|
|
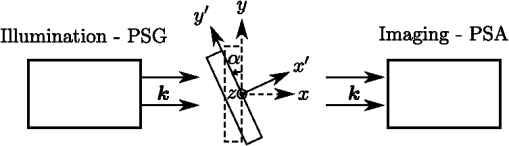
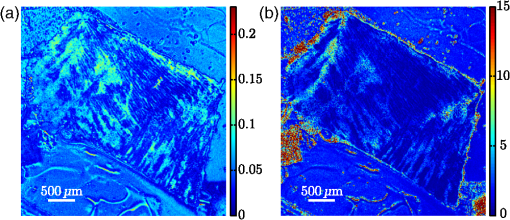
1.IntroductionBiomedical research is experiencing a revolution due to the development of instruments for spectral and spatial characterization. In addition, pulsed lasers are readily available for nonlinear optical applications. Recently, polarization sensitive techniques, known from the thin film community, have been developed for biomedical applications. In the thin film community, the polarization sensitive technique spectroscopic ellipsometry has been successfully used to characterize material properties of many kinds for several decades.1 As a consequence, ellipsometry now plays an important role in the semiconductor industry. More recently, spectroscopic Mueller matrix ellipsometry has been employed to characterize anisotropic nanostructured materials and plasmonic structures.2–6 Due to the turbidity of biological tissue,7 the modeling is more complicated than for more uniform samples such as thin films. In addition, partial depolarization of light in the sample requires acquisition of the full polarization properties, i.e., the Mueller matrix, and not only the ellipsometric parameters (amplitude) and (phase difference). Nevertheless, Mueller matrix ellipsometry has been shown to be a promising technique for the characterization of biological tissue.8–13 Recently, several setups have been developed to acquire Mueller matrix images (MMIs) of biological samples. In particular, Pierangelo et al.13 demonstrated the use of a reflection imaging Mueller matrix ellipsometer to characterize and diagnose colorectal cancer. Furthermore, several broadband Mueller matrix designs for imaging have been proposed14 and some have been implemented.15 By carefully choosing the probing wavelength, it is possible to make the technique sensitive to a certain depth range in the tissue.7 Hence, if MMI is combined with hyperspectral imaging it would be possible to study the depth dependent effects. The MMI technique is, in principle, nondestructive and can, with further development, achieve a submicrometer resolution, as well as being sensitive to structural features smaller than the diffraction limit. There are a range of polarization effects which alters a Mueller matrix, the common being depolarization, diattenuation, birefringence, and optical activity. If a material possesses several of these effects, it is not always easily seen from the Mueller matrix which of the effects are responsible for a certain feature in the measured data. One way of simplifying the analysis is to decompose the measured Mueller matrix. The typical way of decomposing the Mueller matrix has been the forward polar decomposition,16,17 but recently, Ossikovski et al.18 pointed out that polar decomposition assumes the polarization effects to be multiplicative, which is rarely the case for biological media. They suggested that the differential decomposition is a more suitable approach for decomposing such simultaneous effects. The differential decomposition was originally proposed by Azzam19 and later extended to include depolarizing media by Ossikovski20 and has recently been applied on biological tissue by Kumar et al.12 In the latter work, it was shown how to calculate physical properties from the decomposed matrices. Using decomposed MMIs of biological tissue containing collagen fibers, it has been shown possible to extract the in-plane direction of the fibers from their induced birefringence.11 We, here, generalize this idea further in order to find the 3-D direction of collagen fibers in biological tissue. This generalized method is tested and validated by comparing the three-dimensional (3-D) direction image of tendon derived from MMIs, with second-harmonic (SHG) images of the same sample. SHG imaging is well known to be a good diffraction limited technique for imaging collagen fibers.21,22 2.Materials and Methods2.1.Sample PreparationTendon tissue was taken from medial femoral condyle of a chicken’s knee, bought fresh from the supermarket. A small section of the tissue was embedded in a mounting medium for cryo-sectioning (O.C.T., Sakura, Alphen aan den Rijn, The Netherlands). Rapid freezing of the O.C.T. embedded tissue was completed using liquid nitrogen. This frozen section was stored in a freezer () until cut with a cryostat into 50-μm thin tissue sections. The cutting plane was parallel to the collagen fibers. The thin sections were placed on standard microscope glass slides and stored in a freezer (). Before being measured, the tissue samples were brought back to room temperature and covered with a standard cover slip. Edges of the cover slip were sealed with Vaseline to avoid dehydration. Between measurements, the slides were stored at 4°C. 2.2.SHG ImagingSHG images were collected on a Zeiss LSM 510 meta microscope using a Coherent Mira 900 for excitation at 790 nm. Imaging was done with a 1.2 NA objective. A custom built polarization setup, which compensates for any birefringence in the optical path, was used to ensure circular polarization. The average power at the focal plane was approximately 8. 2.3.MMI SetupAfter the samples were imaged with SHG, they were measured in a custom built MMI ellipsometer, shown in Fig. 1. The system uses ferroelectric liquid crystals for both the polarization state generator and analyzer. Details of the system can be found in Aas et al.23 An improvement was made to the illumination, by replacing both the laser and rotating diffuser, with a 940 nm collimated light emitting diode (LED). In addition, a motorized rotation stage for the sample was introduced in order to image the sample at the different projections needed to extract the 3-D direction, as described below. Fig. 1The Mueller matrix imaging (MMI) setup consisting of the polarization state generator (PSG) and the polarization state analyzer (PSA). P1 and P2 are the linear polarizers, are the retarders and are the ferroelectric liquid crystals.  The system was calibrated using the eigenvalue calibration method24 on four reference samples (air, two polarizers, and a retarder), ensuring the correct measurement of the Mueller matrix. By comparing the measurement of air to the identity matrix, an error estimate was made resulting in a measure for the accuracy of the system. 2.4.Decomposition of the Mueller MatrixAs the differential decomposition method is able to decompose simultaneous polarization effects, it was chosen for the decomposition of the measured Mueller matrices presented here. Due to the measurement noise, some of the measured Mueller matrices are slightly unphysical, which was compensated for by using the filtering described by Cloude,25 prior to the decomposition. The differential decomposition results in two matrices, and ,20 where contains the elements used to calculate the retardance and the diattenuation. The uncertainties (standard deviation) in the retardance and the diattenuation can be calculated by using the same matrix elements from as those originally used from . Furthermore, the depolarization can be calculated from . In this study, the relevant properties are the linear retardance , the angle of orientation of the linear retardance , and the depolarization (It is here noted that the depolarization [, Eq. (19) in the paper by Kumar et al.12] has the wrong signs of the exponential, it should be as discussed in a private correspondence with Ossikovski, one of the authors of the original paper.).12 2.5.Directional CalculationIn every day life, we are familiar with determining the orientation of an object just by looking at it. Because our eyes are separated by a distance, they see two different projections of an object which enables deduction of the object’s orientation. The MMI setup can only image one projection, but it is possible to rotate the sample in two different sample rotations , see Figs. 2 and 3, and then use the two resulting images to calculate the direction of the imaged birefringent structure, i.e., the collagen framework. Fig. 3The coordinate systems used for the calculation of the directions. The laboratory frame is identical to the sample frame of reference at . The -axis points along the illumination direction toward the camera and is vertical. As the rotation is around , for any . The lower row of figures shows the projection of a three-dimensional (3-D) fiber onto the imaging plane, for different rotations of the sample (). 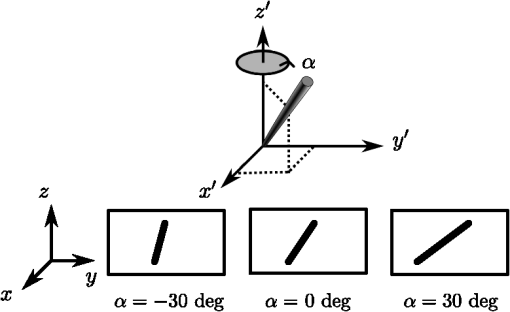 In order to derive the direction of anisotropic structures, it is convenient to start with the Euler transformations.26 They require the definition of two coordinate systems, the laboratory frame of reference and the sample frame of reference. Let describes a vector in the laboratory frame of reference. The frame of reference is defined in such a way that the -axis points along the direction of illumination, the -axis is horizontal, and the -axis is vertical, see Fig. 2. Let be a vector in the sample frame of reference. The sample frame of reference coincides with the laboratory frame for a rotation , see Fig. 2. The sample is only rotated around the axis, see Figs. 2 and 3, resulting in the following Euler rotation matrix By using this transformation, it is possible to transform from the sample frame of reference to the laboratory frame of reference , by . Applying this transformation gives the following relations The goal is to determine by measuring the Mueller matrix at two different sample rotations, and , by looking at the projections into the laboratory frame of reference (the measured image), resulting in the measured and . By choosing , (in our setup due to the direction of rotation) and solving Eq. (2), the components of results in As the MMI measurement only yields the direction of the slow axis, , and not the projected length (the length in the plane) of the fiber, it is not possible to find the absolute value (length) of the vector. In order to resolve this, we define the projected length of the fiber as It is now possible to define the coordinates with respect to the measured angles Using we find that which, when inserted into Eq. (3), givesThese equations depend on the absolute length of , however, as we are only interested in the direction of the fiber, we can set . This solution is limited to only include positive , which is not a problem since all solutions with negative can be represented by the opposite vector located in the positive space. In addition, it will not be possible to get a solution purely in the plane (). A real measurement contains some noise, both from the measurement itself and from numerical noise, ensuring that the angle is never exactly zero. In addition, the rotation around means that it is not possible to see the difference between different vectors in the plane if . The final equations are then When presenting the results derived with Eq. (4), the vector is normalized to its length . Another consideration to make is that the angle of incidence is not the same as the rotation angle of the sample, due to the difference in refractive index. This difference yields a correction for which is , where is the angle of rotation (incidence angle on the glass) and is the refractive index of tissue, assumed here to be 1.4.7 According to Snell’s law, the two glass slides sandwiching the sample do not affect this correction. 2.6.Directional ImagingBy changing the angle of incidence on the sample, i.e., rotating the sample, and using the slow axis direction found from the decomposition, it is possible to calculate and hence make an image of the 3-D direction of the fibers as described in Sec. 2.5. The calculation is done using incidence angles of . The resulting images are resampled, using , such that the stretching due to rotation is counteracted and the pixels are square. is used because the image is seen on the surface of the glass. 3.Results and DiscussionTwo of the recorded Mueller matrices, after cropping and resampling, are shown in Fig. 4. The first element of the Mueller matrix has the intensity image overlaid. These Mueller matrices are the basis for the decomposition and calculating the directions as explained in Sec. 3.2. Fig. 4The normalized, cropped, and resampled MMI for 30 deg (a) and (b), with the intensity image (gray scale image) overlaid the first element in the Mueller matrix as it is 1 in every pixel.  3.1.Depolarization and Linear RetardanceLinear retardance describes how much the polarization in one direction is phase shifted with respect to its orthogonal polarization. Figure 5 shows the linear retardance together with its uncertainty for the tendon sample. The uncertainty is calculated from and represents the standard deviations of the linear retardance. As previously reported,11 a high concentration of collagen results in a high retardance, mainly due to the form birefringence induced by the shape of the fibers in the effective medium. The mentioned study looked at the collagen fibers present in cartilage, which is less ordered than the collagen fibers in tendons. Being less ordered means that the structure should be modeled as a multilayered effective medium (requiring a layered decomposition of the Mueller matrix) with one orientation for every layer. The higher degree of order in the tendon makes the presented results more clear, and thus avoids the need to consider the sample as a multilayered system. Fig. 5Linear retardance (a) and the uncertainty (b) for the tendon sample at normal incidence. Both images are in degrees. 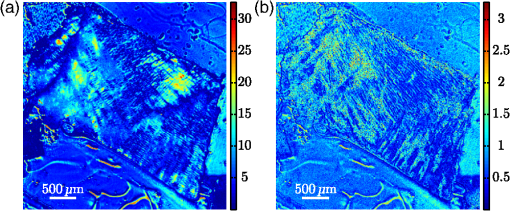 Figure 5(b) shows the uncertainty image (standard deviation) of the linear retardance, which is seen to be above the random noise level in some areas. As the uncertainty in this figure is a result of depolarization effects, it is useful for the analysis of the sample. Previous evaluation of the uncertainty has been limited to the knowledge of the measurement error found from the calibration. Specifically, the error is found from the measurement of air which, here, results in a measurement error on the order of a few percents. The uncertainty image supplies additional information about the sample. It gives a measure for the uncertainty induced by randomness in the sample. This randomness can, for instance, be the fiber orientations and/or sizes, as well as depolarizing effects like multiple scattering and integration over several polarization states in one pixel. This is confirmed by comparing the uncertainty measurement to the depolarization shown in Fig. 6(a). 3.2.Directional ImagingFrom the retardance found in the decomposition, it is not only possible to calculate the linear retardance, but also the direction of the fast axis of the birefringence. This latter property can, together with the correct effective medium model, be used to find the direction of the collagen fibers as explained in Sec. 2. The directional image and the SHG image for the tendon sample are shown in Fig. 7. As the 3-D directional image in Fig. 7(a) shows, the fibers are mostly in the plane, as expected due to the direction of the cryostat cut. The calculated in-plane directions [black lines in Figs. 7(a) and 7(b)] correspond very well with the apparent directions in the SHG images [Figs. 7(c) and 7(e)]. There are some areas that are clearly out-of-plane [red areas in the 3-D directional image, Fig. 7(a)], which in the SHG images are either dark or show some weak structure. The latter can be explained by the SHG signal generation. When considering SHG signal generation, fiber orientations are important, as a fiber in-plane has a much larger cross section for generating SHG, compared to one out-of-plane. This means that the darker parts of the image in Fig. 7(c) probably are due to an out-of-plane orientation of the fibers, in accordance with the 3-D directional image. Figure 7(d) shows an overlay of the out-of-plane part of Fig. 7(a) and the SHG image in Fig. 7(c). Fig. 73-D directional image (a) with a zoomed view (b), an SHG image (c) with a zoomed in view (e) and an overlay of the SHG and the out-of-plane direction of the tendon (d). The colorwheel beside the directional image shows the out-of-plane direction in degrees, with 0 deg being in the image plane and normal to the image plane. is the direction toward the reader (positive ). The overlay (d) (area outside of the sample masked for clarity) shows the SHG and out-of-plane directions overlain. The yellow color represent the negative out-of-plane direction and the cyan the positive direction. No color represents an in-plane direction and increasing color more out-of-plane. 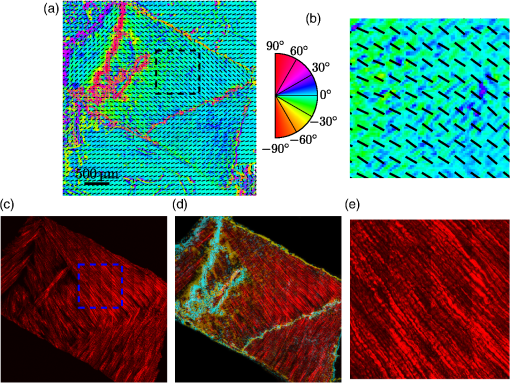 The lower right of the sample shows an offset between the directional image and the SHG. There could be several reasons for this offset, one being that the size of the fibers is around the upper limit of the validity of the effective medium model, which is (here ). In this study, it was necessary that the fibers were sufficiently large to visualize with SHG, which is limited by the diffraction limit. It is expected that the MMI results will be superior for smaller fibers. Another contributing factor to the error in the direction is the depolarization induced uncertainties in the direction of the fast axis. The calculation of this uncertainty was not given by Kumar et al.12 as it cannot be calculated from in the same way as the uncertainty in the other parameters. As a solution to this, we have calculated the uncertainty, shown in Fig. 6(b), using the standard method for propagation of errors, here, the uncertainties found in . If the propagation is made at an incidence angle of 0 deg, the resulting uncertainty in the angle of the fast axis is given by The reasons for choosing to study the uncertainty at an incidence angle of 0 deg, instead of , are due to the approximations used to derive the propagation uncertainty. Propagation of uncertainties uses only the first derivatives and not the higher orders, which for Eq. (4) might be an incorrect approximation. The uncertainty at 0 and is expected to be similar, although the exact relation is hard to predict. A prediction is hard because there could be a reduction in the uncertainty as a result of the increase in the number of data points (two data sets), although the increase in apparent thickness could on the other hand increase the uncertainty. As a consequence, the 0 deg uncertainty will be used as a good indication for the uncertainty. By studying Fig. 6(b), it is possible to see that the uncertainty in the directions is larger in the lower right part of the sample compared to the upper, which is in accordance with the comparison between the 3-D image and the SHG image, shown in Fig. 7. In addition, by comparing with Fig. 6(a), it is seen that they correlate well showing that the uncertainty is a result of the depolarization. As the depolarization is generated by, among others, the randomness of the fiber orientations, it offers a better understanding of the sample structure. The out-of-plane directions in Fig. 7 are seen to correspond well with the same areas in the SHG image. Additionally, by studying Fig. 7(a) and the zoomed in views in Figs. 7(b) and 7(e), it is possible to see that the oscillating structure (the oscillation between green and blue) along the fibers is visible both in the SHG image and the out-of-plane direction image. Both of these results confirm that the 3-D directional imaging finds the correct directions. It is worth noting that the method described above makes it possible to find the fiber directions using MMI, even though the resolution in our system is much poorer than for SHG. Another notable property is that this method only requires the sample to be birefringent, as the calculation is based on the direction of linear retardance. Linear retardance is present in a range of different biologicals,9 colloidals,27 and solid matter systems,23 extending the possible applications for the method. 4.ConclusionWe have presented a method for determining the 3-D direction of collagen fibers embedded in biological tissue from MMIs. The resulting images are shown to be in good agreement for a tendon sample when compared to SHG images. In particular, it is possible to see oscillating structures in the collagen orientation, as well as the out-of-plane directions of the fibers. The possibility to see effects from collagen fibers below the diffraction limit could be an important input to the understanding of how the collagen framework looks. Additionally, the use of the differential decomposition instead of the, until now, most common polar decomposition has provided a good insight into the uncertainties in the calculation of the physical properties. AcknowledgmentsThe authors would like to thank Razvigor Ossikovski for the correspondence on the implementation of the differential decomposition method. ReferencesR. M. A. AzzamN. M. Bashara, Ellipsometry and Polarized Light, North-Holland PublishingAmsterdam,1977). Google Scholar
D. Schmidtet al.,
“Optical, structural, and magnetic properties of cobalt nanostructure thin films,”
J. Appl. Phys., 105
(11), 113508
(2009). http://dx.doi.org/10.1063/1.3138809 JAPIAU 0021-8979 Google Scholar
I. Nerbøet al.,
“Real-time in situ Mueller matrix ellipsometry of GaSb nanopillars: observation of anisotropic local alignment,”
Opt. Express, 19
(13), 571
–575
(2011). http://dx.doi.org/10.1364/OE.19.012551 OPEXFF 1094-4087 Google Scholar
L. Aaset al.,
“Determination of small tilt angles of short GaSb nanopillars using UV-visible Mueller matrix ellipsometry,”
Thin Solid Films, 541 97
–101
(2013). http://dx.doi.org/10.1016/j.tsf.2012.10.136 THSFAP 0040-6090 Google Scholar
T. OatesH. WormeesterH. Arwin,
“Characterization of plasmonic effects in thin films and metamaterials using spectroscopic ellipsometry,”
Prog. Surf. Sci., 86
(11–12), 328
–376
(2011). http://dx.doi.org/10.1016/j.progsurf.2011.08.004 PSSFBP 0079-6816 Google Scholar
L. M. S. Aaset al.,
“Optical properties of biaxial nanopatterned gold plasmonic nanowired gridpolarizer,”
Opt. Express, 21
(25), 30918
–30931
(2013). http://dx.doi.org/10.1364/OE.21.030918 OPEXFF 1094-4087 Google Scholar
V. V. Tuchin, Tissue Optics: Light Scattering Methods and Instruments for Medical Diagnosis, 2nd ed.SPIE Press, Bellingham, WA
(2007). Google Scholar
J. Chunget al.,
“Use of polar decomposition for the diagnosis of oral precancer,”
Appl. Opt., 46
(15), 3038
–3045
(2007). http://dx.doi.org/10.1364/AO.46.003038 APOPAI 0003-6935 Google Scholar
N. Ghoshet al.,
“Mueller matrix decomposition for polarized light assessment of biological tissues,”
J. Biophotonics, 2
(3), 145
–156
(2009). http://dx.doi.org/10.1002/jbio.v2:3 JBOIBX 1864-063X Google Scholar
M.-R. Antonelliet al.,
“Mueller matrix imaging of human colon tissue for cancer diagnostics: how Monte Carlo modeling can help in the interpretation of experimental data,”
Opt. Express, 18
(10), 10200
(2010). http://dx.doi.org/10.1364/OE.18.010200 OPEXFF 1094-4087 Google Scholar
P. G. Ellingsenet al.,
“Quantitative characterization of articular cartilage using Mueller matrix imaging and multiphoton microscopy,”
J. Biomed. Opt., 16
(11), 116002
(2011). http://dx.doi.org/10.1117/1.3643721 JBOPFO 1083-3668 Google Scholar
S. Kumaret al.,
“Comparative study of differential matrix and extended polar decomposition formalisms for polarimetric characterization of complex tissue-like turbid media,”
J. Biomed. Opt., 17
(10), 105006
(2012). http://dx.doi.org/10.1117/1.JBO.17.10.105006 JBOPFO 1083-3668 Google Scholar
A. Pierangeloet al.,
“Multispectral Mueller polarimetric imaging detecting residual cancer and cancer regression after neoadjuvant treatment for colorectal carcinomas,”
J. Biomed. Opt., 18
(4), 046014
(2013). http://dx.doi.org/10.1117/1.JBO.18.4.046014 JBOPFO 1083-3668 Google Scholar
P. A. Letneset al.,
“Fast and optimal broad-band Stokes/Mueller polarimeter design by the use of a genetic algorithm,”
Opt. Express, 18
(22), 23095
–23103
(2010). http://dx.doi.org/10.1364/OE.18.023095 OPEXFF 1094-4087 Google Scholar
L. M. S. Aaset al.,
“Overdetermined broadband spectroscopic Mueller matrix polarimeter designed by genetic algorithms,”
Opt. Express, 21
(7), 8753
–8762
(2013). http://dx.doi.org/10.1364/OE.21.008753 OPEXFF 1094-4087 Google Scholar
S.-Y. LuR. A. Chipman,
“Interpretation of Mueller matrices based on polar decomposition,”
J. Opt. Soc. Am. A, 13
(5), 1106
–1113
(1996). http://dx.doi.org/10.1364/JOSAA.13.001106 JOAOD6 0740-3232 Google Scholar
S. Manhaset al.,
“Mueller matrix approach for determination of optical rotation in chiral turbid media in backscattering geometry,”
Opt. Express, 14
(1), 190
–202
(2006). http://dx.doi.org/10.1364/OPEX.14.000190 OPEXFF 1094-4087 Google Scholar
R. OssikovskiA. De MartinoS. Guyot,
“Forward and reverse product decompositions of depolarizing Mueller matrices,”
Opt. Lett., 32
(6), 689691
(2007). http://dx.doi.org/10.1364/OL.32.000689 OPLEDP 0146-9592 Google Scholar
R. Azzam,
“Propagation of partially polarized light through anisotropic media with or without depolarization: a differential matrix calculus,”
J. Opt. Soc. Am., 68
(12), 1756
–1767
(1978). http://dx.doi.org/10.1364/JOSA.68.001756 JOSAAH 0030-3941 Google Scholar
R. Ossikovski,
“Differential matrix formalism for depolarizing anisotropic media,”
Opt. Lett., 36
(12), 2330
–2332
(2011). http://dx.doi.org/10.1364/OL.36.002330 OPLEDP 0146-9592 Google Scholar
P. Stolleret al.,
“Quantitative second-harmonic generation microscopy in collagen,”
Appl. Opt., 42
(25), 5209
–5219
(2003). http://dx.doi.org/10.1364/AO.42.005209 APOPAI 0003-6935 Google Scholar
G. Coxet al.,
“3-Dimensional imaging of collagen using second harmonic generation,”
J. Struct. Biol., 141
(1), 53
–62
(2003). http://dx.doi.org/10.1016/S1047-8477(02)00576-2 JSBIEM 1047-8477 Google Scholar
L. M. S. AasP. G. EllingsenM. Kildemo,
“Near infra-red Mueller matrix imaging system and application to retardance imaging of strain,”
Thin Solid Films, 519
(9), 2737
–2741
(2011). http://dx.doi.org/10.1016/j.tsf.2010.12.093 THSFAP 0040-6090 Google Scholar
E. CompainS. PoirierB. Drevillon,
“General and self-consistent method for the calibration of polarization modulators, polarimeters, and Mueller-matrix ellipsometers,”
Appl. Opt., 38
(16), 3490
–3502
(1999). http://dx.doi.org/10.1364/AO.38.003490 APOPAI 0003-6935 Google Scholar
S. R. Cloude,
“Conditions for the physical realisability of matrix operations in polarimetry,”
Proc. SPIE, 1166 177
–185
(1990). http://dx.doi.org/10.1117/12.962889 PSISDG 0277-786X Google Scholar
H. GoldsteinC. PooleJ. Safko, Classical Mechanics, 3rd ed.Addison Wesley, San Fransisco, CA
(2002). Google Scholar
M. Kildemoet al.,
“Mueller matrix imaging of nematic textures in colloidal dispersions of Na-fluorohectorite synthetic clay,”
Proc. SPIE, 8082 808221
(2011). http://dx.doi.org/10.1117/12.889569 PSISDG 0277-786X Google Scholar
|