|
|
1.IntroductionDiffuse optical tomography (DOT) is a volumetric optical imaging technique that relies on modeling light transport in tissue using the diffusion approximation, which is generally applicable in scatter dominated systems. The spectral measure of the diffuse transport of near-infrared light through soft tissue can provide the ability to image functional tissue information such as hemoglobin oxygenation and water fraction, which can be useful as a noninvasive means of identifying cancer.1–3 This method has also been proven successful by the use of luminescence probes using, for example, fluorescence markers to allow quantitative molecular imaging of functional exogenous reporters.4,5 Light modeling can be done analytically,6 providing high accuracy and computational speed, but only on simple and dominantly homogeneous geometries. Numerical approaches allow solutions to be computed for more complex geometries, but require more computational time as well as a discrete representation (volume mesh) of the domain.7,8 Due to the generally poor spatial resolution of DOT, the prevailing trend in the field is toward combining it with other imaging modalities and incorporating high-resolution tissue structural information in the image recovery algorithm. Notable examples of this include computed tomography (CT) or magnetic resonance imaging (MRI)-guided DOT, and these techniques provide the potential for increased accuracy.9–12 Although the details of finite-element-based methods for modeling light transport in tissue are well covered in literature,13–21 the computational packages available for such modeling have until now included quite limited mesh creation tools or no mesh creation tools at all. In this work, an integrated and freely available software package is outlined and tested, which allows users to go all the way from import of standard digital imaging and communications in medicine (DICOM) images (and other related formats) to segmentation and meshing, and through to light simulation and property recovery. Image-guided DOT is very dependent on the ability to easily produce high-quality three-dimensional (3-D) volume meshes from medical images, and the process of mesh creation is a significantly underappreciated but complex issue, which is directly solved in many cases by software such as this. The software tool developed at Dartmouth College and University of Birmingham, United Kingdom, called Nirfast, is a finite-element-based package for modeling near-infrared light transport in tissue for medical applications.22,23 It is open source, free, and cross platform as developed under MATLAB (Mathworks Inc.), which also allows user-friendly understanding and modifications. Applications of Nirfast are diverse, including optical modeling for small animal imaging,24–27 breast imaging,3 brain imaging,28,29 and light dose verification in photodynamic therapy of the pancreas. Accurate diffusion modeling in optical tomography requires a 3-D geometry since the photon scattering is in all directions.20 Since the core finite element method (FEM) code of Nirfast is based on MATLAB,22 this has in the past hindered its ability to allow for easy coupling to highly complex 3-D meshing tools. One issue stems from the inability of MATLAB to efficiently visualize large 3-D meshes, while another issue is the necessity for custom image processing tools when dealing with an assortment of different medical image types and formats. Using a visualization toolkit/insight segmentation and registration toolkit-based platform, which itself is an open-source application, for segmentation and meshing has helped to address these issues by providing a seamless coupling within Nirfast. Providing the tools and workflow needed to create an FEM mesh from a variety of different types of medical images and seamlessly using this mesh for light transport modeling are essential to making DOT accessible and useful. The current version of Nirfast includes full-featured segmentation and mesh creation tools for quickly and easily creating high-quality 3-D finite-element meshes from medical images.23 The segmentation tools have been developed in collaboration with Kitware Inc. (Clifton Park, NY). Creating suitable volumetric meshes of complex tissue geometries is a particular challenge for multimodal DOT, due to the variety of contrast characteristics present in different imaging modalities and tissue types/models. Manual manipulation of the segmentation and mesh creation process often requires an overwhelming time investment, and high mesh element quality is notoriously difficult to ensure. It is also very important to retain both the outer and inner region surfaces (internal boundaries) in a mesh to allow the application of prior knowledge for both the forward and inverse models. Segmentation is rarely fully automated because some manual manipulation or input is standard for many complex problems, but by providing a customized collection of semiautomated routines, it is possible to substantially reduce the amount of manual touch-up required. There are various mesh creation tools available either commercially or freely, but each has its own limitations in application to optical tomography. For example, MeshLab is an open-source tool for creating unstructured 3-D triangular meshes, but has no semiautomated segmentation routines30 and is also lacking some workflow features such as the ability to undo the last action. Mimics is a commonly used commercial package designed for medical image processing, but mesh creation requires a great deal of manual input, and it has difficulty with multiple-region problems.31 Netgen is a freely available 3-D tetrahedral mesh generator, but has limitations with multiple-region problems.32 Some other mesh creation tools include DistMesh,33 iso2mesh,34 and quality mesh generation,35 but these are not linked to segmentation tools per se. There is no freely available tool that incorporates all of the workflow elements needed for segmentation and mesh creation in optical tomography in a seamless manner. The new tools in Nirfast help to address these issues, and in this study, their capabilities are tested and quantified in a series of cases which are representative of key application areas. 2.Materials and MethodsThe segmentation and mesh creation tools in Nirfast allow for a variety of different inputs, including standard DICOM formats for medical images, general image formats (stacks of bmp, jpg, png, etc.), and structured geometry formats (vtk, mha, etc.). It can be used for a variety of different medical imaging modalities, such as CT, MR, ultrasound, and microCT. Both automatic and manual means of segmenting these images have been provided, and mesh creation is fully automated with customizable parameters. The capability of these tools is demonstrated on four different cases that are relevant to the modeling of light propagation in tissue and optical tomography: small animal imaging, breast imaging, brain imaging, and light dose modeling in photodynamic therapy of the pancreas. The small animal example used a stack of CT images of the front portion of a mouse, consisting of 30 axial slices of 256 by 256 pixels, with a slice thickness of 0.35 mm. The images were taken on a Phillips MR Achieva medical system, in the form of a DICOM stack. The breast example used a stack of T1-weighted MR images, consisting of 149 coronal slices of 360 by 360 pixels, with a slice thickness of 0.64 mm. The images were taken on a Phillips MR Achieva medical system, in the form of a DICOM stack. The brain example used a stack of T1-weighted MR images, consisting of 256 axial slices of 256 by 256 pixels, with a slice thickness of 1 mm. The images were taken on a Siemens Trio 3T scanner and are stored in .hdr and .img files. The pancreas example used a stack of arterial phase CT images, consisting of 90 axial slices of 512 by 512 pixels, with a slice thickness of 1 mm. The images were taken on a Seimens Sensation 64 CT system, in the form of a DICOM stack. In each case, the appropriate modules were used to maximize the quality of the resulting mesh and increase the speed of the entire process. The general procedure for processing the images follows: First, the medical images are imported into the segmentation interface, shown in Fig. 1. Next, automatic segmentation modules are used to identify different tissue types and regions as accurately as possible. See Table 1 for the steps used in each case, as well as parameter values. The modules and their respective parameters are detailed in Table 2. Explanations of the major segmentation modules are described below. Fig. 1The interface for segmentation of tissue types in medical images is shown, with 3-D orthogonal views at right and histogram information at left. 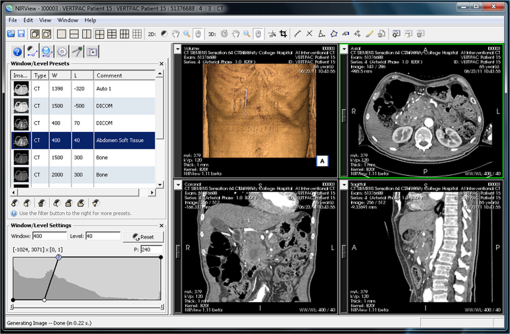 Table 1Time benchmarks for the segmentation and mesh creation of four different imaging cases: brain, pancreas, breast, and small animal, with the key steps in accurate segmentation identified and the time required for each step specified.
Table 2List of automated segmentation modules available for identifying tissue types, including an explanation of the parameters in each case.
The iterative hole-filling algorithm classifies small volumes within a larger region as part of the outer region. In each iteration, a voting algorithm determines whether each pixel is filled based on the percentage of surrounding pixels that are filled, a ratio defined as the majority threshold.36 The number of iterations controls the maximum size of holes and cavities filled. This is useful as a final step for any segmentation, to ensure that each tissue type region is homogeneous and lacking unintended holes. The K-means and Markov random field module performs a classification using a K-means algorithm to cluster grayscale values and then further refines the classification by taking spatial coherence into account.37 The refinement is done with a Markov random field algorithm. The essential minimization function governing this module is given below, where is the set of grayscale values, is the sets of classification groups, and is the set of mean values in each . This method is most relevant to situations where the grayscale values and spatial location of different tissue types show significant differences. It is used as a first step in the segmentation process after any image processing has been applied to the medical images. The MR bias field correction module removes the low-frequency gradient often seen in MR images, using the nonuniform intensity normalization (N3) approach.38,39 This bias is largely caused by the spatial dependence of the receiving coil and can cause other segmentation modules to be ineffective due to the range of grayscale values produced in each region. This module is useful for all MRI data, as well as other imaging modalities that may produce low-frequency gradients in the images. It is applied before any segmentation modules to ensure that the gradient does not adversely affect the segmentation process. The MR breast skin extraction module helps extract the skin in MR breast images, as it is often lumped in with the glandular region by other automatic modules. It is a specially designed module that uses several other filters in performing the skin extraction: cropping, thresholding, dilation, connected component thresholding, hole filling, and Boolean operations. This should be used as a first step after medical image processing for MR breast data where the skin is visible. Thresholding is a fundamental module that identifies a particular range of grayscale values as a single region.40 It is most useful when tissue types have distinct ranges of grayscale values with negligible overlap and is used at many stages in segmentation to identify and separate regions. Region dilation and erosion expand or contract a single region by a specified number of pixels in all directions.41 This can be useful as an alternate method of hole filling, by performing a dilation followed by an erosion of the same magnitude. It can also be used to remove insubstantial components of a volume by performing an erosion followed by a dilation of the same magnitude. An example of this would be removing the ears in a mouse model, due to the extremely small volume. Finally, dilation and erosion can be used to correct region sizing in cases where K-means has produced regions that are too small or large.After automatic segmentation, the regions are manually touched up using a paintbrush in order to fix any remaining issues with the segmentation such as stray pixels or holes. Finally, the segmentation is provided as an input to the meshing routine, which creates a 3-D tetrahedral mesh from the stack of two-dimensional (2-D) masks in a single run. This eliminates intermediate steps such as creating 3-D surfaces and thus requires less mesh preprocessing. The resulting mesh is multiregional and can preserve the structural boundaries of segmented tissues. The user has control over element size, quality, and approximation error. For ease of use, these values are set automatically based on the segmentation and medical image information, and no prior knowledge of mesh generation is required to use the tool. The volume meshing algorithm is unique and based on the computational geometry algorithms library (CGAL)42 and consists of several new features and implementations that are briefly outlined. The CGAL mesh generation libraries are based on a meshing engine utilizing the method of Delaunay refinement.43 It uses the method of restricted Delaunay triangulation to approximate one-dimensional curved features and curved surface patches from a finite set of point samples on a surface44,45 to achieve accurate representation of boundary and subdividing surfaces in the mesh. One very important feature that is of importance is that the domain to be meshed is a region of 3-D space that has to be bounded and the region may be connected or composed of multiple components and/or subdivided in several subdomains. The flexibility of this volume meshing algorithm allows the creation of 3-D volumes consisting of several nonoverlapping regions, allowing the utilization of structural prior information in diffuse optical imaging. The output mesh includes subcomplexes that approximate each input domain feature as defined in the segmented mask described above. During the meshing phase, several parameters can be defined to allow optimization of 3-D meshing, consisting of surface facet settings and tetrahedron size, which are detailed in Table 3. Table 3List of parameters controlling the 3-D tetrahedral mesh creation, including description of the function of each parameter.
Once the volumetric mesh has been created, a new feature has been added to allow the users to further optimize the 3-D mesh utilizing the Stellar mesh improvement algorithm.46 This optimization routine improves tetrahedral meshes so that their worst tetrahedra have high quality, making them more suitable for finite element analysis. Stellar employs a broad selection of improvement operations, including vertex smoothing by nonsmooth optimization, stellar flips and other topological transformations, vertex insertion, and edge contraction. If the domain shape has no small angles, Stellar routinely improves meshes so that the smallest dihedral angle is and the largest dihedral angle is . Reconstructions were performed for the small animal case with meshes created from segmentations in Mimics and Nirfast, using the optimization tools in each. The nude mouse was implanted with tumor cells in the animal’s brain, injected with Licor IRDye-800CW EGF, and imaged at Dartmouth College with an MRI-fluorescence molecular tomography system using a protocol described in a previous publication.47 Fluorescence optical data from eight source and detector locations positioned evenly around the mouse head were calibrated in Nirfast and used in reconstruction. 3.ResultsThe time of each step in segmentation and meshing was recorded for all cases, and the results are shown in Table 1. A visualization of a mesh for each case is shown in Figs. 2Fig. 3Fig. 4–5 for illustration. Using the case of the pancreas, the time taken for segmenting and creating a mesh using the tools in Nirfast was compared with that of the commercial package Mimics, designed for medical image processing.31 As seen in Fig. 6, Nirfast shows drastic improvements in the speed of both segmentation and meshing. It is worth noting that postprocessing mesh improvements have not been applied to the Mimics mesh, other than tools available in Mimics. Fig. 2Original MRI axial slice of the brain (a), segmentation of different tissue types (b), and the 3-D tetrahedral mesh for the brain (c), showing the regions as different colors: red is the skin, yellow is the cerebral spinal fluid, green is the skull, blue is the white matter, and orange is the gray matter. 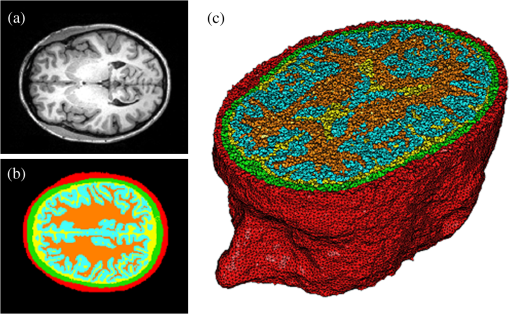 Fig. 3Original CT slice of the pancreas and surrounding tissue (a), segmentation of different tissue types (b), and the 3-D tetrahedral mesh for the pancreas (c), showing the regions as different colors: green indicates the blood vessels and red is pancreas and surrounding tissue. 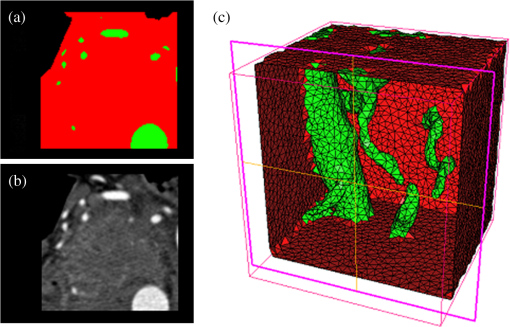 Fig. 4Original MRI slice of the breast (a), segmentation of different tissue types (b), and the 3-D tetrahedral mesh for the breast (c), showing the regions as different colors: red is the glandular tissue and green is other breast tissue. 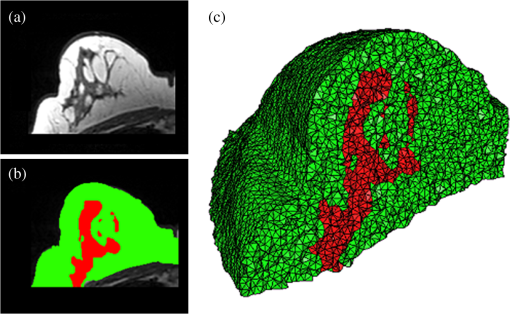 Fig. 5Original CT slice of the mouse (a), segmentation of different tissue types (b), and the 3-D tetrahedral mesh for the mouse (c), showing the regions as different colors: red is general tissue, green indicates the brain, and yellow is the tumor. 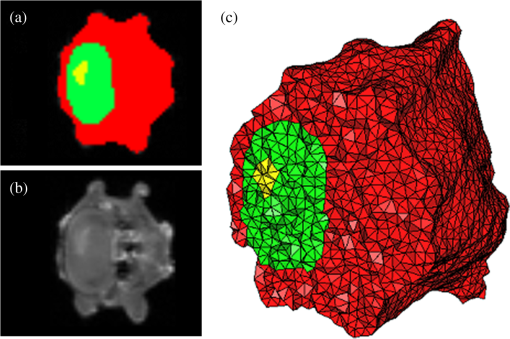 Fig. 6Time comparison of segmentation and mesh creation from pancreas CT between Nirfast and the commercial package Mimics. 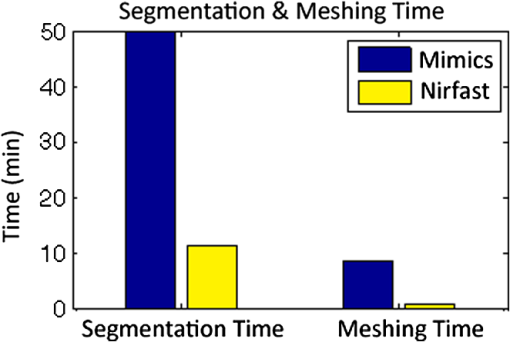 The resulting 3-D tetrahedral meshes from both programs were then analyzed to assess element quality. Also analyzed was a new mesh optimization feature in Nirfast. This is an optional procedure that searches the mesh for poor-quality elements and attempts to fix them to improve quality, as defined below. It can take a significant amount of time for large mesh sizes, but can vastly increase the quality. In this case, optimization took 15 min. The meshes were made to have a similar number of nodes: 224,049 for the Mimics mesh, 224,445 for the Nirfast mesh, and 224,989 for the optimized Nirfast mesh. The metric used for quality criterion is the sine of minimum dihedral angle of each tetrahedron. Values close to zero would indicate an almost flat element. Such elements can cause loss of numerical accuracy as well as make the stiffness matrix in the FEM formulation ill-conditioned. The optimal value of this quality would be for an equilateral tetrahedron; however the upper bound is 1.0. Figure 7 shows the quality histograms using Mimics, Nirfast, and mesh optimization in Nirfast. The minimum quality for each case respectively was 0.06, 0.12, and 0.65, with average quality values of 0.71, 0.73, and 0.77. Fig. 7Histograms comparing mesh element quality between the commercial package Mimics and the tools developed in Nirfast. Also shown is the quality histogram when using the mesh optimization feature in Nirfast. No subsequent postprocessing improvements were applied to the Mimics mesh (but are available in the 3-matic package). 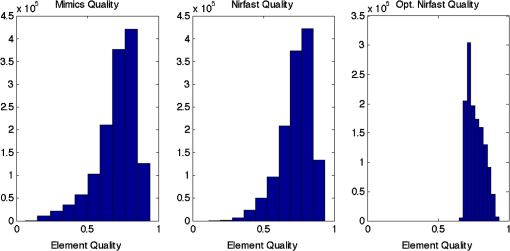 Figure 8 shows the reconstruction results in the mouse head using the Mimics mesh and the Nirfast mesh, displaying fluorescence yield overlaid on the MR images. The recovered fluorescence yield for each region in both cases is reported in Table 4. Fig. 8Reconstructed fluorescence yield overlaid on sagittal MR images of the mouse head, based on reconstructions on a mesh created in Mimics (a) and in Nirfast (b). The fluorescence tomographic reconstructions are based on the segmentation of tissue types and region-based reconstruction on the resulting tetrahedral meshes. 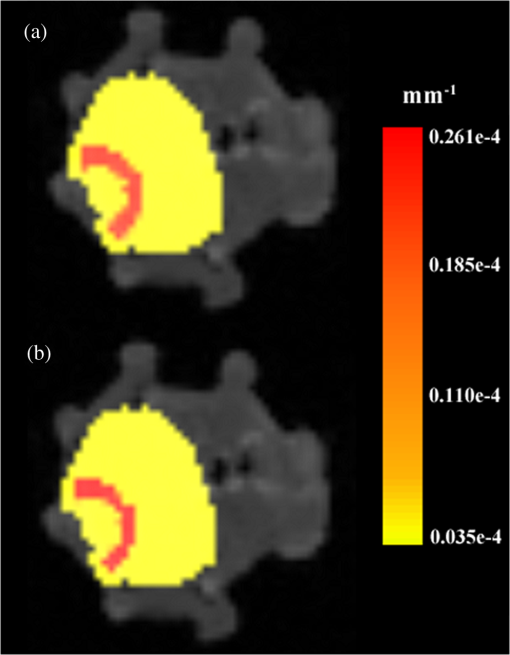 Table 4Recovered fluorescence yield for each region in reconstruction using the mouse head. Results are reported on both the Mimics- and Nirfast-created meshes.
4.DiscussionNew segmentation and mesh creation tools have been implemented in Nirfast, with the ability to work from the variety of medical images encountered in optical tomography. The efficacy of these tools has been compared with the commercial package Mimics in a case study. The minimum and average tetrahedron element quality values are better using Nirfast (especially when using mesh optimization). In particular, the minimum quality is 62% higher relative to the optimal value using Nirfast. Low-quality elements can produce erroneous numerical solutions by several orders of magnitude, or even prevent a solution from being computed, so this improvement in the minimum quality threshold is essential for DOT. There is a large difference in the amount of time spent, with Nirfast being far more efficient by approximately fivefold. In segmentation, this is partly affected by the efficiency of the automatic segmentation methods, and also by the availability of many advanced segmentation tools that are particularly useful for the typical contrast profiles seen in MR/CT. A good example is breast imaging using MR guidance, where low-frequency gradients are often seen in the images. In the past, this has often hindered the ability to segment these images, as grayscale values of the same tissue type will no longer be in the same range.48 These gradients can be easily removed using MR bias removal, thus greatly reducing the amount of manual touch-up needed after automatic segmentation. In meshing, the improved computational time is in part due to the fact that the new meshing tools are completely automatic and do not require any fixing after mesh creation. The metrics used for speed and quality in comparison with Mimics account for all pre- and post-processing done using tools available in Mimics to improve mesh quality, but not any external tools that may be used separate from Mimics. For example, 3-matic is also a tool marketed by Materialise, capable of postprocessing mesh quality improvement, which could significantly improve the quality of meshes produced by Mimics. An advantage of the Nirfast package that is not evident from the time benchmarks is the ease of use in the workflow. Since the entire package has been designed around seamlessly segmenting, creating a mesh, modeling light transport, and then visualizing the result, it is much easier to use than a combination of packages that are not optimized for optical tomography. In reconstruction results, as seen in Fig. 8, the recovered fluorescence yield is very similar between Mimics and Nirfast. In fact, there is a small improvement in the tumor and tumor boundary to background tissue fluorescence yield contrast recovered. This indicates that the new tools do not adversely affect reconstruction, despite saving significant time during segmentation and mesh creation. Furthermore, the higher minimum element qualities ensure that numerical issues do not arise with generating forward data on a poor-quality mesh, which can often cause a reconstruction to fail entirely and terminate before converging upon a solution. The tools have been presented with a focus on optical tomography and the types of medical images often encountered in image-guided optical tomography. However, these tools could certainly be used for other applications in which it is useful to have a 3-D tetrahedral mesh created from 2-D image slices, such as electrical impedance tomography. One of the advantages of the meshing tools presented is the fact that interior region surfaces are maintained in the mesh, as opposed to simply labeling interior elements based on region proximity. This is very important in FEM modeling for optical tomography, as having the boundary of a surface inaccurately represented can lead to poor quantification.49 5.ConclusionTools have been created to allow for segmentation and 3-D tetrahedral mesh creation from a variety of medical images and systems used in optical tomography applications. These tools show promising computational time and element quality benchmarks. The ease and speed of segmentation and meshing is very useful in promoting the use of optical tomography, which has long suffered from long, difficult, and nonrobust meshing procedures. Furthermore, the available automatic segmentation modules provide essential tools for many different types of medical images, particularly in regard to artifacts often seen in MR images. The tools are provided as part of a complete package designed for modeling diffuse light transport in tissue, allowing for a seamless workflow that has never before been available. AcknowledgmentsThis work has been funded by RO1 CA132750 (M.J., H.G., B.W.P., H.D.), P01 CA84203 (B.W.P., M.J.), Department of Defense award W81XWH-09-1-0661 (S.C.D.), and a Neukom Graduate Fellowship (M.J.). The segmentation tools in Nirfast have been developed in collaboration with Kitware Inc. (Clifton Park, NY) under subcontract. Joseph Culver and Adam Eggebrecht of Washington University made available MRI data for the human head. Stephen Pereira and Matthew Huggett of University College London made available CT data for the human pancreas. ReferencesA. P. GibsonJ. C. HebdenS. R. Arridge,
“Recent advances in diffuse optical imaging,”
Phys. Med. Biol., 50
(4), R1
–R43
(2005). http://dx.doi.org/10.1088/0031-9155/50/4/R01 PHMBA7 0031-9155 Google Scholar
S. Srinivasanet al.,
“Interpreting hemoglobin and water concentration, oxygen saturation and scattering measured in vivo by near-infrared breast tomography,”
PNAS, 100
(21), 12349
–12354
(2003). http://dx.doi.org/10.1073/pnas.2032822100 PNASA6 0027-8424 Google Scholar
H. Dehghaniet al.,
“Multiwavelength three-dimensional near-infrared tomography of the breast: initial simulation, phantom, and clinical results,”
Appl. Opt., 42
(1), 135
–145
(2003). http://dx.doi.org/10.1364/AO.42.000135 APOPAI 0003-6935 Google Scholar
S. C. Daviset al.,
“Contrast-detail analysis characterizing diffuse optical fluorescence tomography image reconstruction,”
J. Biomed. Opt., 10
(5), 050501
(2005). http://dx.doi.org/10.1117/1.2114727 JBOPFO 1083-3668 Google Scholar
A. B. Milsteinet al.,
“Fluorescence optical diffusion tomography,”
Appl. Opt., 42
(16), 3081
–3094
(2003). http://dx.doi.org/10.1364/AO.42.003081 APOPAI 0003-6935 Google Scholar
S. R. ArridgeM. Schweiger,
“Direct calculation of the moments of the distribution of photon time-of-flight in tissue with a finite-element method,”
Appl. Opt., 34
(15), 2683
–2687
(1995). http://dx.doi.org/10.1364/AO.34.002683 APOPAI 0003-6935 Google Scholar
S. R. Arridge,
“Optical tomography in medical imaging,”
Inverse Probl., 15
(2), R41
–R93
(1999). http://dx.doi.org/10.1088/0266-5611/15/2/022 INPEEY 0266-5611 Google Scholar
B. Brooksbyet al.,
“Magnetic resonance-guided near-infrared tomography of the breast,”
Rev. Sci. Instrum., 75
(12), 5262
–5270
(2004). http://dx.doi.org/10.1063/1.1819634 RSINAK 0034-6748 Google Scholar
S. C. Daviset al.,
“Comparing implementations of magnetic-resonance-guided fluorescence molecular tomography for diagnostic classification of brain tumors,”
J. Biomed. Opt., 15
(5), 051602
(2010). http://dx.doi.org/10.1117/1.3483902 JBOPFO 1083-3668 Google Scholar
B. Brooksbyet al.,
“Magnetic resonance-guided near-infrared tomography of the breast,”
Rev. Sci. Instrum., 75
(12), 5262
–5270
(2004). http://dx.doi.org/10.1063/1.1819634 RSINAK 0034-6748 Google Scholar
M. SchweigerS. R. Arridge,
“Optical tomographic reconstruction in a complex head model using a priori boundary information,”
Phys. Med. Biol., 44
(11), 2703
–2722
(1999). http://dx.doi.org/10.1088/0031-9155/44/11/302 PHMBA7 0031-9155 Google Scholar
G. Gulsenet al.,
“Congruent MRI and near-infrared spectroscopy for functional and structural imaging of tumors,”
Technol. Cancer Res. Treat., 1
(6), 497
–505
(2002). TCRTBS 1533-0346 Google Scholar
P. K. Yalavarthyet al.,
“Critical computational aspects of near infrared circular tomographic imaging: analysis of measurement number, mesh resolution and reconstruction basis,”
Opt. Express, 14
(13), 6113
–6127
(2006). http://dx.doi.org/10.1364/OE.14.006113 OPEXFF 1094-4087 Google Scholar
M. E. Eameset al.,
“An efficient Jacobian reduction method for diffuse optical image reconstruction,”
Opt. Express, 15
(24), 15908
–15919
(2007). http://dx.doi.org/10.1364/OE.15.015908 OPEXFF 1094-4087 Google Scholar
S. R. ArridgeM. Schweiger,
“Image reconstruction in optical tomography,”
Phil. Trans. R. Soc. London, 352
(1354), 717
–726
(1997). http://dx.doi.org/10.1098/rstb.1997.0054 PTRSAV 0370-2316 Google Scholar
M. SchweigerS. R. ArridgeI. Nissila,
“Gauss-Newton method for image reconstruction in diffuse optical tomography,”
Phys. Med. Biol., 50
(10), 2365
–2386
(2005). http://dx.doi.org/10.1088/0031-9155/50/10/013 PHMBA7 0031-9155 Google Scholar
S. ArridgeJ. Hebden,
“Optical imaging in medicine: II. Modeling and reconstruction,”
Phys. Med. Biol., 42
(13), 841
–854
(1997). http://dx.doi.org/10.1088/0031-9155/42/5/008 PHMBA7 0031-9155 Google Scholar
P. K. Yalavarthyet al.,
“Weight-matrix structured regularization provides optimal generalized least-squares estimate in diffuse optical tomography,”
Med. Phys., 34
(6), 2085
–2098
(2007). http://dx.doi.org/10.1118/1.2733803 MPHYA6 0094-2405 Google Scholar
B. W. Pogueet al.,
“Three-dimensional simulation of near-infrared diffusion in tissue: boundary condition and geometry analysis for finite-element image reconstruction,”
Appl. Opt., 40
(4), 588
–600
(2001). http://dx.doi.org/10.1364/AO.40.000588 APOPAI 0003-6935 Google Scholar
M. SchweigerS. Arridge,
“Comparison of two- and three-dimensional reconstruction methods in optical tomography,”
Appl. Opt., 37
(31), 7419
–7428
(1998). http://dx.doi.org/10.1364/AO.37.007419 APOPAI 0003-6935 Google Scholar
S. C. Daviset al.,
“Image-guided diffuse optical fluorescence tomography implemented with Laplacian-type regularization,”
Opt. Express, 15
(7), 4066
–4082
(2007). http://dx.doi.org/10.1364/OE.15.004066 OPEXFF 1094-4087 Google Scholar
H. Dehghaniet al.,
“Near infrared optical tomography using NIRFAST: algorithm for numerical model and image reconstruction,”
Commun. Numer. Methods Eng., 25
(6), 711
–732
(2009). http://dx.doi.org/10.1002/cnm.v25:6 CANMER 0748-8025 Google Scholar
M. Jermynet al.,
“A user-enabling visual workflow for near-infrared light transport modeling in tissue,”
in Biomedical Optics,
BW1A.7
(2012). Google Scholar
H. Xuet al.,
“MRI coupled broadband near infrared tomography system for small animal brain studies,”
Appl. Opt., 44
(11), 2177
–2188
(2005). http://dx.doi.org/10.1364/AO.44.002177 APOPAI 0003-6935 Google Scholar
S. C. Daviset al.,
“Magnetic resonance-coupled fluorescence tomography scanner for molecular imaging of tissue,”
Rev. Sci. Instrum., 79
(6), 064302
(2008). http://dx.doi.org/10.1063/1.2919131 RSINAK 0034-6748 Google Scholar
D. Kepshireet al.,
“A microcomputed tomography guided fluorescence tomography system for small animal molecular imaging,”
Rev. Sci. Instrum., 80
(4), 043701
(2009). http://dx.doi.org/10.1063/1.3109903 RSINAK 0034-6748 Google Scholar
K. M. Tichaueret al.,
“Computed tomography-guided time-domain diffuse fluorescence tomography in small animals for localization of cancer biomarkers,”
J. Vis. Exp., 65
(4050), e4050
(2012). http://dx.doi.org/10.3791/4050 JVEOA4 1940-087X Google Scholar
J. C. Hebdenet al.,
“Three-dimensional optical tomography of the premature infant brain,”
Phys. Med. Biol., 47
(23), 4155
(2002). http://dx.doi.org/10.1088/0031-9155/47/23/303 PHMBA7 0031-9155 Google Scholar
T. S. Leunget al.,
“Measurement of the absolute optical properties and cerebral blood volume of the adult human head with hybrid differential and spatially resolved spectroscopy,”
Phys. Med. Biol., 51
(3), 703
–717
(2006). http://dx.doi.org/10.1088/0031-9155/51/3/015 PHMBA7 0031-9155 Google Scholar
Materialise: Mimics,
(2010) http://biomedical.materialise.com/mimics September ). 2010). Google Scholar
UC Berkeley: Distmesh,
(2012) http://persson.berkeley.edu/distmesh March ). 2012). Google Scholar
Iso2mesh,
(2011) http://iso2mesh.sourceforge.net/cgi-bin/index.cgi June ). 2011). Google Scholar
Cornell: QMG,
(2000) http://www.cs.cornell.edu/home/vavasis/qmg-home.html March ). 2000). Google Scholar
K. Krishnanet al.,
“An open-source toolkit for the volumetric measurement of CT lung lesions,”
Opt. Express, 18
(14), 15256
–15266
(2010). http://dx.doi.org/10.1364/OE.18.015256 OPEXFF 1094-4087 Google Scholar
J. HartiganM. Wong,
“Algorithm AS 136: a K-means clustering algorithm,”
J. R. Stat. Soc., 28
(1), 100
–108
(1979). 0952-8385 Google Scholar
J. G. SledA. P. ZijdenbosA. C. Evans,
“A nonparametric method for automatic correction of intensity nonuniformity in data,”
IEEE Tran. Med. Imaging, 17
(1), 87
–97
(1998). http://dx.doi.org/10.1109/42.668698 ITMID4 0278-0062 Google Scholar
N. J. Tustisonet al.,
“N4ITK: improved N3 bias correction,”
IEEE Trans. Med. Imaging, 29
(6), 1310
–1320
(2010). http://dx.doi.org/10.1109/TMI.2010.2046908 ITMID4 0278-0062 Google Scholar
M. SezginB. Sankur,
“Survey over image thresholding techniques and quantitative performance evaluation,”
J. Electron. Imaging, 13
(1), 146
–165
(2004). http://dx.doi.org/10.1117/1.1631315 JEIME5 1017-9909 Google Scholar
J. SerraP. Soille,
“Mathematical morphology and its applications to image processing,”
in Proc. 2nd Int. Symp. on Mathematical Morphology,
241
–248
(1994). Google Scholar
Computational Geometry Algorithms Library,
(2012) http://www.cgal.org October ). 2012). Google Scholar
L. P. Chew,
“Guaranteed-quality mesh generation for curved surfaces,”
in SCG Proc. of the 9th Annual Symp. on Computational Geomotry,
274
–280
(1993). Google Scholar
J.-D. BoissonnatS. Oudot,
“Provably good sampling and meshing of surfaces,”
Graph. Models, 67
(5), 405
–451
(2005). http://dx.doi.org/10.1016/j.gmod.2005.01.004 1524-0703 Google Scholar
S. OudotL. RineauM. Yvinec,
“Meshing volumes bounded by smooth surfaces,”
in Proc. 14th Int. Meshing Roundtable,
203
–219
(2005). Google Scholar
M. Klingner,
“Improving tetrahedral meshes,”
University of California, Berkeley
(2008). Google Scholar
S. C. Daviset al.,
“MRI-coupled fluorescence tomography quantifies EGFR activity in brain tumors,”
Acad. Radiol., 17
(3), 271
–276
(2010). http://dx.doi.org/10.1016/j.acra.2009.11.001 1076-6332 Google Scholar
M. Mastandunoet al.,
“Remote positioning optical breast magnetic resonance coil for slice-selection during image-guided near-infrared spectroscopy of breast cancer,”
J. Biomed. Opt., 16
(6), 066001
(2011). http://dx.doi.org/10.1117/1.3587631 JBOPFO 1083-3668 Google Scholar
H. Dehghaniet al.,
“Three-dimensional optical tomography: resolution in small-object imaging,”
Appl. Opt., 42
(16), 3117
(2003). http://dx.doi.org/10.1364/AO.42.003117 APOPAI 0003-6935 Google Scholar
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||

