|
|
|
Stochastic Monte Carlo (MC) modelling is widely used to simulate the peculiarities of light scattering in disperse composite media with complex internal structures including biological tissues.1 An opportunity to imitate directly the influence of varieties of structural and physiologic properties of biological tissues on the photon migration for a particular source-detector configuration makes MC a primary tool in biomedical optics and optical engineering. A number of MC codes have been developed in the past1–5 and applied extensively for various biomedical applications, including light dosimetry modelling for photodynamic therapy treatment planning,6 simulation of reflectance spectra of human skin,7 analysis of fluorescence excitation in skin tissues,8 imitation of photon migration in the brain,9 two-dimensional images of skin by optical coherence tomography (OCT),10 and others. It has been demonstrated that MC technique is able to provide a broad variety of practical solutions in a range of biomedical studies from single cells to the biopsy of specific biological tissues and whole organs. However, simulation time was a significant concern in all of the developed MC models previously mentioned. The situation dramatically changed in 2005 when leading hardware manufactures were faced with fundamental limitations (a rapidly approaching physical limit of transistor size) and were unable to extract additional power from existing architectures, they began offering processors with multiple cores. This progress in technology is known as the “multicore revolution.”11 Today nearly all personal computers have more than one processor core, making truly parallel computing available for developers. With the further developments of multiprocessor architecture a novel compute unified device architecture (CUDA) technology became available,11,12 It has been demonstrated that the multiple-iterative parts of an algorithm can be executed in parallel on CUDA cores bringing up to hundreds of times the speed increase. CUDA is a parallel computing architecture which utilizes both hardware and software levels developed by the world leading graphic accelerators manufacturer NVIDIA Corp. Introduced in 2007, CUDA has undergone significant changes in conformance to a number of futures that NVIDIA’s engineers added to the graphic chip.11 NVIDIA CUDA technology provides an unlimited access to computational resources of graphic cards, processor cores, and different types of memory (can be distinguished by capacity and speed), making it a massive coprocessor in parallel computations.12 Typical CUDA graphic multiprocessors utilize a fundamentally different design philosophy than CPUs and consist of a number of streaming processors logically divided in hundreds of CUDA cores that can execute thousands of threads simultaneously, without context switch performance losses and extremely fast on-chip graphic double data rate memory (GDDR). This philosophy, known as single program multiple data, makes graphics processing units (GPUs) specialized for compute-intensive, highly-parallel computations, such as graphics rendering. Within the development of the CUDA framework (including compiler, debugger, profiler, software development kit, documentation) the ultimate petaflop computational power of GPUs became available to a variety of industries and applications.11,12 Therefore, using GPU acceleration, the time and performance of MC simulation has significantly improved.13–16 Based on the next CUDA generation, so-called architecture code Fermi, introduced in 2010, a new concept of MC model utilizing object-oriented programming (OOP) and GPU acceleration has been proposed.17 The OOP approach allows describing photon packets and the structural components of biological tissues (e.g., layers, vessels, tumors, cells, collagen fiber, etc.) as objects. Thus, the photon migration in the medium is presented as an interaction of objects, photons with the object, medium (or with multiple medium’s objects). Dividing the medium into the objects makes it possible to develop realistic tissue models presenting three-dimensional spatial variations of complex biological structures. Moreover, potentially the OOP approach enables import of the actual structure of medium as an object from a particular imaging modality, e.g., from electron microscopy, magnetic resonance imaging, or OCT. Integrating CUDA acceleration with the modern web technologies, such as Microsoft Silverlight, ASP.NET Framework, the online object-oriented MC (O3MC) computational tool was developed.17 The key idea behind O3MC (Ref. 17) development is the creation of a universal computational tool to simulate the results of real experiments typically used in major applications in biomedical optics and related areas that could provide researchers with practical results nearly in real time. OOP and GPU implementations speeds up the procedure of MC simulation up to times.18 However, due to the multiuser architecture of the online solution, concurrent simulations by multiple clients significantly degrade performance of O3MC. For example, if one user accessing the O3MC can get the results in 4.3 s on TESLA M2090 GPU, 100 users accessing O3MC at the same time can be stacked in a queue and wait for 10 to 15 min. Therefore, in framework of further development of O3MC (available online at: www.biophotonics.ac.nz) to deal with the multiuser access we apply a peer-to-peer (P2P) network. The proposed P2P network consists of a set of computers, called nodes or peers, which communicate and share their GPUs (Fig. 1). The peers in a P2P network are equal among each other, acting both as clients and servers.19 The P2P approach has gained a lot of popularity in recent years, especially in terms of multimedia content delivery and communication (e.g., BitTorrent, Skype, etc.). With current development, for the first time to our knowledge, we apply a hybrid P2P network (Fig. 1) utilizing different types of peers for MC simulation. Fig. 1Schematic presentation of P2P O3MC implementation. Clients interact with the O3MC web interface via a preferred web browser. The server accepts O3MC simulation requests and keeps track of the participating peers. The P2P network consists different computers equipped with the CUDA-supporting GPUs: 1. a workstation with two GeForce GTX 480 GPUs each 480 CUDA cores, 1540 Gigaflops of the peak single-precision Gigaflops double-precision FPP, 1536 GB of GDDR5 memory; 2. Thorlabs OCT imaging system workstation with Quadro FX580 featuring 32 CUDA cores, 512 MB GDDR3 memory; 3. computational server equipped with two Tesla M2090 GPUs each 512 CUDA cores, 1331 Gigaflops of the peak single-precision floating point performance Gigaflops double-precision FPP, 6 GB of GDDR5 memory; and 4. Dell laptop with GeForce GT555M featuring 144 CUDA cores, 3072 GB GDDR3 memory. Images were adopted from manufactures’ websites and/or have a free license. 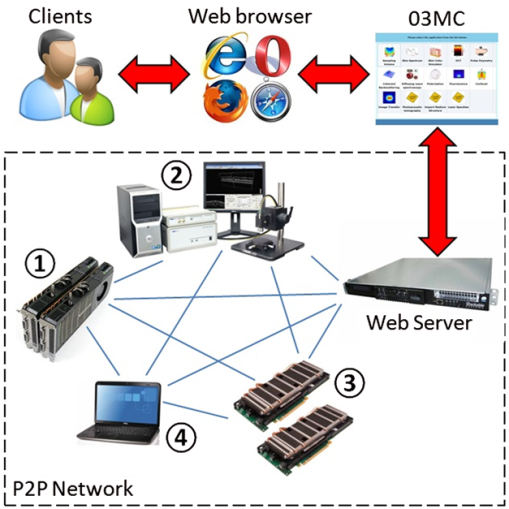 The web server hosts the online MC tool user interface, accepts O3MC simulation requests from clients, and keeps track of the other nodes (Fig. 1). The nodes are responsible for sharing the information about currently queuing MC simulations, processing them on GPUs, uploading, downloading, and hosting the outcomes (presented in a typical journal-paper format) among themselves without the need of the central server. To develop a P2P network (Fig. 1), the recently introduced P2P features of .NET 4.0 Windows Communication Foundation (WCF) were applied.20 This allows integrating the P2P network with the load balancing part of O3MC as both are written in managed code using .NET framework application programming interface.17 Thus, a number of MC simulations can be executed simultaneously on peers without queuing. Following Ref. 16, we consider simulation of fluence rate as a quantitative measure of MC performance (Fig. 2). Fig. 2Gradient of fluence rate counted by P2PMC for photon packets. is the depth and is the radial grid line in cylindrical coordinate system. The power density of the incident beam is . P2PMC available online at www.biophotonics.ac.nz. 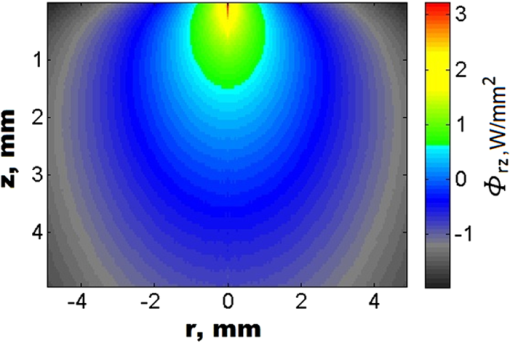 Medium with the following parameters was used in all simulations: the scattering coefficient , absorption coefficient , factor anisotropy , and refractive index . The time required to perform one simulation of photon packets is different at each peer: TESLA M2090 GPU spans 0.253 s for counting photon packets migration; GTX480; 0.567 s; FX580, 7.466 s; and GT555M, 2.134 s. The current network of peers can be slower, e.g., compared with a realization of MC modeling on GPU cluster.14 However, configuring a P2P network with clusters will surpass the performance of a standalone cluster. We also compared P2PMC with O3MC (Ref. 17) and the GPU-based MC codes summarized in Ref. 16. The results are presented in Table 1. Table 1Comparison of P2PMC with other MC codes applied for simulation of fluence rate for 3×107 photon packets.16 The parameters of the medium are: μs=10 mm−1, μa=0.05 mm−1, g=0.9, and n=1.5. Note, in O3MC and P2PMC the time of simulation is not depending on the absorption properties of the medium.
Due to the different nature of peers involved, the current P2P Network operates with Gigaflops of floating-point operations per second for single and with Gigaflops for double-precision computations. Thus, the best speedup of processing multiuser requests has been achieved using single-precision computing, whereas utilizing double precision for floating-point arithmetic operations in O3MC provides the most accurate results of modeling (Table 1). This is explained by the fact that Tesla M2090 GPUs used by a standalone version of O3MC fully support IEEE standards for floating-point arithmetic operations (IEEE 754) revised in 2008 (Ref. 21), whereas FX580 and GT555M GPUs in a P2P network in current study does not have this capability. Having more peers with better double-precision capabilities will increase the double-precision computation capacity of the entire network. In addition, the P2PMC has been also validated by comparing the results of simulation of diffuse reflectance for a semi-infinite scattering medium with known analytical results tabulated by Giovanelli,22 results of adding-doubling method,22 MCML (Ref. 3) and O3MC (Ref. 17). The results of comparison are presented in Table 2. Table 2The results of total diffuse reflectance Rd given by analytical results,22 adding-doubling method,23 MCML,3 O3MC,17 and P2PMC for 5×104 photon packets. The parameters of the medium are: μs=9 mm−1, μa=1 mm−1, g=0, and n=1.5.
The developed MC technique was also comprehensively validated by comparison with the known exact solution by Milne25,26 and with the results of experimental studies of image transfer through the water solution of spherical microparticles of known size and density.27,28 Thus, in this letter, utilizing different types of CUDA GPUs, we consider the possibility of using a P2P network to provide multiuser access for an online MC simulation of photon migration in complex turbid media, such as biological tissues. The multiuser access capability was found to be linearly dependent on the number of peers. The best speedup of processing multiuser requests in a range of 4 to 35 s, depending on the particular peers’ involvement, was achieved using single-precision computing, whereas the double-precision for floating-point arithmetic operations provides the best accuracy. Thus, with the growing number of peers (preferably double-precision), including those distributed worldwide, a P2P solution will allow multiple users to perform accurate reasonably fast MC modeling (i.e., in about 5 s for photon packets, excluding the time for P2P network communication). Further development of such a network worldwide could be the base for a computational platform for biomedical optics and optical diagnostic community. ReferencesF. Martelliet al., Light Propagation through Biological Tissue and Other Diffusive Media: Theory, Solutions, and Software, SPIE Press, Bellingham, WA
(2009). Google Scholar
R. Graaffet al.,
“Condensed Monte Carlo simulations for the description of light transport,”
Appl. Opt., 32
(4), 426
–434
(1993). http://dx.doi.org/10.1364/AO.32.000426 APOPAI 0003-6935 Google Scholar
L. H. WangS. L. JacquesL.-Q. Zheng,
“MCML: Monte Carlo modeling of photon transport in multi layered tissues,”
Comput. Meth. Prog. Bio., 47
(2), 131
–146
(1995). http://dx.doi.org/10.1016/0169-2607(95)01640-F CMPBEK 0169-2607 Google Scholar
J. Ramella-RomanS. PrahlS. Jacques,
“Three Monte Carlo programs of polarized light transport into scattering media: part I,”
Opt. Express, 13
(12), 4420
–4438
(2005). http://dx.doi.org/10.1364/OPEX.13.004420 OPEXFF 1094-4087 Google Scholar
I. V. Meglinskiet al.,
“Monte Carlo simulation of coherent effects in multiple scattering,”
Proc. Roy. Soc. A, 461 43
–53
(2005). http://dx.doi.org/10.1098/rspa.2004.1369 Google Scholar
W. M. Star,
“Light dosimetry in vivo,”
Phys. Med. Biol., 42
(5), 763
–787
(1997). http://dx.doi.org/10.1088/0031-9155/42/5/003 PHMBA7 0031-9155 Google Scholar
I. V. Meglinski,
“Modeling the reflectance spectra of the optical radiation for random inhomogeneous multi-layered highly scattering and absorbing media by the Monte Carlo technique,”
Quantum Electron., 31 1101
–1107
(2001). http://dx.doi.org/10.1070/QE2001v031n12ABEH002108 QUELEZ 1063-7818 Google Scholar
D. Y. ChurmakovI. MeglinskiD. A. Greenhalgh,
“Amending of fluorescence sensor signal localization in human skin by matching of the refractive index,”
J. Biomed. Opt., 9
(2), 339
–346
(2004). http://dx.doi.org/10.1117/1.1645796 JBOPFO 1083-3668 Google Scholar
D. A. Boaset al.,
“Three dimensional Monte Carlo code for photon migration through complex heterogeneous media including the adult human head,”
Opt. Express, 10
(3), 159
–170
(2002). OPEXFF 1094-4087 Google Scholar
M. Kirillinet al.,
“Simulation of optical coherence tomography images by Monte Carlo modeling based on polarization vector approach,”
Opt. Express, 18 21714
–21724
(2010). http://dx.doi.org/10.1364/OE.18.021714 OPEXFF 1094-4087 Google Scholar
D. B. KirkW. W. Hwu, Programming Massively Parallel Processors: A Hands-on Approach, MK Publishers, Burlington, MA
(2010). Google Scholar
J. SandersE. Kandrot, CUDA by Example: An Introduction to General-Purpose GPU Programming, Addison-Wesley, USA
(2010). Google Scholar
E. AlerstamT. SvenssonS. Andersson-Engels,
“Parallel computing with graphics processing units for high-speed Monte Carlo simulation of photon migration,”
J. Biomed. Opt., 13
(6), 060504
(2008). http://dx.doi.org/10.1117/1.3041496 JBOPFO 1083-3668 Google Scholar
W. LoL. Lilge,
“Accelerated 3-D Monte Carlo light dosimetry using a graphics processing unit (GPU) cluster,”
Proc. SPIE, 7376 737609
(2010). http://dx.doi.org/10.1117/12.871592 PSISDG 0277-786X Google Scholar
Q. Fang,
“Mesh-based Monte Carlo method using fast ray-tracing in Plücker coordinates,”
Biomed. Opt. Express, 1 165
–175
(2010). http://dx.doi.org/10.1364/BOE.1.000165 BOEICL 2156-7085 Google Scholar
H. ShenG. Wang,
“A study on tetrahedron-based inhomogeneous Monte Carlo optical simulation,”
Biomed. Opt. Express, 2
(1), 44
–57
(2011). http://dx.doi.org/10.1364/BOE.2.000044 BOEICL 2156-7085 Google Scholar
A. DoroninI. Meglinski,
“Online object oriented Monte Carlo computational tool for the needs of biomedical optics,”
Biomed. Opt. Express, 2
(9), 2461
–2469
(2011). http://dx.doi.org/10.1364/BOE.2.002461 BOEICL 2156-7085 Google Scholar
A. DoroninI. Meglinski,
“Monte Carlo simulation of photon migration in turbid random media based on the object-oriented programming paradigm,”
Proc. SPIE, 7907 790709
(2011). http://dx.doi.org/10.1117/12.879558 PSISDG 0277-786X Google Scholar
K. RagabT. HelmyA. Hassanien,
“Developing advanced web services through P2P computing and autonomous agents: trends and innovations,”
Information Science Reference, IGI Global, Hershey, PA
(2010). Google Scholar
M. Neely,
“Peer fun: a peer-to-peer work processing app with WCF,”
MSDN Magazine, 6 68
–78
(2009). Google Scholar
N. Pathak, Pro WCF 4: Practical Microsoft SOA Implementation, APRESS, Berkely, CA
(2011). Google Scholar
R. Giovanelli,
“Reflection by semi-infinite diffusers,”
Optica Acta, 2
(4), 153
–162
(1955). http://dx.doi.org/10.1080/713821040 OPACAT 0030-3909 Google Scholar
S. Prahl,
“The adding-doubling method,”
Optical-Thermal Response of Laser Irradiate Tissue, 101
–125 Plenum Press, New York
(1995). Google Scholar
NVIDIA Corporation,
(2011). Google Scholar
V. L. KuzminI. V. Meglinski,
“Coherent effects of multiple scattering for scalar and electromagnetic fields: Monte-Carlo simulation and Milne-like solutions,”
Opt. Commun., 273 307
–310
(2007). http://dx.doi.org/10.1016/j.optcom.2007.01.025 OPCOB8 0030-4018 Google Scholar
V. L. KuzminI. V. MeglinskiD. Y. Churmakov,
“Stochastic modeling of coherent phenomena in strongly inhomogeneous media,”
J. Exp. Theor. Phys., 101
(1), 22
–32
(2005). http://dx.doi.org/10.1134/1.2010658 JTPHES 1063-7761 Google Scholar
E. Berrocalet al.,
“Image transfer through the complex scattering turbid media,”
Laser Phys. Lett., 3
(9), 464
–468
(2006). http://dx.doi.org/10.1002/(ISSN)1612-202X 1612-2011 Google Scholar
E. Berrocalet al.,
“Imaging through the turbid scattering media: part I: experimental and simulated results for the spatial intensity distribution,”
Opt. Express, 15 10649
–10665
(2007). http://dx.doi.org/10.1364/OE.15.010649 OPEXFF 1094-4087 Google Scholar
|

