. The method presented in this work has proven completely acceptable in this regard, and superior to full stacks in particular, because of the more rapid throughput. For the identification of labeled cells within a relatively large subject, the approach presented here provides an image adequate to identify the presence or absence of a fluorescently labeled cell in large numbers of C. elegans adults (where image throughput is important) without first having to identify the proper plane of focus. The need to first identify such a plane would be detrimental for automated screening of large populations of worms. 5.Linear Versus Nonlinear DeconvolutionIt is often argued that for 2-D deconvolution of microscopic images, it is unnecessary to use nonlinear methods, such as the EM-ML described earlier or the Jansson-van Cittert method of repeated convolution,9, 10, 11 the argument being that the 2-D optical transfer function (OTF) does not suffer from the missing cone that affects the 3-D OTF of wide-field microscopes. In fact, the 2-D OTF corresponding to either (for thin specimens) or to given in Eq. 4, are nonzero over a circle of radius in the spatial frequency domain, and there are no regions inside this circle where the OTF is zero. However, because the OTF is exactly zero outside this circle, linear deconvolution methods (whether iterative or not) cause artifacts in the estimated specimen function, the main artifact being pixels with negative values that result from enhancing frequency components within the passband of the OTF without obtaining the out-of-band frequency components necessary to obtain a non-negative specimen function estimate. We tested two of the most widely used linear deconvolution algorithms, namely the Moore-Penrose pseudo-inverse (sometimes referred to as the linear least squares algorithm12) and a filter based on the Wiener filter (or, more properly, the Wiener-Helstrom filter. See, for example, pages 206 to 210 of Ref. 13). In either case, the Fourier transform of the estimated specimen function is calculated as where upper case functions denote the 2-D Fourier transform of the corresponding lower-case function; is the 2-D coordinate in the spatial frequency domain; and is the filter function, given byfor the Moore-Penrose pseudo-inverse (MPPI) and byfor the Wiener-Helstrom (WH) filter. In Eqs. 7, 8, is the OTF corresponding to the PSF in Eq. 4 [the Fourier transform of the latter normalized so that ]; is a small positive number related to the signal-to-noise ratio in the image; the superscript denotes complex conjugate; and the bars denote the modulus of the complex quantity within. Figure 3 shows the results of deconvolving the image in Fig. 1a using the MPPI and the WH filters for different values of . Figure 3a and 3b are the results from the MPPI for and , respectively. Values of larger than 0.03 result in even blurrier images, whereas values smaller than 0.01 result in noisier images. Figures 3c and 3d show the results of deconvolving Fig. 1a with a Wiener-Helstrom filter with and , respectively. As with the MPPI, values of larger than result in blurrier images, whereas values smaller than produce noisier images with little or no gain in resolution. Thus, although both the WH and MPPI filters are much faster than the EM-ML and other constrained ML algorithms, the results afforded by the latter are vastly superior. This is because is dominated by low frequencies (see Fig. 4 ). In fact, components with frequencies larger than are greatly attenuated in the 2-D projection. Thus, the MPPI and WH filters enhance only a very limited band of spatial frequencies. On the other hand, it has been shown by us4 and by others14 that the EM-ML algorithm uses the frequency components passed by the imaging system to obtain the frequency components that are either blocked or severely attenuated by the imaging system. This capability is of great help in the current application where the practical cut-off frequency is severely reduced by the projection in .6.Sensitivity to Point Spread Function-Depth MismatchWe obtain the PSF we use for deconvolution from the theoretical model of Gibson and Lanni6 at a representative depth of the specimen under the coverslip. For example, if a -thick specimen extends from 15 to under the coverslip, ideally we would use a PSF calculated for a depth of . Unfortunately, it is impossible to know a priori the depth spanned by the specimen, and thus the depth at which we calculate the PSF—the mid-depth through the volume collected—is unlikely to be halfway through the specimen and might even be outside the specimen depth. We assessed the sensitivity of the deconvolved image to the mismatch between the depth of the specimen and the depth at which the PSF is calculated with a simulation. We generated images at different depths and then deconvolved with PSFs calculated also at different depths. For this simulation, we generated 3-D synthetic images using PSFs that vary with the depth under the coverslip as where is the plane in the image space conjugate to the plane at which the microscope is focused, and , , , , and are defined as before. To implement Eq. 9 in the computer, we generated numerically a 3-D specimen and a sequence of 3-D PSFs. The specimen consisted of nine ellipsoids of different brightness and sizes. All the ellipsoids have their two major axes equal and parallel and perpendicular to the optical axis (so they look like as circles in the 2-D image). The major axes ranged from 1.6 to , and the minor axes from 0.6 to . The ellipsoids do not overlap in 3-D space, but some of their 2-D projections do overlap. The specimen was assumed to be in an aqueous medium with refractive index . The synthetic specimen has cubic pixels with per side. To generate the images according to Eq. 9, we generated a sequence of 64 3-D PSFs covering the depth of the specimen (e.g., from 0 to for the image shown in Fig. 5 ). Each of the PSFs was calculated over a grid with pixels. For the PSF, we assumed a oil-immersion objective and a fluorescent wavelength of . After generating the image numerically using a sampled version of Eq. 9, we added all the planes in the image to obtain a 2-D image equivalent to the one we would obtain with the through-focus sweep. We then deconvolved the resulting 2-D image using 2-D PSFs calculated at different depths. Figures 5a and 5b show the 2-D projection of a simulated specimen function and a through-focus image whose central plane is about under the coverglass. Figures 5c and 5d show the deconvolution of this image with PSFs calculated at the correct depth and away from the correct depth, respectively.Even though the projection PSF is used to obtain the deconvolution in Fig. 5c, the restored image is very similar to the synthetic specimen function [Fig. 5a]. Also, although the image restored with a PSF at a wrong depth is somewhat artifactual, it is still possible to accurately determine the location of the different ellipsoids. Figure 6 shows brightness traces along the diagonal of the box shown in Fig. 5b. There is little difference between the deconvolved image obtained with the correct PSF (circles) and with PSFs up to about mismatch (squares). However, a PSF-depth mismatch of about results in an artifactual deconvolution. In these simulations, the image covered a depth of approximately , and therefore using a PSF calculated anywhere within the recorded volume gives results very similar to those obtained with the correct PSF.Fig. 5Sensitivity to PSF-depth mismatch. (a) 2-D projection of the synthetic specimen function. (b) Simulated through-focus image for a oil immersion lens focusing into an aqueous medium. (c) Deconvolution using the correct PSF. (d) Deconvolution using a PSF depth wrong by about . (Scale bar .)  Fig. 6Brightness traces along deconvolved images. Traces along the diagonal of the box in Fig. 5. Up to about depth mismatch (squares), the deconvolution is very similar to that obtained with the correct PSF (circles). However, for a mismatch of about (triangles), the deconvolution departs from the correct one. 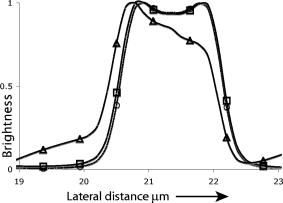 7.Earlier DevelopmentsA method to render images with EDOF was proposed by Holmes, 15 in which a 3-D stack of images is collected and its Fourier transform is computed. From the Fourier transform, the central slice is extracted. Likewise, the central slice of the OTF is extracted. From these central slices, they calculate the Fourier transform of the estimated specimen function where is the Wiener-Helstrom filter described before [Eq. 8]. Although the method proposed here and the one proposed by Holmes 15 are mathematically equivalent, their approach requires the collection of a 3-D stack that greatly increases the data acquisition time and is one of the main reasons for using an EDOF image instead of the full 3-D image.Häusler16 and Häusler and Körner17 suggest a method in which an image is acquired in the way we propose, that is, by keeping the camera shutter open while the specimen moves through focus. Häusler16 processes the resulting image with an optical filter. That is, the recorded image is projected through a lens that has a mask in its back-focal plane. The optical density of this mask is proportional to the function . This processing, however, assumes that the image was formed under coherent illumination, a situation that does not apply to fluorescence microscopy. Häusler and Körner17 suggest processing the image by analog electronics using the so-called -filter. That is, a filter whose amplitude is proportional to . The selection of this filter is based on the observation that the projection OTF (see Fig. 4) is proportional to except for . The -filter, however, is known to greatly amplify high-frequency noise. Häusler and Körner suggest using homomorphic filtering to reduce the noise. Even with the noise reduction, their method is limited by the use of a linear filter for deconvolution, which at best enhances components up to about 1/4 of the cut-off frequency of the imaging system. Neither the method proposed by Holmes 15 nor that of Häusler 16, 17 can render an EDOF image with the resolution afforded by the method we propose. We have similarly acquired through-focus fluorescent image data using a spinning disk confocal microscope. In this application, fluorescent signals above and below the volume do not interfere with the signals of interest, deconvolution is not required and, by translating along as well as during two overlapping acquisitions (positively along for the first image, negatively along for the second), have acquired stereo pair images. However, this approach is not as sensitive as the method described before, and the focus and translation movements must be relatively slow to avoid streaking introduced by the moving pinholes. 8.Conclusions and Future WorkWe presented a method to obtain extended depth-of-focus images that combines a new way to acquire images and a constrained deconvolution algorithm. The method allows for fast data acquisition by collecting the whole image volume within a single image, thus eliminating the dead time that results from stopping and starting focus changes and opening and closing the illumination and camera shutters. There are, however, several issues to investigate to optimize the throughput of the method. For example, how far above and below the specimen is it necessary to collect light? If the recorded volume is too small, then Eq. 4 is no longer independent of the axial coordinates and , and the 2-D recorded image can no longer be approximated as a 2-D convolution. On the other hand, a thick volume requires longer exposure time that increases photobleaching and risks saturating the CCD camera. Another important issue is the choice of the deconvolution algorithm. The EM-ML we currently used is flexible and robust, but slow. We have developed deconvolution algorithms that are significantly faster but less robust (in particular to truncated data).5 For the routine application of our method, we now use these faster algorithms. Finally, we will determine rapid and high resolution methods for producing stereo pairs of images. AcknowledgmentsThe authors thank Danny J. Smith for his help setting up and maintaining the computers used for this research; Ben Fowler for his expert help with the microscopes; and Mary Flynn and Adrienne Gidley for their help with the everyday running of our laboratories. The support of NIH grants R01 GM49798, R01 GM55708, and R01 EB002344; NSF grant EPS-0132534; the College of Engineering of the University of Oklahoma, and the Oklahoma Medical Research Foundation is gratefully acknowledged. ReferencesE. R. Dowski Jr., W. T. Cathey,
“Extended depth of field through wave-front encoding,”
Appl. Opt., 34
(11), 1859
–1866
(1995). 0003-6935 Google Scholar
M. E. Dresser and
J. A. Conchello,
“Extended depth of focus microscopy,”
(2003) Google Scholar
A. P. Dempster,
N. M. Laird, and
D. B. Rubin,
“Maximum likelihood from incomplete data via the EM algorithm,”
J. R. Stat. Soc. Ser. B (Methodol.), 39
(1), 1
–38
(1977). 0035-9246 Google Scholar
J. A. Conchello,
“Super-resolution and convergence properties of the expectation maximization for maximum-likelihood deconvolution of incoherent images,”
J. Opt. Soc. Am. A, 15
(10), 2609
–2619
(1998). 0740-3232 Google Scholar
J. Markham and
J. A. Conchello,
“Fast maximum-likelihood image restoration algorithms for three-dimensional fluorescence microscopy,”
J. Opt. Soc. Am. A, 18
(5), 1062
–1071
(2001). https://doi.org/10.1364/JOSAA.18.001062 0740-3232 Google Scholar
S. F. Gibson and
F. Lanni,
“Experimental test of an analytical model of aberration in an oil-immersion objective lens used in three-dimensional light microscopy,”
J. Opt. Soc. Am. A, 8
(10), 1601
–1613
(1991). 0740-3232 Google Scholar
D. P. Laurie,
“Calculation of Gauss-Kronrod quadrature rules,”
Math. Comput., 66
(219), 1133
–1145
(1997). https://doi.org/10.1090/S0025-5718-97-00861-2 0025-5718 Google Scholar
D. Calvetti,
G. H. Golub,
W. B. Gragg, and
L. Reichel,
“Computation of Gauss-Kronrod quadrature rules,”
Math. Comput., 69
(231), 1035
–1052
(2000). https://doi.org/10.1090/S0025-5718-00-01174-1 0025-5718 Google Scholar
J. A. Conchello and
E. W. Hansen,
“Enhanced 3D reconstruction from confocal scanning microscope images I: Deterministic and maximum likelihood reconstructions,”
Appl. Opt., 29
(26), 3795
–3804
(1990). 0003-6935 Google Scholar
D. A. Agard,
“Optical sectioning microscopy,”
Annu. Rev. Biophys. Bioeng., 13 191
–219
(1984). https://doi.org/10.1146/annurev.bb.13.060184.001203 0084-6589 Google Scholar
D. A. Agard,
Y. Hiraoka,
P. J. Shaw, and
J. W. Sadat,
“Fluorescence microscopy in three dimensions,”
Methods Cell Biol., 30 353
–377
(1989). 0091-679X Google Scholar
C. Preza,
M. I. Miller, L. J. Thomas Jr., J. G. McNally,
“Regularized linear method for reconstruction of three-dimensional microscopic objects from optical sections,”
J. Opt. Soc. Am. A, 9
(2), 219
–228
(1992). 0740-3232 Google Scholar
B. R. Frieden, Probability, Statistical Optics, and Data Testing, Springer-Verlag, Berlin, Germany (1982). Google Scholar
P. J. Sementilli,
B. R. Hunt, and
M. S. Nadar,
“Analysis if the limit to superresolution in incoherent imaging,”
J. Opt. Soc. Am. A, 10
(11), 2265
–2276
(1994). 0740-3232 Google Scholar
T. J. Holmes,
Y. H. Liu,
D. Kosla, and
D. A. Agard,
“Increased depth of field and steropairs of fluorescence micrographs via inverse filtering and maximum-likelihood estimation,”
J. Microsc., 164
(part 3), 217
–237
(1991). 0022-2720 Google Scholar
G. Häusler,
“A method to increase the depth of focus by two step image processing,”
Opt. Commun., 61
(1), 33
–42
(1972). https://doi.org/10.1016/0030-4018(87)90119-2 0030-4018 Google Scholar
G. Häusler and
E. Körner,
“Imaging with expanded depth of focus,”
Zeiss Inf., 29 9
–13
(1986-1987). Google Scholar
|