|
|
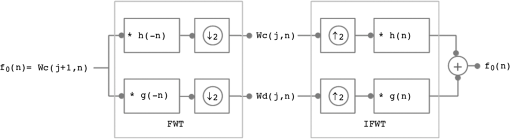
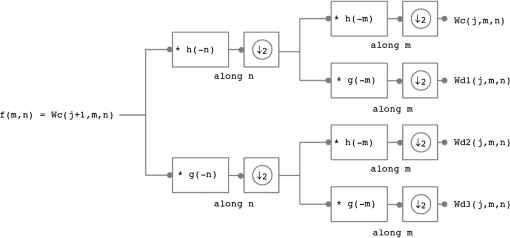
1.IntroductionFluorescence diffuse optical tomography (fDOT) is an imaging modality that aims at reconstructing three-dimensional (3-D) distributions of fluorescent markers embedded within biological tissues.1 This technology has facilitated monitoring of molecular activity, tumor growth, response to drug therapy, etc., mostly in small animals. In fDOT, a near-infrared (NIR) light source at excitation wavelength is illuminated onto the subject under study at different positions. Fluorophores absorb the excitation light and re-emit part of the energy at a longer wavelength. In most modern noncontact fDOT systems, light intensity measurements (CW) at the emission, and possibly, excitation wavelengths are collected by a charged-coupled device (CCD) camera placed opposite to the light source, which is rotated around the object of study.2 The image reconstruction consists of the inversion of a linear operator (sensitivity matrix), mapping the fluorescence yield of the fluorophore, which is linearly related to its concentration, to the measured data. However, due to the diffusive nature of light propagation in biological tissue the image reconstruction is an ill-posed inverse problem, meaning that noise in the data may give rise to significant errors in the reconstructed image. The ill-posedness of the problem can be reduced by using large data sets.3,4 The use of CCD cameras produces large amounts of data, typically of the order of . However, this makes the problem large scale and more challenging to solve. Furthermore, the parameters to reconstruct are so numerous, , that the sensitivity matrix is extremely large. The large matrix size poses a problem for storage, as well as computation time required for inversion. Therefore, specialized algorithms that can handle problems with large dimensions are required. Markel and Schotland3,5 proposed a series of algorithms for the reconstruction of images from extremely large data sets, which exploit symmetry properties of simple geometries. Konecky et al.6 reconstructed diffuse optical tomography (DOT) images using a fast inversion method, which combines plane wave illumination7 with analytic image reconstruction methods. The reconstruction method exploits the block structure of the linear operator that couples the measurements to the optical properties of the object of study, and hence, instead of inverting a large matrix, the problem is reduced to inverting a set of relatively small matrix blocks. Lukic et al.8 used a similar method, but replaced point sources by structured illumination (Fourier encoding), and hence, data are directly measured in the Fourier space. Since DOT suffers from low spatial resolution, only a few low Fourier components retain relevant information. In this approach, the inverse problem is fully Fourier encoded, i.e., sources, detectors, and solution space are represented in the Fourier space. Ducros et al.9 used a similar approach, but based on wavelet encoding methods. Ripoll10 proposed a hybrid approach, where only the measurements are Fourier encoded, while the sources and solution space are kept in the real space. However, most of these approaches are based on analytical expressions (Green’s functions) for the slab geometry. For complex geometries, an alternative is to use a matrix-free Krylov-subspace approach, in which the storage of the sensitivity matrix is replaced by vector-matrix products, i.e., solving the forward and adjoint problems where the solutions are computed numerically using the finite element method (FEM).11 The matrix-free method relies on iterative Krylov-subspace methods, such as generalized minimal residual method, which may suffer from slow convergence. Therefore, the method overcomes the storage problems of large data sets at the expense of computational efficiency. Multigrid methods offer the capability of reconstructing images at lower spatial scales by solving smaller instances of the problem, which allows approximate solutions to be computed quickly at low resolution and progressively refined. Zhu et al.12 developed a wavelet-based multigrid approach. The rows and columns of the sensitivity matrix are wavelet transformed and the system at a coarser resolution is solved. Solutions at coarser resolutions are used as the initial guess for solving systems at finer resolutions. It is also possible to select regions of interest at smaller scales based on coarse level reconstructions. However, in order to maintain reconstruction resolution, this approach requires reconstructions at many scales, and thus, does not result in significant reduction in computation time. Furthermore, it requires the full sensitivity matrix to be calculated and stored. Another approach is to use the wavelet-Galerkin method to generate the forward model, instead of using the more time consuming FEM (Ref. 13). However, unlike FEM, this method is only applicable to simple geometries, but the fictitious domain method is used to overcome this limitation.14 In the wavelet-Galerkin method, the wavelet scaling functions are used as a basis on a regular grid. This allows a more compact representation of the problem and the use of multigrid techniques. As before, solutions can be obtained at progressively higher wavelet resolution scales, terminating when appropriate. Although this is an efficient way to compute the full matrix for large solution spaces, it is not suitable for large data sets. Wavelets were also used in Ref. 15 to reduce the fDOT forward model computation time by projecting the original FEM matrices into a series of wavelet bases. This paper proposes a method that reduces significantly the size of the sensitivity matrix by compressing its rows and columns using wavelets, allowing it to be stored explicitly while maintaining the spatial resolution and accuracy of the reconstructed images and low-computational time. This method is an extension of the work in Ref. 16, where wavelet compression is applied to a very large data sets in order to reduce data space dimensions. Here, a similar approach is used to reduce the solution space dimensions. Therefore, given the reduced dimensions of the image reconstruction problem, the solution can be found using direct inversion methods. The performance of the method is assessed using simulations and experimental phantom data. The effect of incorporating a priori structural information into the compressed image reconstruction is also analyzed. 2.Methods2.1.Forward ProblemIn CW-fDOT, the forward model is described by a set of coupled diffusion equations in a domain :17 with boundary conditions on where is the excitation source flux, is the photon density, is a boundary term that incorporates the refractive index mismatch, is the boundary measurement on , and is the outer normal vector. The diffusion coefficient is given by , where and are the absorption and reduced scattering coefficients, respectively. The superscript and indicates the excitation and emission wavelengths and , respectively. The fluorescence yield coefficient is related to the quantum yield of the fluorophore and its absorption coefficient at . For CCD camera based measurements we have where is a measurement operator that gives the data, is the unknown source and detector coupling coefficients and the operator represents the projection from the domain boundary to the camera .The forward problem is computed numerically using the FEM on a tetrahedral mesh based on the geometry being considered, and then mapped into a Cartesian voxel grid. In the FEM, the domain is divided into elements, joined at nodes. The solution of the diffusion equations is approximated by the piecewise function , where is the basis functions. In the FEM framework, Eqs. (1)–(5) can be expressed as where Therefore, the solution can be found by inverting the matrix .Normalizing the measured fluorescence photon density by the measured excitation photon density reduces the effects of .18 The normalized forward problem in fDOT is given by where is the solution to the adjoint diffusion equation for a source located on the boundary ,19 is the fluorescence yield coefficient, and is the Jacobian or sensitivity matrix. For a set of source-detector positions, where the detector is a camera with pixels, it follows that the measurements are vectors of size and is a matrix of dimensions , where is the dimension of the solution space that, once mapped into a regular grid, has dimensions .2.2.Image Reconstruction ProblemThe image reconstruction in fDOT consists in solving the problem where is the regularization parameter and is a regularizing functional that represents a priori information. The previous equation can be solved using zero-order Tikhonov regularization, i.e., , or iteratively using the split operator method with anisotropic diffusion regularization and structural a priori information (ADSP) introduced in Ref. 20: where the first step is the Levenberg–Marquardt method, where is the damping factor that changes at each iteration; and the second step is the nonlinear anisotropic diffusion method,21 where is the time step and is the anisotropic diffusion function given by . Here, is the exceedance function, which is the probability of an edge of interest being present and can be calculated using the normalized cumulative histogram (NCH) of the image gradient.20 The NCH indicates the probability of a gradient taking on a value less than or equal to the value that the bin represents, i.e., . In a multimodality framework, is a weighting factor related to the structural information.20 It is an edge detection function that stops the diffusion process across edges. The additive operator splitting scheme is used to discretize the nonlinear anisotropic diffusion equation.202.3.Fast Wavelet TransformThe fast wavelet transform (FWT) decomposes a signal or function into different frequency subbands.22 It uses a low-pass filter and a high-pass filter to obtain the approximation Wc and detail Wd coefficients. The approximation coefficients represent the approximation of the signal at a resolution , where is an integer that specifies the resolution level. The detail coefficients contain the details of the original signal, i.e., the high-frequency information. For a signal , with samples and at a starting scale Thus, the FWT consists of a convolution, followed by downsampling by a factor of 2 (), i.e., keeping the even index samples. The wavelet transform can be implemented as a decomposition filter bank, where the initial signal goes through a series of filters. The synthesis filter bank can be used to perform the inverse transform (IFWT) and reconstruct the signal . First, the signal is upsampled by a factor of 2 (), i.e., adding a zero between samples, followed by convolution with the inverse filter. Note that the filters are related to each other by . Figure 1 shows a one-level decomposition and synthesis filter bank. The separability property of the two-dimensional (2-D) wavelet transform means that performing a 2-D wavelet transform is equivalent to performing two one-dimensional (1-D) transforms, i.e., one 1-D transform along with the columns of the image (-axis) and another 1-D transform along with the rows of (-axis) (Fig. 2). Analogously, performing a 3-D wavelet transform is equivalent to performing three 1-D transforms. The same applies to the IFWD in higher dimensions. 2.3.1.Data compressionData compression is used to reduce the dimensions of the data for computational efficiency,16 while simultaneously reducing the redundancy of the data. We apply a four-level 2-D FWT () to the projection image captured by a CCD camera, where . The obtained wavelet coefficients are , where Wd1, Wd2, and Wd3 are the detail coefficients that passed through a high-pass filter at least once. We keep the coefficients that are larger than a threshold , set to the largest coefficients (i.e., ), which contain most of the relevant information, and hence, the dimensions of the vectorized compressed data are reduced to . For example, if is the total number of wavelet coefficients, then , where are the wavelet coefficients sorted in descending order and is the percentage of coefficients to be removed. If , then and the compressed data has dimensions . Similarly, the size of the row compressed Jacobian is reduced to . The resulting compressed forward problem is . 2.3.2.Solution compressionIn this paper, we extend the work in Ref. 16 by applying a similar wavelet compression to each row of the Jacobian, thus reducing the dimensions of the solution space. Each row of the Jacobian represents the sensitivity of one measurement (matrix ) or compressed measurement (matrix ) to changes in . Both and represent 3-D images of dimensions , and therefore, a 3-D FWT () is performed on the rows of the sensitivity matrix. Two methods are investigated, one where the sensitivity matrix is sparse and another where the matrix is fully compressed. Sparse matrix: A highly sparse matrix can be obtained by keeping the wavelet coefficients larger than a threshold , set to the largest coefficient in each row, and setting the remaining coefficients to zero where the subscript is the row index and is the column index. For example, if is the total number of wavelet coefficients, then , where are the wavelet coefficients sorted in descending order. If we consider , then .The resulting forward problem, using data and solution compression, is . The solution is a sparse image of size and nonzero elements. The matrix has dimensions and nonzero elements. The solution is obtained by solving the inverse problem and performing a 3-D IFWT () to . Fully compressed matrix: Alternatively, if only wavelet coefficients are kept, then the size of the compressed Jacobian is reduced to . The forward problem is , where is a vector of size . Therefore, the solution is a compressed image of size , which is converted into a sparse image and inversely wavelet transformed. Consider and (average along the rows), the wavelet coefficients kept correspond to the location of the largest elements: . Unlike the sparse matrix method, where the location corresponds to the location of the largest coefficients in each row and, therefore, the locations are different for each row, in the fully compressed method the locations are the same in all the rows of the wavelet transformed matrix . 2.4.Computation Procedure for Compression of the Solution SpaceThe FWT is used to efficiently compute the compressed matrix , without explicitly computing the matrix or a wavelet transform matrix. The matrix is calculated row by row; and hence, an FWT is performed on each row of the sensitivity matrix and only the largest components are kept. To compute the fully compressed matrix , one needs to first explicitly compute the wavelet transformed matrix of size . The computational procedure is described in Algorithms 1 and 2 for the sparse matrix and fully compressed matrix, respectively. Algorithm 1Sparse matrix.
Algorithm 2Fully compressed matrix.
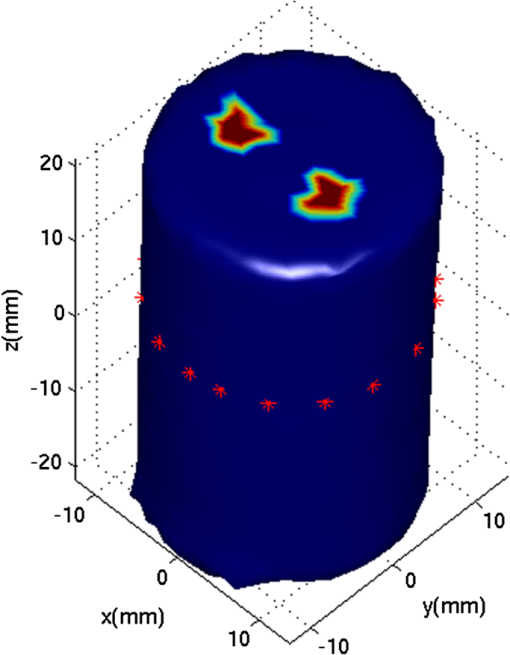
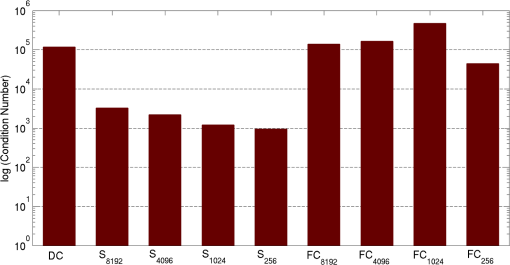
2.5.EvaluationThe performance of our data and solution compression method is evaluated through simulations and a phantom study. The matrix is calculated using TOAST, which is an FEM based software developed at University College London (UCL). Images are reconstructed (a) using zero-order Tikhonov regularization and solving the linear problem directly and (b) iteratively using our ADSP method.20 Solutions are computed on a tetrahedral mesh and mapped into a regular grid covering the domain of the mesh. In the following sections, we refer to the data compression method as DC, sparse matrix method as and the fully compressed matrix method as , where represents the number of coefficients . Wavelet filters: After experimenting different wavelet filters, we choose to use the Battle–Lemarié wavelet filters, which appear to be suitable for this type of application. The Battle–Lemarié wavelets are symmetric, orthonormal, and smooth. These wavelets are attractive since they have good time and frequency localization properties and can approximate smooth solutions.22 Other popular wavelets, such Daubechies, are not smooth enough and do not possess good frequency localization.23 Battle–Lemarié wavelet filters are built from polynomial splines of order . Let be the Fourier transform of the wavelet scaling function . The low-pass quadrature mirror filter can be obtained from the following relation in the Fourier domain22 Lemarié has shown that the scaling function can be written as and where Alternatively, one can calculate by computing the derivative of the formula:For an approximation built from cubic splines, i.e., , and thus, , it follows from the previous equations that Figures of merit: The relative error (RE) and contrast-to-noise ratio (CNR) are used to evaluate the quality of the reconstructed images. The RE between the true image and the reconstructed image is defined as . The CNR is calculated as follows:24 where is the mean value of the region of interest (ROI); is the mean value of the background (B); and are the standard deviations of the ROI and background, respectively; and and are noise weights, where and are the ROI and B areas, respectively. The ROI is defined as the region where voxels have attained at least half maximum intensity and the remaining voxels represent the background B.2.5.1.SimulationThe geometry used in the simulation is similar to the phantom geometry. The matlab package Iso2mesh25 was used to generate a tetrahedral mesh from the X-ray computed tomography (XCT) image of a cylindrical phantom (see Sec. 2.5.2 and Fig. 3). The FEM mesh has 8449 nodes, 49,797 elements and dimensions . Similar to the phantom, the optical properties are and , and the fluorescent targets are two vertical cylinders placed parallel to each other (Fig. 3) with contrast . Projection images of size pixels are calculated for 18 evenly spaced source-camera positions over a full 360 degrees range at (Fig. 3). Data consist of fluorescence and excitation projections with 2% Gaussian random noise. The data space is compressed by setting and solutions are computed with . For the reconstructions using zero-order Tikhonov regularization the damping factor is initially set to and for reconstructions using ADSP regularization it is set to , where and are the different types of compressed Jacobian matrices. 2.5.2.PhantomData are acquired using the rotating fDOT–XCT system described in Ref. 2. The optical and XCT images are acquired sequentially. The solid phantom consists of a mixture of Agar, Intralipid, and ink. The stock solution is 4.56 ml Intralipid 20% (Sigma–Aldrich Co. LLC, St. Louis, USA), 0.256 ml India ink, 100 ml deionized water and 2.8 g Agar (Sigma–Aldrich Co. LLC, St. Louis, USA). The phantom is a cylinder with diameter , height and optical properties and , which were measured using a spectrometer (USB4000-VIS-NIR, Ocean Optics, London, UK). Two translucent tubes with an inside diameter of 3 mm (and outside diameter 4 mm) are inserted parallel to its long axis and filled with an optically matched fluid containing a fluorescent dye (Alexa 750) at 500 nM (molar) concentration. Excitation and fluorescence data images of size (resized to ) are recorded at 18 angular positions (same than the simulations). The FEM mesh is the same than the one used in the simulation study. We use the compression and ( when ADSP regularization is used). The damping factor is initially set to and , where . 3.ResultsProfile plots across the reconstructed fluorescence targets ( and ) are shown in Figs. 4 and 5 for images obtained using Tikhonov regularization for the sparse and fully compressed matrix cases, respectively. Similarly, Figs. 6 and 7 show the profile plots for images obtained using ADSP regularization. Figures 4 and 6 also show the profile plots for the data compression case. Fig. 4Profile plots across the fluorescence images ( and ) reconstructed from simulated data using Tikhonov regularization and data compression, a sparse matrix with different compression levels, and the true solution. 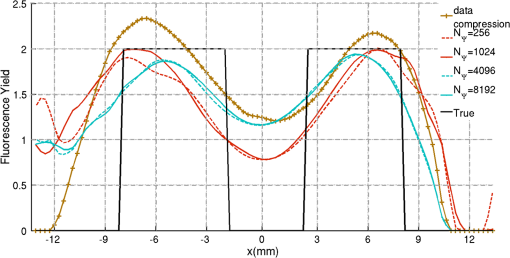 Fig. 5Profile plots across the fluorescence images ( and ) reconstructed from simulated data using Tikhonov regularization and a fully compressed matrix with different compression levels, and the true solution. 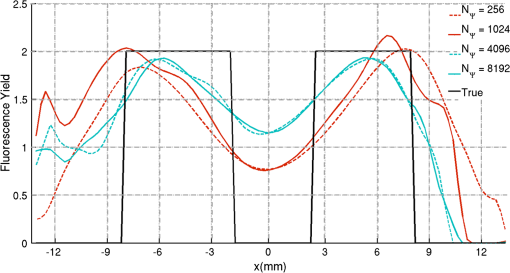 Fig. 6Profile plots across the fluorescence images ( and ) reconstructed from simulated data using ADSP regularization and data compression, a sparse matrix with different compression levels, and the true solution. 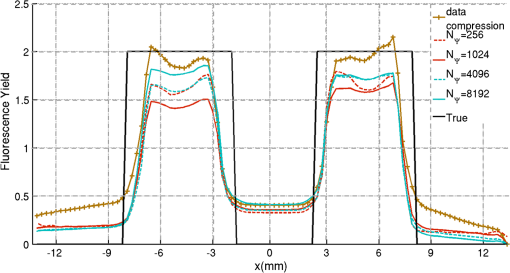 Fig. 7Profile plots across the fluorescence images ( and ) reconstructed from simulated data using ADSP regularization and a fully compressed matrix with different compression levels, and the true solution. 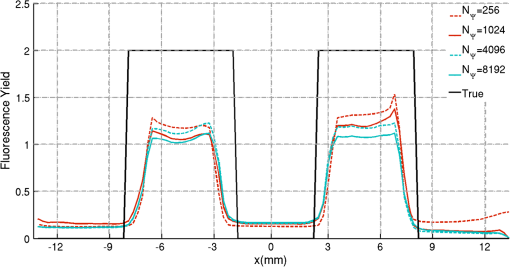 Since the profiles across the fluorescent inclusions do not differ significantly for the different types of compression, particularly when ADSP regularization is used, only results obtained without solution compression and high levels of solution compressions are shown. Therefore, reconstructions obtained using ADSP are shown for data compression and solution compression with . Reconstructions obtained using Tikhonov regularization and solution compression with are displayed for comparison with the case where only data compression is used, to show the effect of the solution compression on the noise in the reconstructions. The fluorescence distribution images reconstructed from simulated data using zero-order Tikhonov regularization are shown in Figs. 8Fig. 9Fig. 10Fig. 11–12. The reconstructions are displayed as 3-D isosurfaces (70% of the maximum value) together with the structural volumetric image. Additionally, a 2-D cross section of the reconstructed volume is taken at and thresholded at 70% of the maximum value. The thresholded images are overlaid onto the structural image. Figures 13Fig. 14–15 show the images reconstructed using ADSP regularization and thresholded at 50% of the maximum value. The threshold value is lower since these images are not as contaminated with noise as the previous ones. Fig. 8Fluorescence yield reconstructed from simulated data using Tikhonov regularization and data compression in (left) 3-D and (right) 2-D cross section at . 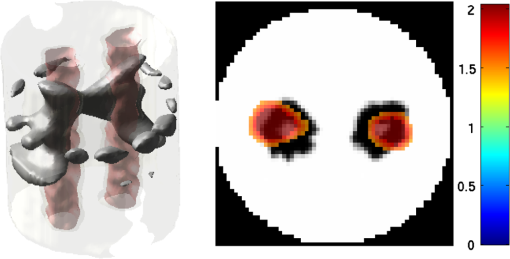 Fig. 9Fluorescence yield reconstructed from simulated data using Tikhonov regularization and in (left) 3-D and (right) 2-D cross section at . 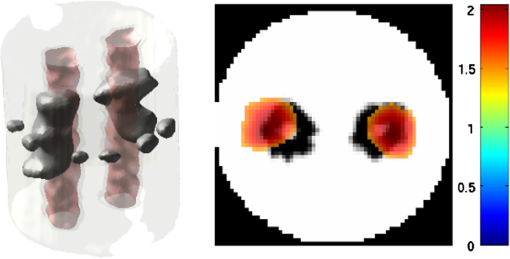 Fig. 10Fluorescence yield reconstructed from simulated data using Tikhonov regularization and in (left) 3-D and (right) 2-D cross section at . 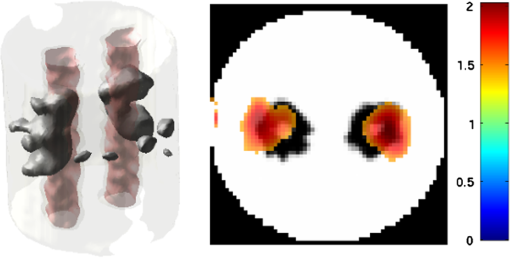 Fig. 11Fluorescence yield reconstructed from simulated data using Tikhonov regularization and in (left) 3-D and (right) 2-D cross section at . 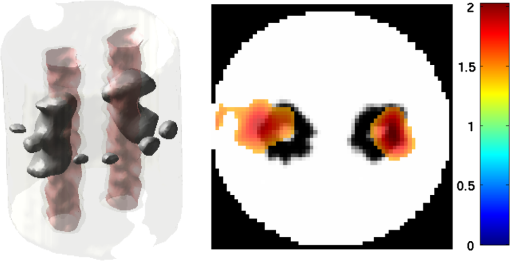 Fig. 12Fluorescence yield reconstructed from simulated data using Tikhonov regularization and in (left) 3-D and (right) 2-D cross section at .  Fig. 13Fluorescence yield reconstructed from simulated data using ADSP regularization and data compression in (left) 3-D and (right) 2-D cross section at . 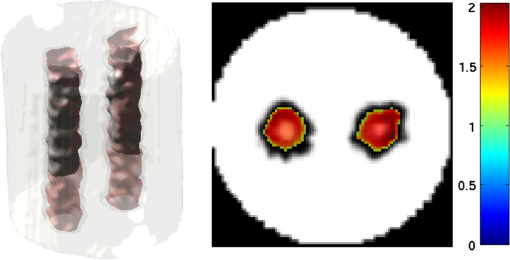 Fig. 14Fluorescence yield reconstructed from simulated data using ADSP regularization and in (left) 3-D and (right) 2-D cross section at . 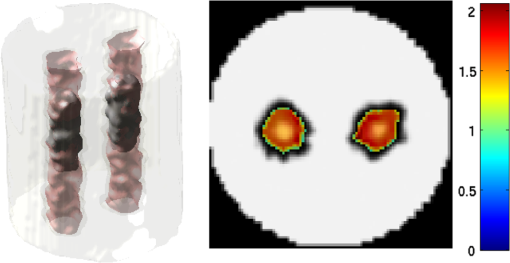 Fig. 15Fluorescence yield reconstructed from simulated data using ADSP regularization and (left) 3-D and (right) 2-D cross section at . 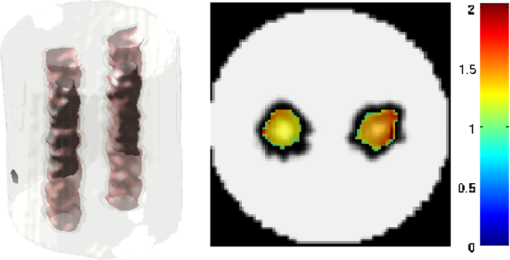 Similarly, the images reconstructed from phantom data using zero-order Tikhonov regularization are shown in Figs. 16Fig. 17Fig. 18Fig. 19–20 and using ADSP regularization are shown in Figs. 21Fig. 22–23. Their respective profile plots are shown in Figs. 24 and 25. Fig. 16Fluorophore concentration (nM) reconstructed from phantom data using Tikhonov regularization and data compression in (left) 3-D and (right) 2-D cross section at . 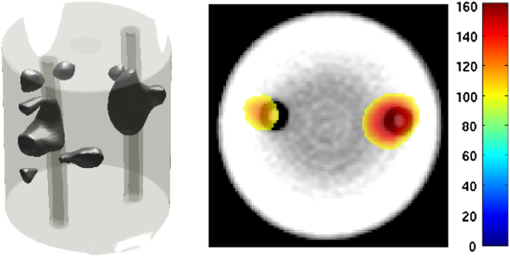 Fig. 17Fluorophore concentration (nM) reconstructed from phantom data using Tikhonov regularization and in (left) 3-D and (right) 2-D cross section at . 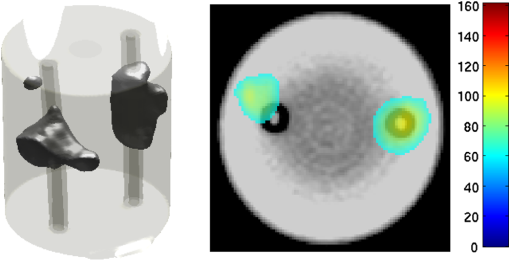 Fig. 18Fluorophore concentration (nM) reconstructed from phantom data using Tikhonov regularization and in (left) 3-D and (right) 2-D cross section at . 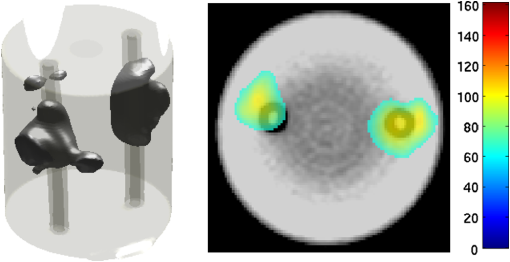 Fig. 19Fluorophore concentration (nM) reconstructed from phantom data using Tikhonov regularization and in (left) 3-D and (right) 2-D cross section at . 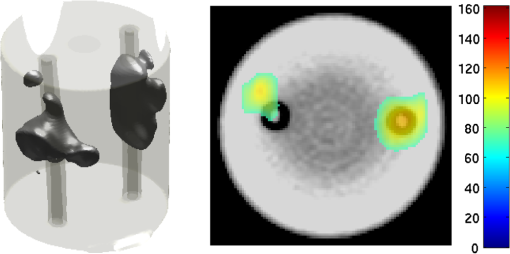 Fig. 20Fluorophore concentration (nM) reconstructed from phantom data using Tikhonov regularization and in (left) 3-D and (right) 2-D cross section at . 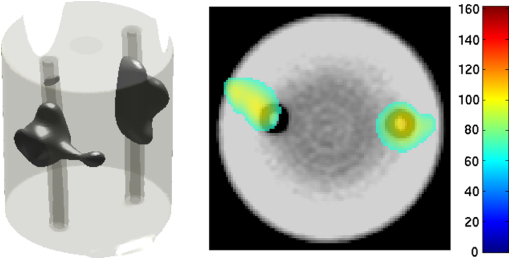 Fig. 21Fluorophore concentration (nM) reconstructed from phantom data using ADSP regularization and data compression in (left) 3-D and (right) 2-D cross section at . 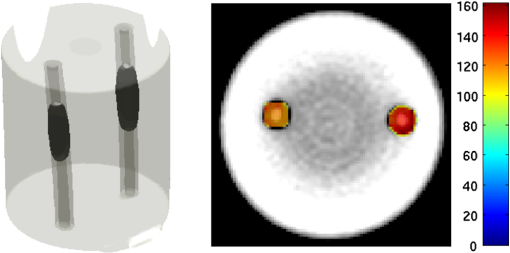 Fig. 22Fluorophore concentration (nM) reconstructed from phantom data using ADSP regularization and in (left) 3-D and (right) 2-D cross section at .  Fig. 23Fluorophore concentration (nM) reconstructed from phantom data using ADSP regularization and in (left) 3-D and (right) 2-D cross section at . 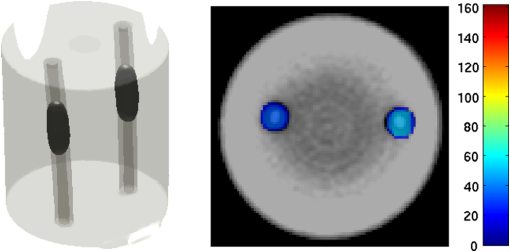 Fig. 24Profile plots across the fluorescence images ( and ) reconstructed from phantom data using Tikhonov regularization and DC, , , and the true solution. 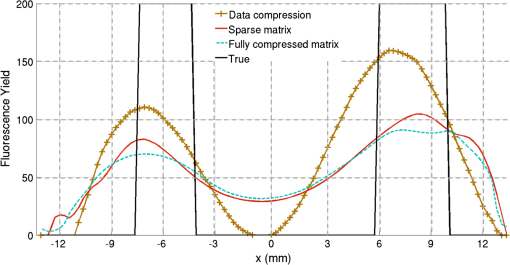 Fig. 25Profile plots across the fluorescence images ( and ) reconstructed from phantom data using ADSP regularization and DC, , , and the true solution. 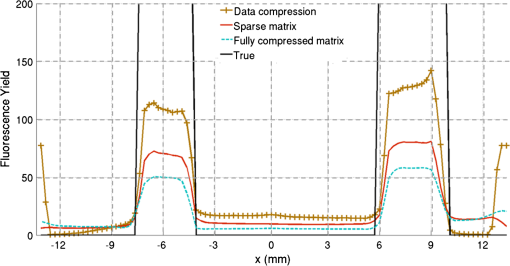 Figures 26 and 27 show the RE and CNR, respectively, of the images reconstructed from simulated and phantom data using Tikhonov and ADSP regularization for the different compression methods. Figure 28 shows the condition number of the matrices in the data compression, compressed sparse, and fully compressed format. Fig. 26RE of images reconstructed from (left column) simulated data and (right column) phantom data using (top row) Tikhonov regularization and (bottom row) ADSP regularization, for the different compression cases.  Fig. 27CNR of images reconstructed from (left column) simulated data and (right column) phantom data using (top row) Tikhonov regularization and (bottom row) ADSP regularization, for the different compression cases. 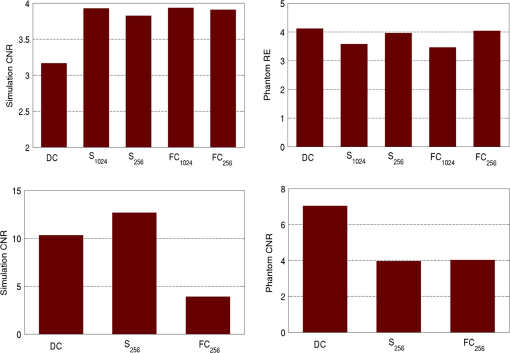 The reconstructions are faster when a fully compressed matrix is used, and the sparse matrix method is faster than using data compression only, with a ratio of . 4.Discussion and ConclusionIn fDOT, the data and solution spaces can be quite large, and hence, the sensitivity matrix may be too large to store. Even if this matrix can be stored, it may not be possible to directly invert it, and thus, iterative methods must be employed. However, these are computationally expensive and may take a long time to converge. The method we proposed reduces the dimensions of the sensitivity matrix allowing the storage and direct inversion of the matrix and, for the sparse matrix case, without requiring the construction of the full matrix. Both sparse and fully compressed matrix representations allow storage of large amounts of data generated from CCD-based fDOT systems, rather than keeping redundant data and solution components. The performance of our method was tested on simulated and experimental phantom data. Our simulations show that our solution compression approach can help to suppress noise in the reconstructed images (Figs. 8Fig. 9Fig. 10Fig. 11–12). This result is not surprising, since wavelet transforms are a common and efficient technique for denoising. The use of structural a priori information in the image reconstruction greatly improved the quality of the images (Figs. 6, 7, and 13Fig. 14–15). The images reconstructed using the different types of compression are qualitatively very similar. However, the fluorescence yield was underestimated when both data and solution compression were used with ADSP regularization. The images reconstructed from experimental data are similar (Figs. 16Fig. 17Fig. 18Fig. 19–20). The target locations and dimensions were accurately estimated by all the methods when structural information was incorporated into the image reconstruction (Figs. 21Fig. 22–23). The profiles plots in Figs. 24 and 25 also show that the use of structural information can greatly improve the quality of the reconstructions. Our results show that the sensitivity matrix can be effectively compressed into a wavelet space by selecting the most significant wavelet coefficients. The sensitivity matrix, which in our studies has dimensions , can effectively be represented in a fully compressed form with dimension or sparse form with size and nonzero elements. Figure 26 shows that the RE decreases with increasing level of compression, i.e., the quality of the reconstruction improves, which is presumably a consequence of the decrease of the condition number (Fig. 28). The fully compressed matrix with has a higher condition number, and hence, one should expect a higher RE. Nevertheless, the denoising characteristic of the wavelets seems to compensate for this effect. Figure 27 shows that the CNR of the images reconstructed from simulated data improves when solution compression is used, whereas the CNR of the images obtained from phantom data decreases. Overall, the simulation results using Tikhonov regularization with data and solution compression are quite similar, but the lowest RE was achieved with the method and the highest CNR using . For the phantom study, the best results were obtained with the . The images reconstructed using ADSP and had the lowest RE and highest CNR. The sparse matrix compression method has the advantage that the problem becomes less ill-conditioned and it does not require the explicit construction of the full matrix. The fully compressed matrix method provides images that are visually quite similar, but it underestimates the fluorescence yield more than the latter when ADSP is included. This method requires the construction of the full matrix and the wavelet coefficients kept in the compressed form are all in the same locations in all the rows of the matrix. These are not necessarily the most relevant coefficients, which may explain the lower qualitative accuracy. Nevertheless, this method allows a faster inversion process and, when Tikhonov regularization is used, provides images with the lowest RE and highest CNR. The disadvantage of using ADSP regularization is that the solution depends on the parameter value and the number of iterations until convergence is reached, i.e., it depends on the convergence criteria, and thus, there is still potential for improvement. Nevertheless, the results obtained using data and sparse matrix compression are quite satisfactory. In this work, we map from the FEM mesh to a regular grid, which is then transformed into the wavelet space, with no regard to the irregular boundary of the object. In future work, we will attempt to apply a similar compression directly on the FEM mesh, so that the reconstruction is performed on the mesh and to avoid error propagation resulting from these mappings. Furthermore, the boundary of the object introduces sharp edges in the images, which may lead to artifacts in the compressed representation. There is a large diversity of available wavelet filters, and hence, we performed a simple qualitative comparison of the suitability of some of the most popular ones (e.g., Haar, Daubechies, Coiflet, Symmlets, Battle–Lemarié, etc.) for use in fDOT. However, improved results can be expected if wavelet filters more adequate for the fDOT problem are designed and the ideal number of wavelet coefficients to keep is identified. In summary, image reconstruction using data and solution compression can greatly reduce the dimensions and ill-conditioning of the problem, allowing fast and less noisy reconstructions. AcknowledgmentsThis work has also been supported by funding from ECs seventh framework programme FMT-XCT under Grant Agreement No. 201792. ReferencesV. Ntziachristos,
“Fluorescence molecular imaging,”
Annu. Rev. Biomed. Eng., 8 1
–33
(2006). http://dx.doi.org/10.1146/annurev.bioeng.8.061505.095831 ARBEF7 1523-9829 Google Scholar
R. B. Schulzet al.,
“Hybrid system for simultaneous fluorescence and X-ray computed tomography,”
IEEE Trans. Med. Imag., 29
(2), 465
–473
(2010). http://dx.doi.org/10.1109/TMI.2009.2035310 ITMID4 0278-0062 Google Scholar
V. MarkelJ. Schotland,
“Symmetries, inversion formulas and image reconstruction for optical tomography,”
Phys. Rev. E, 70 056616
–056619
(2004). http://dx.doi.org/10.1103/PhysRevE.70.056616 PLEEE8 1063-651X Google Scholar
Z. Wanget al.,
“Experimental demonstration of an analytic method for image reconstruction in optical tomography with large data sets,”
Optic. Lett., 30
(24), 3338
–3340
(2005). Google Scholar
V. MarkelJ. Schotland,
“The inverse problem in optical diffusion tomography: III. Inversion formulas and singular value decomposition,”
J. Opt. Soc. A, 20
(5), 890
–902
(2003). http://dx.doi.org/10.1364/JOSAA.20.000890 JOSAAH 0030-3941 Google Scholar
S. D. Koneckyet al.,
“Imaging complex structures with diffuse light,”
Opt. Express, 16
(7), 5048
–5060
(2008). http://dx.doi.org/10.1364/OE.16.005048 OPEXFF 1094-4087 Google Scholar
M. XuM. LaxR. Alfano,
“Time-resolved Fourier optical diffuse tomography,”
J. Opt. Soc. Am. A, 18
(7), 1535
–1542
(2001). http://dx.doi.org/10.1364/JOSAA.18.001535 JOAOD6 0740-3232 Google Scholar
V. LukicV. MarkelJ. Schotland,
“Optical tomography with structured illumination,”
Opt. Lett., 34
(7), 983
–985
(2009). http://dx.doi.org/10.1364/OL.34.000983 OPLEDP 0146-9592 Google Scholar
N. Ducroset al.,
“Full-wavelet approach for fluorescence diffuse optical tomography with structured illumination,”
Opt. Lett., 35
(21), 3676
–3678
(2010). http://dx.doi.org/10.1364/OL.35.003676 OPLEDP 0146-9592 Google Scholar
J. Ripoll,
“Hybrid Fourier-real space method for diffuse optical tomography,”
Opt. Lett., 35
(5), 688
–690
(2010). http://dx.doi.org/10.1364/OL.35.000688 OPLEDP 0146-9592 Google Scholar
A. Zacharopouloset al.,
“A matrix-free algorithm for multiple wavelength fluorescence tomography,”
Opt. Express, 17
(5), 3025
–3035
(2009). http://dx.doi.org/10.1364/OE.17.003042 OPEXFF 1094-4087 Google Scholar
W. Zhuet al.,
“A wavelet-based multiresolution regularized least squares reconstruction approach for optical tomography,”
IEEE Trans. Med. Imag., 16
(2), 210
–217
(1997). http://dx.doi.org/10.1109/42.563666 ITMID4 0278-0062 Google Scholar
A. Landragin-Frassatiet al.,
“Application of a wavelet-Galerkin method to the forward problem resolution in fluorescence diffuse optical tomography,”
Opt. Express, 17
(21), 18433
–18448
(2009). http://dx.doi.org/10.1364/OE.17.018433 OPEXFF 1094-4087 Google Scholar
J. BaccouJ. Liandrat,
“On coupling wavelets with fictitious domain approaches,”
Appl. Math. Lett., 18
(12), 1325
–1331
(2005). http://dx.doi.org/10.1016/j.aml.2005.02.028 AMLEEL 0893-9659 Google Scholar
A. Landragin-Frassatiet al.,
“Model reduction using wavelet multiresolution technique applied to fluorescence diffuse optical tomography,”
Appl. Opt., 48
(36), 6878
–6892
(2009). http://dx.doi.org/10.1364/AO.48.006878 APOPAI 0003-6935 Google Scholar
T. J. RudgeV. Y. SolovievS. R. Arridge,
“Fast image reconstruction in fluorescence optical tomography using data compression,”
Opt. Lett., 35
(5), 763
–765
(2010). http://dx.doi.org/10.1364/OL.35.000763 OPLEDP 0146-9592 Google Scholar
S. R. ArridgeJ. C. Shotland,
“Optical tomography: forward and inverse problems,”
Inverse Probl., 25
(12), 123010
(2009). http://dx.doi.org/10.1088/0266-5611/25/12/123010 INPEEY 0266-5611 Google Scholar
A. SoubretJ. RipollV. Ntziachristos,
“Accuracy of fluorescent tomography in the presence of heterogeneities: study of the normalized Born ratio,”
IEEE Trans. Med. Imag., 24
(10), 1377
–1386
(2005). http://dx.doi.org/10.1109/TMI.2005.857213 ITMID4 0278-0062 Google Scholar
S. R. Arridge,
“Optical tomography in medical imaging,”
Inverse Probl., 15
(2), R41
–R93
(1999). http://dx.doi.org/10.1088/0266-5611/15/2/022 INPEEY 0266-5611 Google Scholar
T. Correiaet al.,
“Split operator method for fluorescence diffuse optical tomography using anisotropic diffusion regularisation with prior anatomical information,”
Biomed. Opt. Exp., 2
(9), 2632
–2648
(2011). http://dx.doi.org/10.1364/BOE.2.002632 BOEICL 2156-7085 Google Scholar
P. PeronaJ. Malik,
“Scale-space and edge detection using anisotropic diffusion,”
IEEE Trans. Pattern Anal. Mach. Intell., 12
(7), 629
–639
(1990). http://dx.doi.org/10.1109/34.56205 ITPIDJ 0162-8828 Google Scholar
S. Mallat,
“A theory for multiresolution signal decomposition: the wavelet representation,”
IEEE Trans. Pattern Anal. Mach. Intell., 11
(7), 674
–693
(1989). http://dx.doi.org/10.1109/34.192463 ITPIDJ 0162-8828 Google Scholar
I. Daubechies, Ten Lectures on Wavelets, SIAM, Philadelphia
(1992). Google Scholar
X. Songet al.,
“Automated region detection based on the contrast to noise ratio in near-infrared tomography,”
Appl. Opt., 43
(5), 1053
–1062
(2004). http://dx.doi.org/10.1364/AO.43.001053 APOPAI 0003-6935 Google Scholar
Q. FangD. Boas,
“Tetrahedral mesh generation from volumetric binary and gray-scale images,”
in Proc. IEEE Int. Symposium on Biomedical Imaging,
1142
–1145
(2009). Google Scholar
|