|
|
1.IntroductionAnimal models allow us to recapitulate many of human pathologies including cancer and, although they do not perfectly mimic all aspects of cancer development, they greatly contributed to our understanding of many tumorigenesis mechanisms.1 Initial studies consisted of implantation of human cancer cells or tumor biopsies into nude mice and evaluation of tumor progression by palpation and/or measurement of tumor sizes by devices such as a caliper.2 However, these manipulations could not be applied to study deep tumors or metastatic deposits, unless the animal was dissected to inspect for tumor lesions in several tissues and organs, therefore rendering these approaches laborious and animal consuming.2 Hence, there was a need to find out alternative methodologies that would ease early tumor detection and evaluation of cancer-linked events such as neoangiogenesis, invasion and metastatic dissemination.3 Accordingly, interest has increased within the past decade in sensitive noninvasive techniques such as fluorescence-based imaging with proteins of the enhanced green fluorescent protein (EGFP) family,4 the Discosoma sp. DsRed or their shifted wavelength spectra variants5 as well as luciferase-based bioluminescence.3 Yet, for bioluminescence to happen, it is obligatory to inject the substrates luciferin or coelentarazin prior to imaging in order to elicit luciferase reaction.6 Fluorescence imaging of deep tumors using EGFP, DsRed or their mutants is hampered by the limited light penetration in the body, scattering, and tissue absorption.7 Another problem encountered with EGFP/DsRed-based imaging is autofluorescence which mostly occurs in the green part of the spectrum.8 Recently, a series of very bright red-shifted fluorescent proteins, such as mKate,9 E2-Crimson and mNeptune,7 with an average excitation/emission in the range of , have been developed with significantly increased tissue penetration and reduced autofluorescence. With regard to these limitations, probes with the wavelengths in the near-infrared (NIR) to infrared (IR) ranges (i.e., 650 to 900 nm), would be superior to others for deep-tissue imaging. Recently, two NIR fluorescent proteins, IFP1.4 (Ref. 10) and iRFP (Ref. 11), have emerged with fluorescence characteristics laying within an NIR transparency of mammalian tissues.12 The first described in 2009 was the bacteriophytochrome IFP1.4 followed, 2 years later by iRFP, cloned from the Rhodopseudomonas palustris and engineered through a series of mutations.11 Although fluorescence spectra of the two molecules are quite similar, IFP1.4 must incorporate its chromophore biliverdin (which needs to be exogenously supplied) to further emit fluorescence, whereas the physiological level of biliverdin13 is sufficient to fully activate iRFP. This property, added to the high brightness, prompted us to use iRFP as a tool to track primary tumors and metastasis in deep tissues and behind natural barriers such as bones. We, herein, used in vivo NIR optical imaging together with a human immunodeficiency virus (HIV)-based lentiviral vector to deliver the iRFP gene into different tumor cell models including glioblastoma, osteosarcoma, and melanoma cells. We then used these cells to explore intracranial and intratibial primary tumor growth as well as metastatic dissemination of melanoma cells. 2.Material and Methods2.1.Vector ConstructionThe HIV-derived recombinant lentivirus vector expressing iRFP was made by replacing the deoxyribose nucleic acid (DNA) of EGFP in the lentiviral vector pRRLSIN.cPPT.PGK-GFP.WPRE (referred to as pRRL, kindly provided by Dr. Trono, University of Geneva, Switzerland) by the iRFP DNA from the pShuttle-CMV-iRFP plasmid (a gift of Dr. Verkhusha, Addgene plasmid # 31856). To this end, iRFP DNA was excised with BglII and SalI, gel purified and ligated into BamHI/SalI-digested pRRL resulting in pRRL/iRFP construct [Fig. 1(a)]. Fig. 1(a) Schematic diagram of pRRL/iRFP construct obtained by swapping deoxyribose nucleic acid (DNA) of EGFP in pRRLSIN.cPPT.PGK-GFP.WPRE vector by iRFP DNA. The expression of the transgene is under the control of the human phosphoglycerate kinase promoter (PGK). The central polypurine tract (cPPT) and the posttranscriptional Woodchuck hepatitis virus (WPRE) are located upstream and downstream of the transgene, respectively. RSV, rous sarcoma virus; SD, splice donor; SA, splice acceptor; Gag, deleted gag region; RRE, rev-responsive element; LTR, long terminal repeat; EGFP, enhanced green fluorescent protein; iRFP, infrared fluorescent protein. (b) Phase contrast and iRFP-based fluorescence of tumor cells transduced with pRRL/iRFP and observed with the Cy5.5 filter at excitation/emission of (magnification ). (c and d) Sequential images and the deduced growth curve after subcutaneous injection of iRFP-expressing U87 MG cells. The color bar indicates the total fluorescence radiant efficiency. 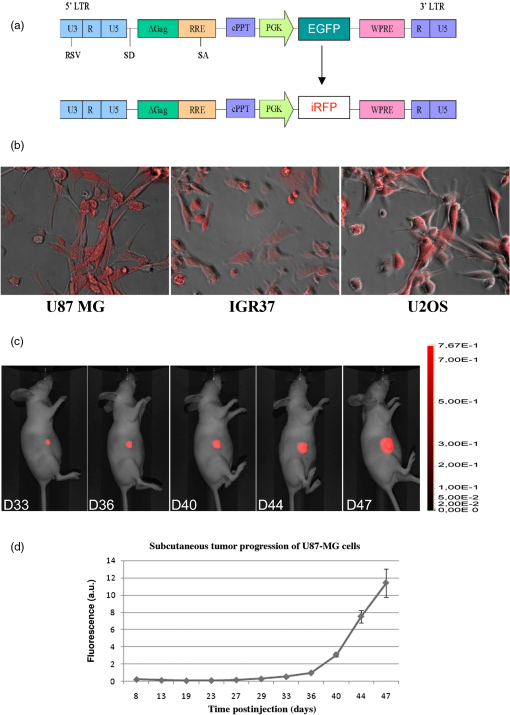 2.2.Cells and Lentiviral TransductionThe human embryonic kidney cells HEK293T, the glioblastoma U87 MG and the melanoma IGR37 cells were cultured in Dulbecco's modified Eagle medium (Cambrex, Verviers, Belgium). The osteosarcoma U2OS cells were cultured in Roswell Park Memorial Institute (RPMI) medium 1640 (Cambrex). All media were supplemented with 10% fetal calf serum and antibiotics penicillin and streptomycin (Eurobio, Les Ullis, France). Lentiviral particles were produced according to Mathieu et al.14 2.3.Animals and Tumor ImplantationsFour- to six-week old male athymic Naval Medical Research Institute nude mice (Harlan, Gannat, France) were maintained in a barrier facility on high efficiency particules arresting (HEPA)-filtered racks and fed with autoclaved alfalfa-free rodent diet to avoid digestive tract autofluorescence.15 Animal experiments were conducted in accordance with the European Directive EEC/2010/63, under the supervision of authorized investigators. Subcutaneous (s.c) and intracranial (i.c) injections of U87 MG glioblastoma cells were performed as described earlier16,17 using cells (s.c) or cells (i.c), respectively. For the experimental metastases, we injected melanoma cells IGR37 through the tail veins. For intratibial xenografts, mice were anesthetized and U2OS cells were injected into the tibia with a needle introduced at the top of the tibial metaphysics. Immunohistochemistry of melanoma tumors was performed according to Ref. 18, using S100, HBM-45, and Melan-A as markers. 2.4.In Vivo Imaging and AnalysesUnless otherwise indicated, animals were subjected to NIR imaging every 3 to 4 days with a Pearl® Impulse Small Animal Imaging System Li-Cor® Biosciences (LI-COR Biosciences GmbH, Bad Homburg, Germany). Excitation and emission wavelengths were fixed at 690 and 710 nm, respectively. The fluorescence signal was overlaid with the white channel light-emitting diode [(LED) light] and detected by a cooled CCD camera of the imager. During the in vivo imaging process, mice were kept on the imaging bed by isoflurane inhalation (Abbott Laboratories, Rungis, France) and warmed at 37°C. In some experiments, organs or tumor deposits were retrieved with the minimum surrounding tissue from mice sacrificed by cervical dislocation and subjected to ex vivo imaging. Excised tissues were then placed on the stage and their fluorescence captured as indicated above. Image analysis was performed according to Nakayama et al.19 In brief, regions of interest (ROIs) were targeted on iRFP fluorescent sites and circles with the minimum size were drawn around lesions on each image. The pixel density in the selected ROI was quantitated using the software for image analysis provided with the Pearl® Imager. The fluorescence signal on the plots was normalized to the background and presented as arbitrary units (a.u.) according to Filonov et al.11 3.Results3.1.Visualization of iRFP-Expressing Tumor Xenografts in MiceTumor cells expressing iRFP were produced as described and analyzed for the NIR fluorescence before inoculation to animals [Fig. 1(b)]. As shown in Fig. 1(c), fluorescence of iRFP-expressing U87 MG glioma cells,20 not only clearly distinguishes the tumor from the host but also allows a real-time monitoring of subcutaneous tumor growth, without any additive [Fig. 1(d)]. By subcutaneously inoculating increasing numbers of iRFP-labeled cells, we found that the minimal number of detectable cells by our NIR imaging device is 50,000 cells (data not shown). Filonov et al. advocated the use of iRFP for imaging of deep organs such as the liver.11 We, therefore, wanted to ask whether iRFP would uncover intraosseous tumors using intracranial and intratibial inoculated iRFP-expressing tumor cells. As shown in Fig. 2(A), the growth of an intracranial implant of iRFP-expressing U87 MG cells can be easily detected and monitored in real-time in vivo [Fig. 2(A)] and ex vivo, after the brain of this mouse has been retrieved [Fig. 2(Aa)]. The deduced growth curve shows a good correlation with the images [Fig. 2(Ab)]. Fig. 2(A) Serial whole-body images of mice harboring intracranial tumors derived from U87 MG cells. Shown also is a retrieved brain from mice at day 33 (a) and the deduced growth curve from A. The color bar indicates the total fluorescence radiant efficiency. (B) Intratibial tumors of U2OS cells and images of retrieved tibias from mice at days 43 (a) and 70 (b). The color bar indicates the total fluorescence radiant efficiency. The deduced growth curve of U2OS growth is shown in (c). 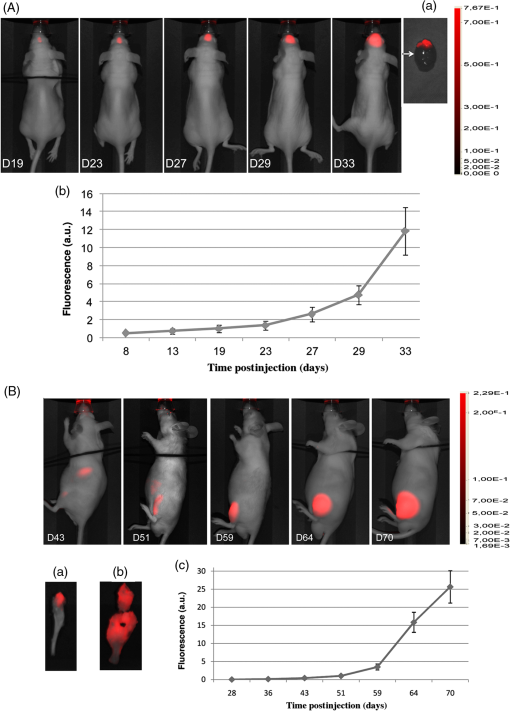 Intratibial growth of U2OS cells is shown in Fig. 2(B). Note that the presence of additional NIR fluorescent spots on the abdomen of mice between days 43 and 51, which could be due to traces of alfalfa in this animal food;15 a problem that we subsequently solved. To further analyze the intrabone tumor progression of iRFP-expressing U2OS cells, two animals from a parallel series and corresponding to those of days 43 and 70 were sacrificed and their tibias retrieved. As shown in Fig. 2(Ba), the tumor that initially localized to the head of the bone at day 43, progressively invaded the rest of the organ a week later (at day 51), eventually leading to fracture of the bone and spreading over surrounding soft tissue at day 70 [Fig. 2(Bb)]. Therefore, the iRFP-emitted fluorescence crosses the bone barrier, thus allowing us to reliably estimate tumor burden without sacrificing the animal [see also Fig. 2(Bc)]. 3.2.Visualization of MetastasisThese data being acquired, we then asked whether iRFP-based imaging could be applied to the study of metastatic spread. For this purpose, we used melanoma cells IGR37, originating from a metastatic spread of the primary human melanoma cells IGR39.21 As shown in Fig. 3(A), injection of iRFP-expressing IGR37 cells in the tail vein resulted in a tumor deposit in the liver [see m1 series and after dissection of the animal Fig. 3(Aa)], consistent with a previous work showing the liver to be a potential niche for melanoma metastasis.22 Surprisingly, injection of the same cells to two other mice led to tumors that appeared, not in the liver, but in the space between omoplates [Fig. 3(B), see m2 series]. Dissection of the animal revealed the presence of well-defined tumors that strongly adhered to the cervical vertebrae (data not shown) but which were positive for melanoma immunological markers such as S100, HBM-45, and Melan-A [Fig. 3(C)]. These data show that whole-body NIR imaging with iRFP uncovers different metastatic targets for melanomas. Fig. 3Melanoma-derived metastatic lesions were formed after tail vein injection of IGR37 cells to three series of mice. One series developed a liver metastasis (A, m1 series) and the others developed a dorsal tumor (B, only the m2 series is shown). The color bar indicates the total fluorescence radiant efficiency. (C) The tumor from the mouse m2 was retrieved and subjected to hematoxylin/eosin (HE) and immunohistochemistry using S100, HBM-45 and Melan-A markers (magnification ). 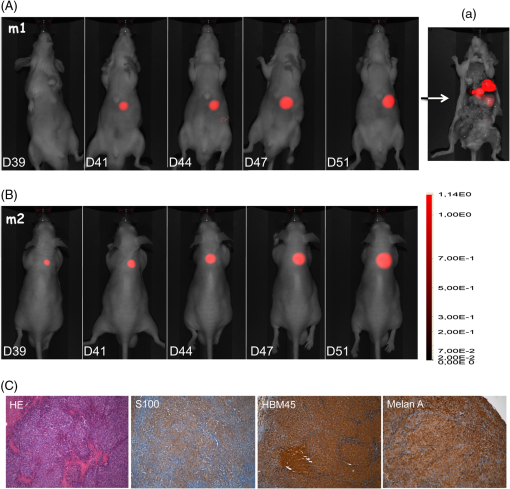 4.DiscussionConsiderable progress has been made in recent years to cure cancer. Besides surgery, drug-based therapies are more and more efficient and already allow us to save lives or at least to meaningfully extend patient survival.23 Yet, it is still highly desirable to afford new molecules with an improved bioactivity and decreased toxicity. In order to meet this need, preclinical assays involving small animals and genetic tools for whole-body imaging and tumor-emitted signal quantification would significantly improve anticancer therapeutic strategies. Several previous studies have used fluorescent proteins to track tumor lesions in mice,24 including those related to EGFP, DsRed, or their variants.6 However, the wavelengths of these tracers are in a range of low signal-to-noise background and low coefficient of penetration in the animal body,7 particularly within the green part of the spectrum.8,25 Autofluorescence, scattering, and absorbance are crucial parameters, particularly when imaging organs that lie millimeters deep under the skin layers.7 With the NIR light, however, these parameters become less important.25 Another problem encountered with many previous studies is the use of cells that have been transfected and selected with drugs such as neomycin.4,24 In addition to the possible clone-to-clone variability, the main challenge with such approaches is whether the transgene expression would be kept stable throughout the study. In fact, once the drug-selected cells are deposited in the animal body, they must grow in a drug-free environment and therefore, might lose the gene of interest due to the lack of selection pressure. To overcome these difficulties, we herein used a lentivirus-mediated delivery system to permanently express the NIR fluorescent protein iRFP in cells. Another attractive characteristic of this system is that this human immunodeficiency virus type 1 (HIV-1)-based lentiviral vector can infect virtually all kinds of cells including the nondividing ones.26 Using this genetically encoded probe and an NIR fluorescence imaging device, we are herein reporting the usefulness of this system for the detection of deeply buried tumor lesions or those hidden behind the bone barrier. Most importantly, metastatic deposits can be easily detected and quantified [see Figs. 3(A) and 3(B)], thereby avoiding cumbersome and tedious dissection steps. Although the detection of tumors by iRFP in deep organs has been demonstrated by Filonov et al.11 and confirmed in the present study, there is no data available using such a tool for visualization of intraosseous-implanted tumors. Intrabone emitted signals are bright enough to be detected and quantified without any contrast agent or chromophore previously administered to the animal [see Figs. 1(c) and 2(A) and 2(B)]. It is noteworthy that attempts to detect glioblastoma cell growth in the brain using IFP1.4 and biliverdin supplementation have been unsuccessful (data not shown). In conclusion, this iRFP- and lentiviral-based system present many other characteristics that combine the permanent and strong expression of iRFP with the low scattering and absorption by biological liquids and bones. Altogether, these features would allow this HIV-based iRFP-expression system to validly support the emerging therapeutic strategies in cancer treatment. AcknowledgmentsThe authors thank S. Salas for critical discussion of the paper, P. Weber for assistance with the fluorescence, and Y. Dauchy for help with the cloning experiments. ReferencesB. HannA. Balmain,
“Building ‘validated’ mouse models of human cancer,”
Curr. Opin. Cell Biol., 13
(6), 778
–784
(2001). http://dx.doi.org/10.1016/S0955-0674(00)00283-0 COCBE3 0955-0674 Google Scholar
K. Garber,
“Realistic rodents? Debate grows over new mouse models of cancer,”
J. Natl. Cancer Inst., 98
(17), 1176
–1178
(2006). http://dx.doi.org/10.1093/jnci/djj381 JJIND8 0198-0157 Google Scholar
C. P. Klerket al.,
“Validity of bioluminescence measurements for noninvasive in vivo imaging of tumor load in small animals,”
Biotechniques, 4330
(S1), 7
–13
(2007). http://dx.doi.org/10.2144/000112515 BTNQDO 0736-6205 Google Scholar
M. Bouvetet al.,
“Real-time optical imaging of primary tumor growth and multiple metastatic events in a pancreatic cancer orthotopic model,”
Cancer Res., 62
(5), 1534
–1540
(2002). CNREA8 0008-5472 Google Scholar
N. C. Shaneret al.,
“Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein,”
Nat. Biotechnol., 22
(12), 1567
–1572
(2004). http://dx.doi.org/10.1038/nbt1037 NABIF9 1087-0156 Google Scholar
R. M. Hoffman,
“The multiple uses of fluorescent proteins to visualize cancer in vivo,”
Nat. Rev. Cancer, 5
(10), 796
–806
(2005). http://dx.doi.org/10.1038/nrc1717 NRCAC4 1474-175X Google Scholar
J. LecoqM. J. Schnitzer,
“An infrared fluorescent protein for deeper imaging,”
Nat. Biotechnol., 29
(8), 715
–716
(2011). http://dx.doi.org/10.1038/nbt.1941 NABIF9 1087-0156 Google Scholar
Y. T. Limet al.,
“Selection of quantum dot wavelengths for biomedical assays and imaging,”
Mol. Imaging, 2
(1), 50
–64
(2003). http://dx.doi.org/10.1162/153535003765276282 MIOMBP 1535-3508 Google Scholar
D. Shcherboet al.,
“Bright far-red fluorescent protein for whole-body imaging,”
Nat. Methods, 4
(9), 741
–746
(2007). http://dx.doi.org/10.1038/nmeth1083 1548-7091 Google Scholar
X. Shuet al.,
“Mammalian expression of infrared fluorescent proteins engineered from a bacterial phytochrome,”
Science, 324
(5928), 804
–807
(2009). http://dx.doi.org/10.1126/science.1168683 SCIEAS 0036-8075 Google Scholar
G. S. Filonovet al.,
“Bright and stable near-infrared fluorescent protein for in vivo imaging,”
Nat. Biotechnol., 29
(8), 757
–761
(2011). http://dx.doi.org/10.1038/nbt.1918 NABIF9 1087-0156 Google Scholar
K. Konig,
“Multiphoton microscopy in life sciences,”
J. Microsc., 200
(2), 83
–104
(2000). http://dx.doi.org/10.1046/j.1365-2818.2000.00738.x JMICAR 0022-2720 Google Scholar
C. MancusoE. Barone,
“The heme oxygenase/biliverdin reductase pathway in drug research and development,”
Curr. Drug Metab., 10
(6), 579
–594
(2009). http://dx.doi.org/10.2174/138920009789375405 CDMUBU 1389-2002 Google Scholar
S. Mathieuet al.,
“Transgene expression of alpha(1,2)-fucosyltransferase-I (FUT1) in tumor cells selectively inhibits sialyl-Lewis x expression and binding to E-selectin without affecting synthesis of sialyl-Lewis a or binding to P-selectin,”
Am. J. Pathol., 164
(2), 371
–383
(2004). http://dx.doi.org/10.1016/S0002-9440(10)63127-6 AJPAA4 0002-9440 Google Scholar
Y. Inoueet al.,
“Diet and abdominal autofluorescence detected by in vivo fluorescence imaging of living mice,”
Mol. Imaging, 7
(1), 21
–27
(2008). MIOMBP 1535-3508 Google Scholar
C. Berengueret al.,
“Adrenomedullin, an autocrine/paracrine factor induced by androgen withdrawal, stimulates ‘neuroendocrine phenotype’ in LNCaP prostate tumor cells,”
Oncogene, 27
(4), 506
–518
(2008). http://dx.doi.org/10.1038/sj.onc.1210656 ONCNES 0950-9232 Google Scholar
A. Tchoghandjianet al.,
“A2B5 cells from human glioblastoma have cancer stem cell properties,”
Brain Pathol., 20
(1), 211
–221
(2010). http://dx.doi.org/10.1111/bpa.2010.20.issue-1 BRPAE7 1015-6305 Google Scholar
B. C. GleasonA. F. Nascimento,
“HMB-45 and Melan-A are useful in the differential diagnosis between granular cell tumor and malignant melanoma,”
Am. J. Dermatopathol., 29
(1), 22
–27
(2007). http://dx.doi.org/10.1097/01.dad.0000249888.41884.6c AJODDB 0193-1091 Google Scholar
H. Nakayamaet al.,
“Evaluation by bone scintigraphy of osteogenic activity of commercial bioceramics (porous beta-TCP and HAp particles) subcutaneously implanted in rats,”
J. Biomater. Appl., 24
(8), 751
–768
(2010). http://dx.doi.org/10.1177/0885328209341845 JBAPEL 0885-3282 Google Scholar
C. DaiE. C. Holland,
“Glioma models,”
Biochim. Biophys. Acta, 1551
(1), M19
–M27
(2001). BBACAQ 0006-3002 Google Scholar
R. Taghizadehet al.,
“CXCR6, a newly defined biomarker of tissue-specific stem cell asymmetric self-renewal, identifies more aggressive human melanoma cancer stem cells,”
PLoS One, 5
(12), e15183
(2010). http://dx.doi.org/10.1371/journal.pone.0015183 1932-6203 Google Scholar
V. Gray-SchopferC. WellbrockR. Marais,
“Melanoma biology and new targeted therapy,”
Nature, 445
(7130), 851
–857
(2007). http://dx.doi.org/10.1038/nature05661 NATUAS 0028-0836 Google Scholar
C. W. HongQ. Zeng,
“Awaiting a new era of cancer immunotherapy,”
Cancer Res., 72
(15), 3715
–3719
(2012). http://dx.doi.org/10.1158/0008-5472.CAN-12-0063 CNREA8 0008-5472 Google Scholar
A. Miyawaki,
“Fluorescence imaging in the last two decades,”
J. Electron Microsc., 62
(1), 63
–68
(2013). http://dx.doi.org/10.1093/jmicro/dfs130 JELJA7 0022-0744 Google Scholar
R. WeisslederV. Ntziachristos,
“Shedding light onto live molecular targets,”
Nat. Med., 9
(1), 123
–128
(2003). http://dx.doi.org/10.1038/nm0103-123 1078-8956 Google Scholar
W. QasimC. A. VinkA. J. Thrasher,
“Hybrid lentiviral vectors,”
Mol. Ther., 18
(7), 1263
–1267
(2010). http://dx.doi.org/10.1038/mt.2010.76 MTOHCK 1525-0016 Google Scholar
|

