|
|
|
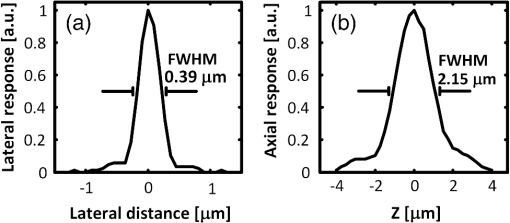
Mapping neural connectivity of the whole brain is one of the key steps to understanding the brain in neuroscience. To map the neural networks of the entire brain, one is required to obtain sub-micron neural connection information on a centimeter scale.1 Recent progresses of micro-optical sectioning tomography (MOST) and fluorescence micro-optical sectioning tomography (fMOST)2,3 have achieved sub-micron voxel resolution imaging of the whole brain. But total data acquisition times still were 242 and 447 h, respectively, which was too slowly for neuroscience applications. Structured illumination microscopy (SIM), which was first demonstrated by Neil et al., has optical sectioning ability comparable to confocal microscopy.4 In this technique, SIM projects a predefined high-frequency cosine illumination pattern on the specimen. Only in-focus information is modulated by this illumination fringe, owing to the rapid attenuation of high spatial frequency pattern with defocus. SIM acquires at least three images with different phase shifts to obtain an optical section. The imaging speed of SIM depends on the process of shifting the cosine illumination pattern. Various methods of cosine fringe projection have been demonstrated in SIM systems, including mechanically moving grating,5,6 two beam interference illumination,7 micro-stripe array light-emitting diodes (LED),8 and liquid crystal spatial light modulators.9 Despite these efforts, an illumination pattern transition time of several tens of milliseconds still limits the net frame rate of SIM to a few hertz. A digital mirror device (DMD)-based SIM has been reported to achieve a cost-effective optical sectioning at an imaging speed of .10 However, because SIM needs three raw images to get a section image, the real net SIM sectioning speed had been reduced to approximately , similar to ones of MOST and fMOST. Relatively low imaging speed is a serious impediment to the high-throughput neuroanatomy application. In this study, by employing the fast modulation ability of DMD in binary mode and the fast imaging ability of scientific complementary metal-oxide semiconductor (CMOS) camera (sCMOS), we demonstrate a SIM image net rate of , an order of magnitude faster than existing high-throughput neural networks imaging methods.2,3,11,12 We used a DMD in binary mode to make full use of the fast modulation ability of DMD. Patterns on the DMD plane can be defined as where and represent pixel coordinates on the DMD. Selecting different values of can obtain various fringe periods. By shifting the pixel values of the DMD to and , respectively, the phase shift steps of and on the specimen plane would then be executed, respectively. After acquiring three images, we can reconstruct the optical section image by4Our experimental setup is shown in Fig. 1. A DMD (XD-ED01N, X-digit) was placed in a conjugate image plane of the excitation optical path. The DMD was a array with a size of . Excitation light from a mercury lamp (X-cite Exacte, Lumen Dynamics) was collimated and oriented toward the DMD. The specimen was fixed on the XY stage (Aerotech), and axial scanning was achieved by a piezoelectric translational stage (P-721, Physik instrumente). Emission light from the sample was captured by a sCMOS camera (ORCA-Flash4.0, Hamamatsu), which was synchronized with the DMD pattern transition. Fig. 1Schematic of the optical setup. LLG, liquid light guide; CL, collimating lens; M, mirror; DMD, digital micro-mirror; TL, tube lens; EX, excite filter; DM, dichroic mirror; PZT, piezoelectric translational stage; OBJ, objective lens; and EM, emission filter. 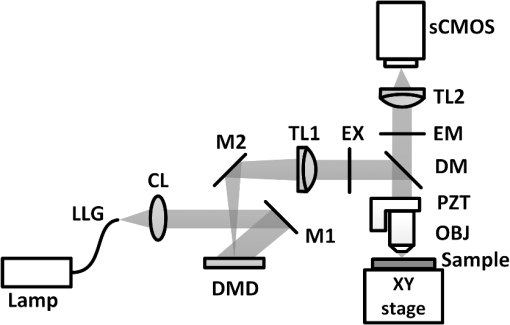 To test the performance of our system, we used fluorescence beads with a diameter of 170 nm (, Molecular Probes) to measure imaging resolution. By setting the pattern period on the DMD plane to 164.16 μm (), a three-dimensional (3-D) stack of the fluorescence beads was acquired with a step of 250 nm using a water-immersion objective (0.8 NA, LUMPLFLN 40XW, Olympus). Figure 2 shows that the measured lateral and axial FWHM values from a selected bead were 0.39 and 2.15 μm, respectively. Figure 3(a) and 3(b) shows the conventional wide field image and corresponding SIM optical section image of fluorescence microspheres (10-μm diameter), demonstrating the optical sectioning ability of SIM. The image was captured by a water-immersion objective (1.0 NA, XLUMPLFLN 20XW, Olympus), and excited light exposure power on the sample was measured to be 2.9 mW. SIM suppressed most of the out-of-focus blurring information. To demonstrate that it was capable of imaging the dynamic process, we developed an optical flow phantom model that consists of 1-mm diameter glass tubing and a micro-syringe pump. By using a micro-syringe pump, we could control the flow speed of fluorescent microspheres inside the glass tubing. The illumination grating is oriented perpendicular to the flow. Figure 3(c)–3(g) shows SIM images that were taken under different frame rates for five different moving fluorescent microspheres. The imaging pixel format of the sCMOS was set to , and binning by camera was used to enhance the signal. These microspheres could be tracked and the mean flowing rate was calculated to be . When the net frame rate of SIM was 133 Hz, there were no movement artifacts in the SIM section image. Once the frame rate decreased to 50 Hz, movement artifacts began to appear in the section image. The artifacts became increasingly severe as the frame rate decreased further, until the imaged fluorescence microsphere completely separated into three consecutive structured light modulated raw images. The reconstructed SIM section image would look like a superposition of three microspheres [Fig. 3(g)]. Fig. 3Images of fluorescent microspheres. (a) Conventional wide-field image of motionless fluorescent microspheres; (b) corresponding optically sectioned image captured by structured illumination microscopy (SIM); (c to g) structured illumination sectioned images of moving microspheres taken at different SIM imaging frame rates: faster recording rate (c) shows clear shape of the microsphere, while slower recording rate (d to g) shows deformed shape. Scale bars: 40 μm (a, b) and 15 μm (c to g), respectively. 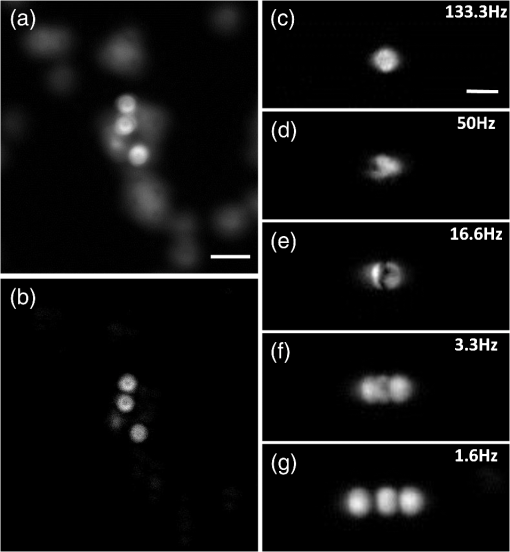 Finally, we demonstrate that the video-rate SIM is a fast imaging method which can be applied to large tissue imaging. The XY stage moves the sample under the objective to extend the field of view of SIM. Using a mosaic technique, we imaged the coronal section of a 100-μm thick formaldehyde-fixed mouse (Thy1-eYFP-H mouse) brain slice using a objective (0.8 NA, LUMPLFLN 40XW, Olympus). The exposure power at the sample was measured to be 1.7 mW. The whole mosaic included 1040 3-D image volumes. Each image volume contained 30 images at 2-μm intervals in the direction, that is, 60-μm thick in total, which was the maximum imaging depth without significant degradation of images for fixed brain tissue. Figure 4 shows the maximum intensity projection of the stitched coronal section. We can readily identify the dendritic spine in the enlarged view of the apical dendrite region of the cortex [Fig. 4(c)]. Fig. 4(a) Coronal section of a mouse brain. (b) Enlarged views of the region marked in (a), pyramid cells in the cortex. (c) Enlarged views of the region marked in (b), white arrows mark the individual spine on dendrite. Scale bars: 1 mm (a), 150 μm (b), and 15 μm (c), respectively. 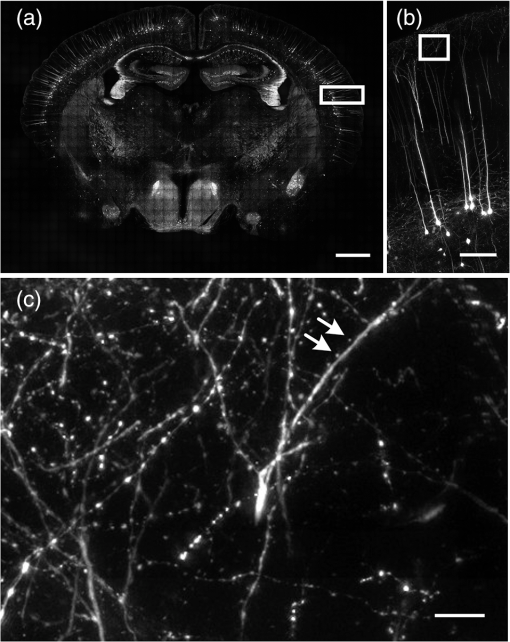 The lateral field of view of each mosaic image was . The refresh rate of the DMD was set as 50 Hz and each 60-μm thick mosaic 3-D volume was acquired in approximately 3.5 s. It took approximately 1 h to acquire the whole coronal 3-D data volume by objective. The depth discrimination ability of SIM makes block-face imaging available for the resin-embedded whole brain samples.2,3 A whole mouse was approximately , and the imaging field of view was when using a objective. mosaics were needed to cover a coronal section, and each 60-μm thick 3-D mosaic took 3.5 s to image. We could estimate that the imaging time for a whole brain would be (60 s for sectioning). It indicates that whole brain data acquisition at sub-micron resolution would require no more than three days. The imaging speed of this video-rate SIM here is limited by the exposure time and readout speed of the camera. The full frame rate of sCMOS is limited to 50 Hz in external trigger mode. Without any compromise in the field of view, 16.6 Hz is the fastest SIM net frame rate we can currently achieve. Further acceleration of the imaging process depends on the use of a brighter light source and a faster camera with a shorter readout time. In conclusion, by taking full advantage of the high-speed binary modulation capability of the DMD and fast capture ability of sCMOS, we demonstrate a video-rate structured illumination microscope for fast optical sectioning. At present, the imaging throughput of video-rate SIM can reach ( at a 50 Hz imaging frame rate). Meanwhile, the system enables one to resolve the fine structures of neurons, such as spine in thick brain tissue. In combination with appropriate brain tissue preparation and mechanical sectioning methods, this video-rate SIM has the potential to map neuron connectivity information for entire brain in a relatively short time. AcknowledgmentsThis work was supported by the National Natural Science Foundation of China (Grant Nos. 81127002 and 91232306) and Science Fund for Creative Research Group of China (Grant No. 61121004). We thank Ming Lei for introducing the concept of SIM. ReferencesJ. W. LichtmanW. Denk,
“The big and the small: challenges of imaging the brain’s circuits,”
Science, 334
(6059), 618
–623
(2011). http://dx.doi.org/10.1126/science.1209168 SCIEAS 0036-8075 Google Scholar
A. Liet al.,
“Micro-optical sectioning tomography to obtain a high-resolution atlas of the mouse brain,”
Science, 330
(6009), 1404
–1408
(2010). http://dx.doi.org/10.1126/science.1191776 SCIEAS 0036-8075 Google Scholar
H. Gonget al.,
“Continuously tracing brain-wide long-distance axonal projections in mice at one-micron voxel resolution,”
Neuroimage, 74
(1), 87
–98
(2013). http://dx.doi.org/10.1016/j.neuroimage.2013.02.005 NEIMEF 1053-8119 Google Scholar
M. A. A. NeilR. JuskaitisT. Wilson,
“Method of obtaining optical sectioning by using structured light in a conventional microscope,”
Opt. Lett., 22
(24), 1905
–1907
(1997). http://dx.doi.org/10.1364/OL.22.001905 OPLEDP 0146-9592 Google Scholar
J. Siegelet al.,
“Whole-field five-dimensional fluorescence microscopy combining lifetime and spectral resolution with optical sectioning,”
Opt. Lett., 26
(17), 1338
–1340
(2001). http://dx.doi.org/10.1364/OL.26.001338 OPLEDP 0146-9592 Google Scholar
F. ChaslesB. DubertretA. C. Boccara,
“Optimization and characterization of a structured illumination microscope,”
Opt. Express, 15
(24), 16130
–16140
(2007). http://dx.doi.org/10.1364/OE.15.016130 OPEXFF 1094-4087 Google Scholar
M. A. A. NeilR. JuškaitisT. Wilson,
“Real time 3D fluorescence microscopy by two beam interference illumination,”
Opt. Commun., 153
(1–3), 1
–4
(1998). http://dx.doi.org/10.1016/S0030-4018(98)00210-7 OPCOB8 0030-4018 Google Scholar
V. Poheret al.,
“Optical sectioning microscopes with no moving parts using a micro-stripe array light emitting diode,”
Opt. Express, 15
(18), 11196
–11206
(2007). http://dx.doi.org/10.1364/OE.15.011196 OPEXFF 1094-4087 Google Scholar
N. Bozinovicet al.,
“Fluorescence endomicroscopy with structured illumination,”
Opt. Express, 16
(11), 8016
–8025
(2008). http://dx.doi.org/10.1364/OE.16.008016 OPEXFF 1094-4087 Google Scholar
D. Danet al.,
“DMD-based LED-illumination Super-resolution and optical sectioning microscopy,”
Sci. Rep., 3 1116
(2013). http://dx.doi.org/10.1038/srep01116 SRCEC3 2045-2322 Google Scholar
T. Raganet al.,
“Serial two-photon tomography for automated ex vivo mouse brain imaging,”
Nat. Methods, 9
(3), 255
–258
(2012). http://dx.doi.org/10.1038/nmeth.1854 1548-7091 Google Scholar
L. Silvestriet al.,
“Confocal light sheet microscopy: micron-scale neuroanatomy of the entire mouse brain,”
Opt. Express, 20
(18), 20582
–20598
(2012). http://dx.doi.org/10.1364/OE.20.020582 OPEXFF 1094-4087 Google Scholar
|