|
|
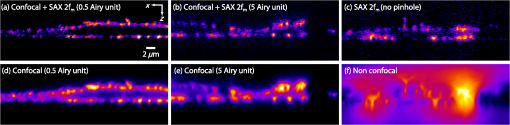
1.IntroductionOptical microscopy plays an important role in the observation of microscale structures or molecular activities in biological cells and tissues. Confocal fluorescence microscopy enables us to visualize a variety of subcellular structures or biological functions with spatial resolution in three-dimensional (3-D) and high-image contrast.1–4 A pinhole placed in front of a light detector is a key component in confocal microscopy; this pinhole eliminates the fluorescence from the out-of-focus planes that contributes a background signal. The depth discrimination granted by the pinhole allows us to observe the inside of a specimen without physically destroying the specimen. Furthermore, the spatial filtering effect of the pinhole helps increase the spatial resolution in the lateral directions. Recently, the spatial resolution of optical microscopy has been improved by manipulating the emission of fluorescent probes,5–19 and the role of optical microscopy has expanded to various different applications. To investigate cellular morphology and physiology of a living biological specimen, such as tissue-specific architectures, gene/protein expression, and drug metabolism, it is important to observe volumetric samples such as tissue and cell clusters.20,21 Multiphoton microscopy is one of the most well-known techniques used for observing such thick specimens.22–25 Multiphoton excitation inherently suppresses the fluorescence emissions from the out-of-focus positions,22 and the fluorescence detection through a pinhole provides further enhancement of the spatial resolution and allows finer structures to be imaged with high-image contrast.26,27 A technique to estimate the background fluorescence signal by introducing aberrations in the focus of the excitation laser has been demonstrated to improve the depth discrimination property of multiphoton excitation microscopy.28,29 For single-photon excitation, the use of excitation-intensity modulation has been demonstrated to limit the area of fluorescence detection to a focal volume; this technique is known as focal modulation microscopy and has been applied to image thick biological samples.30,31 A similar technique was also reported for multiphoton excitation.32 In this study, we report the depth imaging properties of saturated excitation (SAX) microscopy, in which SAX effectively suppresses the background fluorescence signal primarily from the out-of-focus planes, and demonstrate the fluorescence imaging of 3-D-cultured cells with improved spatial resolution. Recently, studies using 3-D cell cultures have emerged, since specimens in 3-D culture can more closely mimic important natural cellular functions and structures. There have been a number of reports about how cellular behaviors differ between cultures in two-dimensional (2-D) and 3-D. As examples, differentiated functions,33 gene expressions,34,35 and cellular morphology36–38 depend on the geometry of the culture. From these facts, high-resolution imaging techniques in volumetric samples will no doubt be in demand in the future for investigating details of cellular structures and activities. Volumetric samples, where light is scattered and aberrated by the refractive index distribution, substantially detract from the ideal imaging conditions. In this report, we describe the imaging property of the SAX microscopy when it is used in observation of cellular interiors and in cell clusters mainly by discussing the background suppression in SAX imaging. The SAX microscopy exploits the saturation fluorescent molecules’ excited state population to improve the spatial resolution.39–42 As the SAX is induced by light illumination at a high-excitation intensity, the nonlinear fluorescence signals are localized in the excitation focus spot, resulting in the improvement of spatial in 3-D. Since SAX microscopy detects the fluorescence signals’ nonlinear response to the excitation intensity, depth imaging properties can be improved in a manner similar to two-photon excitation microscopy. 2.Depth Imaging Property of SAX MicroscopyTo observe the details of thick volumetric specimens with high-spatial resolution and image contrast, background rejection capability is significantly important because thick samples produce strong out-of-focus fluorescence signals. For the evaluation of the depth discrimination properties in SAX microscopy, we estimated the optical transfer functions (OTFs) with different pinhole sizes and compared them with those used in typical confocal microscopy. The effective point-spread function (PSF) of confocal microscopy is given by multiplying the excitation PSF with the detection PSF.43 When the same objective lens is used for both excitation and fluorescence detection, the detection PSF is given by the 2-D convolution of the excitation PSF and the pinhole aperture function, if the wavelength of the fluorescence is similar to the wavelength of the excitation.44 The effective PSF for SAX microscopy is also given by the product of the detection and the excitation PSF, which can be estimated as the spatial distribution of the fluorescence signal that is detectable after harmonic demodulation.39,45 Equations (1) and (2) show the effective PSFs for typical confocal microscopy and for SAX microscopy with the ’th harmonic demodulation . where and denote the excitation PSFs for confocal and SAX microscopies, respectively. In addition, and are the detection PSF and the aperture function for a detection pinhole, respectively. The OTFs were calculated by applying Fourier transforms to the effective PSFs. Figure 1 shows the OTFs of the axial frequency calculated for different pinhole sizes that correspond to 0.17, 0.25, 0.5, 0.83, and 1.66 Airy units at the pinhole. We assumed that Rhodamine 6G was excited at 532 nm through an objective lens with an numerical aperture (NA) of 1.49. For SAX microscopy, the frequency of the excitation modulation was assumed to be 10 kHz, and the fluorescence signal was demodulated at the second- and fourth-harmonic frequencies.Fig. 1Optical transfer functions (OTFs) for (a) confocal and saturated excitation (SAX) microscopies with demodulation at frequencies of (b) and (c) for the axial direction at different pinhole sizes. The calculation was performed for different pinhole diameters that correspond to 0.17, 0.25, 0.5, 0.83, and 1.66 Airy units. 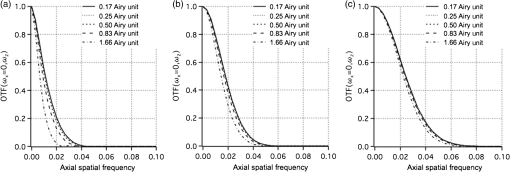 The calculated results show that introducing SAX in confocal microscopy expands the OTF in the axial direction. A comparison of the calculation results indicates that the resolvable spatial frequency is slightly larger in SAX microscopy, and the broadening of the bandwidth by SAX results in higher spatial resolution and image contrast than in confocal microscopy. Interestingly, the shape of the OTF does not greatly depend on the pinhole size in SAX microscopy, and this result indicates that the nonlinear relation between the excitation and fluorescence intensities plays a major role in the improvement of the response. The effect of the pinhole size becomes smaller at higher demodulation frequencies, because the background fluorescence is effectively eliminated through the detection of the higher-order nonlinear fluorescence response. To compare the depth discrimination properties in thick specimens, we simulated the edge responses when a thick fluorescent layer is observed. The images of the fluorescent layer were calculated by convolving a step function and the effective PSF.46 The background rejection properties can be determined by the steepness of the edge response curve at the bottom (baseline response). In the calculation results (Fig. 2), the sharper edge response in the SAX microscopy is clearly evident, which indicates that the fluorescence signal generated at the out-of-focus planes can be effectively eliminated. As in the OTFs in Fig. 1, the pinhole size does not strongly affect the edge response in the SAX microscopy, which indicates that the nonlinear relation between the excitation and fluorescence in the SAX predominantly contributes to the reduction in the background fluorescence. This effect is more significant in the result with the fourth-harmonic demodulation. Fig. 2Edge responses for (a) confocal and SAX microscopies with demodulations of (b) and (c) that were calculated for the measurement at a border between a Rhodamine 6G solution and a glass substrate. The calculation was performed for different pinhole diameters that correspond to 0.17, 0.25, 0.5, 0.83, and 1.66 Airy units. 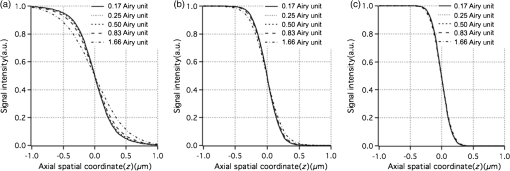 In addition to simulations, we experimentally examined the discrimination properties of both confocal and SAX microscopes. In the experiment, a 532-nm CW laser was focused by an objective lens with an NA of 1.2 into a Rhodamine 6G solution, which served as the thick fluorescence layer. The fluorescence emission from the solution was collected by the same objective lens and was introduced into a pinhole for confocal detection through a dichroic mirror, which removed the excitation wavelength. A photomultiplier tube (Hamamatsu, H7422-40, Iwata, Shizuoka) was used to detect the fluorescence signal. To measure the axial responses, the sample was scanned in the -direction with a piezoelectric translation stage (Physik Instrumente, P-561.3CD, Karlsruhe, Baden-Württemberg) with fluorescence detection. For the SAX imaging, the excitation intensity was modulated at 10 kHz with a set of acousto-optic modulators (IntraAction, AOM-402AF1, Bellwood, Illinois), and a lock-in amplifier (NF Corp., Yokohama, Kanagawa, LI5640 for Fig. 3Fig. 4–5, Zurich Instruments, Zurich, HF2LI for Fig. 6) was used to demodulate the signal from the photomultiplier tube at the second-harmonic frequency (20 kHz). We used 30 and 100 μm pinholes (which correspond to 0.5 and 1.66 Airy units, respectively) to compare the effects of the pinhole size on the axial response. Figure 3 shows the edge responses measured with confocal and SAX microscopies. As predicted by the theoretical investigation, the border between the fluorescent solution and cover slip was observed to be steeper in the SAX images. The steepness of the edge is also less dependent on the pinhole size in SAX microscopy, as indicated by the theoretical calculation, and the profiles of the responses in the experimental results match the calculation well. Fig. 3Experimental results for the edge response measured at the border between a Rhodamine 6G solution and a cover glass by (a) confocal and (b) SAX microscopies with harmonic demodulation of (20 kHz). The edge responses were measured with pinhole sizes of (a) 30 μm and (b) 100 μm, which correspond to 0.5 and 1.66 Airy units, respectively. 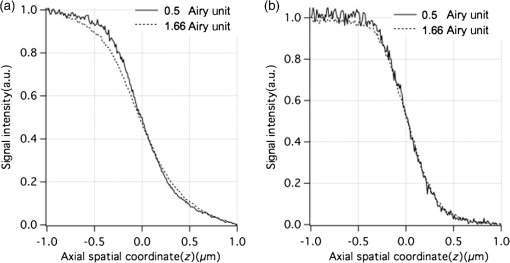 As demonstrated experimentally and theoretically, the pinhole size has less effect in the SAX microscopy. For comparison, the axial response in confocal microscopy is shown in Fig. 3(a), and this response is degraded significantly by increasing the pinhole size. However, no clear difference is observed in the edge responses for the different pinhole sizes in the SAX microscopy, as shown in Fig. 3(b). In this experiment, since we used a water-immersion objective lens and the sample was illuminated through a cover slip, the laser focus is formed correctly when the laser is focused in the solution layer. Therefore, the effect of the pinhole size in the edge response matches the calculation result well when the laser focus exists in the solution (). 3.X-Z Cross-Section Imaging of CellsTo confirm the improvement of depth discrimination properties in the imaging of biological samples, we obtained cross-sectional images of HeLa cells with stained actin filaments. The sample was observed using the above-mentioned optical setup with a different objective lens (an Olympus, Shinjuku, Tokyo NA 1.49 with oil immersion). The HeLa cells were stained with ATTO-Rho6G phalloidin (ATTO-TEC GmbH, Siegen, North Rhine-Westphalia) and were embedded in a Prolong Gold antifade mounting medium, which has a refractive index of 1.43 to 1.45. We observed the same position in the sample in both the confocal and SAX modes, and pinhole sizes corresponding to 0.5 to 5 Airy units were used to examine the differences in the imaging properties. Furthermore, we performed fluorescence imaging without a pinhole to determine the contribution of the nonlinear excitation in the SAX microscopy to the background elimination and to the improvement of the spatial resolution, which we examined in the axial direction. The excitation intensities of the laser were and for the confocal and SAX imaging, respectively. In Figs. 4(a) and 4(d), as expected from the theoretical considerations, any blurring of the image along the -direction is effectively suppressed, and the contrast in all the structures is improved. As the pinhole size increased, the spatial resolution became slightly lower for both the confocal and SAX images, as shown in Figs. 4(b) and 4(e); however, the spatial resolution of the SAX microscopy in the axial direction is still higher than the resolution in the confocal image with a pinhole of 0.5 Airy units. In the fluorescence images observed without a confocal pinhole [Figs. 4(c) and 4(f)], the spatial resolution in the axial direction is not preserved in the nonconfocal image, yet it is still maintained in the SAX image. 4.X-Z Cross-Section Imaging of Cell ClustersTo confirm the improvement of the spatial resolution for imaging thick samples, we observed a thick cell cluster by the confocal and SAX microscopies. The cell clusters were prepared by culturing HeLa cells in a 3-D matrix (BD bioscience, BD matrigel, San Jose, California), and the actin filaments were stained with ATTO488 phalloidin (ATTO-TEC GmbH). The sample was observed with a silicone oil-immersion objective lens (Olympus, NA 1.3) and a pinhole corresponding to 0.5 Airy units. Figure 5(a) shows cross-sectional images of the actin filaments in the HeLa cells that were obtained by the SAX microscopy. The magnification of the boxed area in Fig. 5(a) is shown in Fig. 5(b). For comparison, we also observed the same sample with the confocal microscopy, as shown in Figs. 5(c) and 5(d). The comparison of the magnified images clearly shows that SAX microscopy substantially suppresses the blurring of the image along the -direction. Figures 5(e) and 5(f) are line profiles of the structures indicated by the arrows in Figs. 5(b) and 5(d), respectively. From the profiles, we can confirm the improvement of the spatial resolution at deep inside the specimen in the SAX image as well as the improvement near the surface. Fig. 5Fluorescence images of HeLa cells cultured in a three-dimensional (3-D) matrix and the line profiles of the structures. (a) SAX and (c) confocal images of actin filaments in HeLa cells in the plane. (b and d) Magnifications of the boxed area in (a and c). The line profiles are shown in (e and f) of the structures indicated by the arrows in the adjacent images. The sample was stained with ATTO488 phalloidin, and the pixel size was 175 nm. The pixel numbers were for (a and c) and for (b and d), the pixel dwell time was 500 μs, and the excitation intensities for SAX and confocal modes were 12 and , respectively. A silicone-oil immersion 1.3 NA objective lens was used with a pinhole size corresponding to 0.5 Airy units. In addition, low-pass filtering was applied to the images. 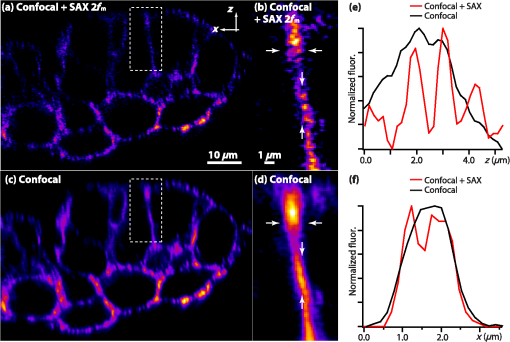 5.Volumetric Imaging of Cell ClustersFinally, we performed volumetric imaging of cell clusters to demonstrate the improved depth discrimination of the SAX microscopy in observations of thick specimens. Here, we observed alpha-tubulin in a 3-D cell cluster, which was stained with Rhodamine 6G goat anti-mouse IgG (Active motif, Carlsbad, California). The sample was mounted with antifade reagent (Invitrogen, Prolong Gold, Carlsbad, California) and observed with an oil-immersion objective lens (Olympus, NA 1.4). The pinhole size in this experiment corresponded to 0.75 Airy units. In this observation, we recorded images of the sample at each imaging depth and constructed the volumetric images by the 3-D projection function in Image J (National Institute of Health). Figure 6(a) shows the volumetric image of alpha-tubulin which was obtained by the SAX mode. The magnification of the boxed areas in Fig. 6(a) is shown in Fig. 6(b). We also observed the same area with the confocal microscopy [Fig. 6(c)] for comparison. From the results, we confirmed that SAX microscopy suppresses the detection of the out-of-focus fluorescence signals, and the effective background rejection enables us to visualize more detailed structures of the sample with high-image contrast. Here, we also observed alpha-tubulin in HeLa cells on a glass substrate [Fig. 6(d)] to determine the difference between the sample in 3-D culture and on the glass substrate. As a result, we confirmed that the distribution of alpha-tubulin is significantly different in 3-D culture and on the glass substrate. In these experiments, to reduce photobleaching effects during the observation, we increased the modulation frequency of the excitation intensity to 50 kHz for reducing the pixel dwell time and speeding up the image acquisition. Fig. 63-D projection of HeLa cells in 3-D culture and on a glass substrate. (a and Video 1, MOV, 3.33 MB) [URL: http://dx.doi.org/10.1117/1.JBO.18.12.126002.1] SAX image of alpha-tubulin in the cell cluster. (b) The magnification of the boxed area in (a). (c) Confocal image of the same area in (b). (d) SAX image of alpha-tubulin in HeLa cells on the glass substrate. The SAX images were obtained with demodulation at the second-harmonic frequency (100 kHz). (e–h and Video 2, MOV, 4.66 MB) [URL: http://dx.doi.org/10.1117/1.JBO.18.12.126002.2] and (i–l and Video 3, MOV, 6.09 MB) [URL: http://dx.doi.org/10.1117/1.JBO.18.12.126002.3] are the same series of data for volumetric images of mitochondria and actin filaments, respectively. The volumetric images are composed of (a) 160, (d) 52, (e) 151, (h) 43, (i) 110, and (l) 25 images. The voxel size was . The pixel dwell time was 50 μs. For the observations of the cell clusters, the excitation intensity was increased with the imaging depth to maintain a sufficient signal-to-noise ratio (SNR) for SAX imaging. The excitation intensities were at the layer of the cell clusters closest to the objective and rising to (a) 180, (e) 150, and (i) at the deepest sections. For imaging of HeLa cells on the glass substrate, the excitation intensity was . 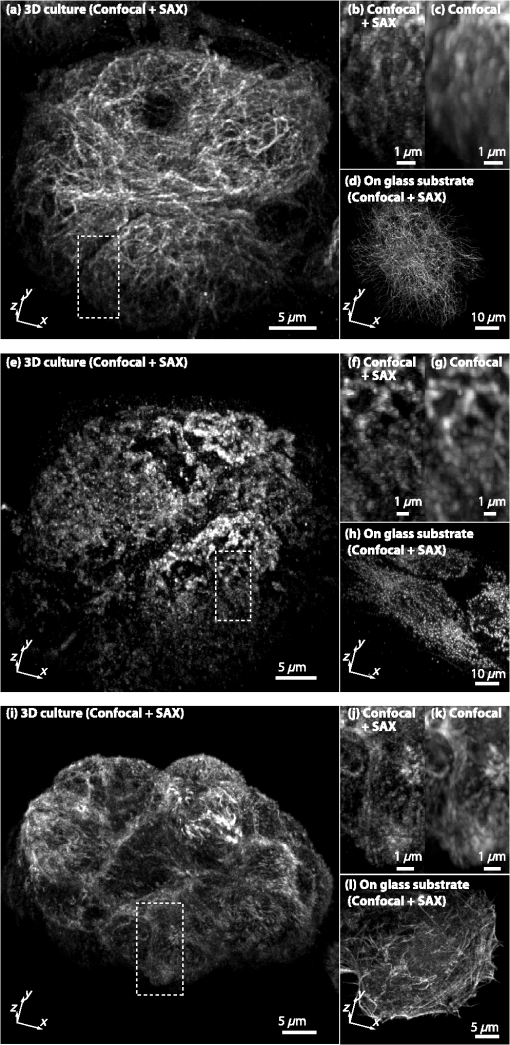 To check the distribution of other cellular structures in cell clusters, we also performed volumetric observations of mitochondria and actin filaments in 3-D culture and on the glass substrate, as shown in Figs. 6(e)–6(l). Mitochondria and actin filaments were stained with Rhodamine 6G goat anti-mouse IgG and ATTO Rhod6G phalloidin, respectively. The other experimental conditions were the same as in Figs. 6(a)–6(d). Here, we again confirmed that the SAX mode visualizes finer structures compared with the confocal, and the cellular morphology is obviously different between cells in 3-D culture and on the glass substrate. 6.ConclusionIn this article, we described the depth imaging properties of the SAX microscopy in comparison to confocal microscopy and showed fluorescence images of an approximately 40-μm-thick cell cluster with higher spatial resolution than conventional confocal microscopy. In practice, in confocal microscopy, the size of the pinhole is determined by considering a balance of the spatial resolution and the amount of fluorescence signal at the detector, and is typically set to be an Airy unit. In this configuration, it is difficult to completely eliminate strong fluorescence emissions from the out-of-focus planes, and the contrast of the fine structures in the sample can be significantly degraded. Even in these samples, it is expected that SAX and harmonic demodulation can suppress the strong fluorescence background and preserve the contrast of the structures in the focal planes. In addition, the high-depth-discrimination property of SAX microscopy can be useful in observations of samples with high degrees of light scattering, because one can choose a slightly larger pinhole to collect the fluorescence signal after the scattering. Furthermore, the spatial resolution can be maintained at a level appropriate for the ideal confocal detection. In the SAX microscopy, higher spatial resolution is achieved by harmonic demodulation of fluorescence signals at higher-order harmonic frequencies. However, in observations of biological samples stained by organic dyes or fluorescent proteins, the achieved spatial resolution is limited due to photobleaching and the limitations of signal-to-noise ratio (SNR) in the fluorescence detection. So far, using photostable fluorescence probes such as fluorescent nanodiamonds, third- and fourth-harmonic demodulations have been demonstrated for a further increase of the spatial resolution. With a 1.4 NA objective lens and an excitation wavelength of 532 nm, the expected axial resolution for confocal is 470 nm and for SAX microscopy with second-, third-, and fourth-harmonic frequencies are 353, 290, and 267 nm, respectively.41 To apply these higher-order harmonic signals to observe intracellular structures, the development of fluorophores with higher photostability and high SNR detection systems is required. In this article, we also demonstrated high-resolution fluorescence imaging of cell clusters in 3-D culture. The results show that the background elimination by SAX microscopy allows us to visualize the smaller details in the cell clusters with improved spatial resolution and image contrast. In comparison with 3-D images of HeLa cells on the glass substrate, we confirmed that the cellular morphology in 3-D culture and on the glass substrate is substantially different, as also indicated in other reports.36–38 Recently, it has been recognized that 3-D culture techniques enable us to mimic the in vivo environment and to preserve more closely important natural cellular functions and morphology. Therefore, volumetric observations of thick specimens in 3-D culture will become more important for biological studies in the future. For the observation of thick specimens, the observable depth is limited by the SNR of fluorescence detection, which decreases with the observation depth. Since SAX microscopy requires a high SNR in fluorescence detection to extract the nonlinear fluorescence response, the loss of fluorescence signal by scattering is the main limitation on the observable depth. Since the typical thickness of specimens in 3-D culture is several tens to hundreds of micometers,22,47,48 the current imaging depth in SAX microscopy may not be sufficient for all future requirements. The combination of SAX and two-photon excitation with near-infrared light could then also be employed to realize deeper imaging with high-spatial resolution. AcknowledgmentsThis study was supported by the Next Generation World-Leading Researchers (NEXT Program) of the Japan Society for the Promotion of Science (JSPS) and the Network Joint Research Center for Materials and Devices. ReferencesT. Wilson, Confocal Microscopy, Academic Press, London
(1990). Google Scholar
T. WilsonC. J. R. Sheppard, Theory and Practice of Scanning Optical Microscopy, Academic Press, London
(1984). Google Scholar
A. Diaspro, Confocal and Two-Photon Microscopy: Foundations, Applications, and Advances, John Wiley & Sons, Inc., Hoboken, New Jersey
(2002). Google Scholar
S. W. HellJ. Wichmann,
“Breaking the diffraction resolution limit by stimulated emission:stimulated-emission-depletion fluorescence microscopy,”
Opt. Lett., 19
(11), 780
–782
(1994). http://dx.doi.org/10.1364/OL.19.000780 OPLEDP 0146-9592 Google Scholar
V. Westphalet al.,
“Video-rate far-field optical nanoscopy dissects synaptic vesicle movement,”
Science, 320
(5873), 246
–249
(2008). http://dx.doi.org/10.1126/science.1154228 SCIEAS 0036-8075 Google Scholar
E. Betziget al.,
“Imaging intracellular fluorescent proteins at nanometer resolution,”
Science, 313
(5793), 1642
–1645
(2006). http://dx.doi.org/10.1126/science.1127344 SCIEAS 0036-8075 Google Scholar
S. T. HessT. P. K. GirirajanM. D. Mason,
“Ultra-high resolution imaging by fluorescence photoactivation localization microscopy,”
Biophys. J., 91
(11), 4258
–4272
(2006). http://dx.doi.org/10.1529/biophysj.106.091116 BIOJAU 0006-3495 Google Scholar
M. J. RustM. BatesX. Zhuang,
“Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM),”
Nat. Methods, 3
(10), 793
–796
(2006). http://dx.doi.org/10.1038/nmeth929 1548-7091 Google Scholar
S.-H. Shimet al.,
“Super-resolution fluorescence imaging of organelles in live cells with photoswitchable membrane probes,”
Proc. Natl. Acad. Sci. U. S. A., 109
(35), 13978
–13983
(2012). http://dx.doi.org/10.1073/pnas.1201882109 PNASA6 0027-8424 Google Scholar
M. Heilemannet al.,
“Super-resolution imaging with small organic fluorophores,”
Angew. Chem. Int. Ed., 48
(37), 6903
–6908
(2009). http://dx.doi.org/10.1002/anie.v48:37 ACIEF5 1433-7851 Google Scholar
M. G. Gustafsson,
“Nonlinear structured-illumination microscopy: wide-field fluorescence imaging with theoretically unlimited resolution,”
Proc. Natl. Acad. Sci. U. S. A., 102
(37), 13081
–13086
(2005). http://dx.doi.org/10.1073/pnas.0406877102 PNASA6 0027-8424 Google Scholar
R. HeintzmannT.M. Jovin,
“Saturated patterned excitation microscopy—a concept for optical resolution improvement,”
J. Opt. Soc. Am. A, 19
(8), 1599
–1609
(2002). http://dx.doi.org/10.1364/JOSAA.19.001599 JOAOD6 0740-3232 Google Scholar
R. Heintzmann,
“Saturated patterned excitation microscopy with two-dimensional excitation patterns,”
Micron, 34
(6–7), 283
–291
(2003). http://dx.doi.org/10.1016/S0968-4328(03)00053-2 MICNB2 0047-7206 Google Scholar
E. H. Regoet al.,
“Nonlinear structured-illumination microscopy with a photoswitchable protein reveals cellular structures at 50-nm resolution,”
Proc. Natl. Acad. Sci. U. S. A, 109
(3), E135
–E143
(2012). http://dx.doi.org/10.1073/pnas.1107547108 PNASA6 0027-8424 Google Scholar
N. T. Urbanet al.,
“STED nanoscopy of actin dynamics in synapses deep inside living brain slices,”
Biophys. J., 101
(5), 1277
–1284
(2011). http://dx.doi.org/10.1016/j.bpj.2011.07.027 BIOJAU 0006-3495 Google Scholar
F. C. Zanacchiet al.,
“Live-cell 3D super-resolution imaging in thick samples,”
Nat. Methods, 8
(12), 1047
–1744
(2011). http://dx.doi.org/10.1038/nmeth.1744 1548-7091 Google Scholar
P. Kneret al.,
“Super-resolution video microscopy of live cells by structured illumination,”
Nat. Methods, 6
(5), 339
–343
(2009). http://dx.doi.org/10.1038/nmeth.1324 1548-7091 Google Scholar
R. Fiolkaet al.,
“Time-lapse two-color 3D imaging of live cells with doubled resolution using structured illumination,”
Proc. Natl. Acad. Sci. U. S. A, 109
(14), 5311
–5315
(2012). http://dx.doi.org/10.1073/pnas.1119262109 PNASA6 0027-8424 Google Scholar
F. PampaloniE. G. ReynaudE. H. Stelzer,
“The third dimension bridges the gap between cell culture and live tissue,”
Nat. Rev. Mol. Cell. Biol., 8
(10), 839
–845
(2007). http://dx.doi.org/10.1038/nrm2236 NRMCBP 1471-0072 Google Scholar
J. DebnathJ. S. Brugge,
“Modeling glandular epithelial cancers in three-dimensional cultures,”
Nat. Rev. Cancer, 5
(9), 675
–688
(2005). http://dx.doi.org/10.1038/nrc1695 NRCAC4 1474-175X Google Scholar
W. DenkJ. H. StricklerW. W. Webb,
“Two-photon laser scanning fluorescence microscopy,”
Science, 248
(4951), 73
–76
(1990). http://dx.doi.org/10.1126/science.2321027 SCIEAS 0036-8075 Google Scholar
R. Kawakamiet al.,
“Visualizing hippocampal neurons with in vivo two-photon microscopy using a 1030 nm picosecond pulse laser,”
Sci. Rep., 3 1
–7
(2012). SREOA7 0474-781X Google Scholar
N. G. Hortonet al.,
“In vivo three-photon microscopy of subcortical structures within an intact mouse brain,”
Nat. Photonics, 7
(3), 205
–209
(2013). http://dx.doi.org/10.1038/nphoton.2012.336 1749-4885 Google Scholar
P. TheerW. Denk,
“On the fundamental imaging-depth limit in two-photon microscopy,”
J. Opt. Soc. Am. A, 23
(12), 3139
–3149
(2006). http://dx.doi.org/10.1364/JOSAA.23.003139 JOAOD6 0740-3232 Google Scholar
C. J. R. SheppardM. Gu,
“Image formation in two-photon fluorescence microscopy,”
Optik, 86
(3), 104
–106
(1990). OTIKAJ 0030-4026 Google Scholar
O. Nakamura,
“A two-photon scanning fluorescence microscope with deep UV excitation and near UV detection,”
Optik, 100
(4), 167
–170
(1995). OTIKAJ 0030-4026 Google Scholar
A. LerayJ. Mertz,
“Rejection of two-photon fluorescence background in thick tissue by differential aberration imaging,”
Opt. Express, 14
(22), 10565
–10573
(2006). http://dx.doi.org/10.1364/OE.14.010565 OPEXFF 1094-4087 Google Scholar
A. LerayK. LillisJ. Mertz,
“Enhanced background rejection in thick tissue with differential-aberration two-photon microscopy,”
Biophys. J., 94
(4), 1449
–1458
(2008). http://dx.doi.org/10.1529/biophysj.107.111476 BIOJAU 0006-3495 Google Scholar
N. ChenmC.-H. WongC. J. R. Sheppard,
“Focal modulation microscopy,”
Opt. Express, 16
(23), 18764
–18769
(2008). http://dx.doi.org/10.1364/OE.16.018764 OPEXFF 1094-4087 Google Scholar
K. Siet al.,
“Enhanced background rejection in thick tissue using focal modulation microscopy with quadrant apertures,”
Opt. Commun., 284
(5), 1475
–1480
(2011). http://dx.doi.org/10.1016/j.optcom.2010.11.007 OPCOB8 0030-4018 Google Scholar
K. Isobeet al.,
“Background-free deep imaging by spatial modulation nonlinear optical microscopy,”
Biomed. Opt. Express, 3
(7), 1594
–1608
(2012). http://dx.doi.org/10.1364/BOE.3.001594 BOEICL 2156-7085 Google Scholar
M. Lechonet al.,
“Long-term expression of differentiated functions in hepatocytes cultured in three-dimensional collagen matrix,”
J. Cell. Physiol., 177
(4), 553
–562
(1998). http://dx.doi.org/10.1002/(ISSN)1097-4652 JCLLAX 0021-9541 Google Scholar
M. J. BissellH. G. HallG. Parry,
“How does the extracellular matrix direct gene expression?,”
J. Theor. Biol., 99
(1), 31
–68
(1982). http://dx.doi.org/10.1016/0022-5193(82)90388-5 JTBIAP 0022-5193 Google Scholar
M. DelcommenneC. H. Streuli,
“Control of integrin expression by extracellular matrix,”
J. Biol. Chem., 270
(45), 26794
–26801
(1995). http://dx.doi.org/10.1074/jbc.270.45.26794 JBCHA3 0021-9258 Google Scholar
C. P. Soareset al.,
“2D and 3D-organized cardiac cells shows differences in cellular morphology, adhesion junctions, presence of myofibrils and protein expression,”
PLoS ONE, 7
(5), e38147
(2012). http://dx.doi.org/10.1371/journal.pone.0038147 1932-6203 Google Scholar
L. C. E. Winduset al.,
“In vivo biomarker expression patterns are preserved in 3D clusters of prostate cancer,”
Exp. Cell. Res., 318
(19), 2507
–2519
(2012). http://dx.doi.org/10.1016/j.yexcr.2012.07.013 ECREAL 0014-4827 Google Scholar
A. NygaU. CheemaM. Loizidou,
“3D tumour models: novel in vitro approaches to cancer studies,”
J. Cell Commun. Signal., 5
(3), 239
–248
(2011). http://dx.doi.org/10.1007/s12079-011-0132-4 1873-9601 Google Scholar
K. Fujitaet al.,
“High-resolution confocal microscopy by saturated excitation of fluorescence,”
Phys. Rev. Lett., 99
(22), 228105
(2007). http://dx.doi.org/10.1103/PhysRevLett.99.228105 PRLTAO 0031-9007 Google Scholar
M. Yamanakaet al.,
“Beyond the diffraction-limit biological imaging by saturated excitation microscopy,”
J. Biomed. Opt., 13
(5), 050507
(2008). http://dx.doi.org/10.1117/1.2992595 JBOPFO 1083-3668 Google Scholar
M. Yamanakaet al.,
“SAX microscopy with fluorescent nanodiamond probes for high-resolution fluorescence imaging,”
Biomed. Opt. Express, 2
(7), 1946
–1954
(2011). http://dx.doi.org/10.1364/BOE.2.001946 BOEICL 2156-7085 Google Scholar
M. Yamanakaet al.,
“Saturated excitation (SAX) of fluorescence proteins for sub-diffraction-limited imaging of living cells in three dimensions,”
Interface Focus, 3
(5), 20130007
(2013). http://dx.doi.org/10.1098/rsfs.2013.0007 20428898 Google Scholar
M. Gu, Principle of Three-Dimensional Imaging in Confocal Microscopes, World Scientific, Singapore
(1996). Google Scholar
K. SiW. GongC. J. R. Sheppard,
“Three-dimensional coherent transfer function for a confocal microscope with two D-shape pupils,”
Appl. Opt., 48
(5), 810
–817
(2009). http://dx.doi.org/10.1364/AO.48.000810 APOPAI 0003-6935 Google Scholar
S. Kawanoet al.,
“Determination of the expanded optical transfer function in saturated excitation imaging and high harmonic demodulation,”
Appl. Phys. Express, 4
(4), 042401
(2011). http://dx.doi.org/10.1143/APEX.4.042401 1882-0778 Google Scholar
M. GuT. TannousC. J. R. Sheppard,
“Improved axial resolution in confocal fluorescence microscopy using annular pupils,”
Opt. Commun., 110
(5–6), 533
–539
(1994). http://dx.doi.org/10.1016/0030-4018(94)90245-3 OPCOB8 0030-4018 Google Scholar
J. Debnathet al.,
“The role of apoptosis in creating and maintaining luminal space within normal and oncogene-expressing mammary acini,”
Cell, 111
(1), 29
–40
(2002). http://dx.doi.org/10.1016/S0092-8674(02)01001-2 CELLB5 0092-8674 Google Scholar
G. Y. Leeet al.,
“Three-dimensional culture models of normal and malignant breast epithelial cells,”
Nat. Protocol., 4
(4), 359
–365
(2007). http://dx.doi.org/10.1038/nmeth1015 1548-7091 Google Scholar
|