|
|
1.IntroductionFunctional brain imaging techniques that have ill-posed inverse problems often rely on structural brain images to provide anatomical priors to constrain tomographic reconstructions.1,2 Creating subject-specific anatomical head models suitable for functional neuroimaging modalities generally requires segmentation of structural images and assignment of tissue properties. The physical properties of the tissues are dependent on the imaging modality used, or the head models can include more than one type of physical property and therefore be suitable for multimodal neuroimaging source reconstructions. Usually, structural magnetic resonance imaging (MRI) scans are used to form the head models in noninvasive brain imaging because the images have generally good tissue contrast and do not require ionizing radiation. Different MRI pulse sequences generate different patterns of tissue contrast. For functional MRI (fMRI), high-resolution T1 images with good gray matter/white matter contrast such as those generated by the magnetization-prepared rapid gradient-echo (MPRAGE) pulse sequence3 are often used to generate better anatomical visualization than could be achieved with the low-resolution functional images. Source reconstruction for diffuse optical tomography (DOT) and electroencephalography (EEG) is particularly reliant on appropriate head models, due to the interaction of brain activity and the intermediating head tissues. DOT forward and inverse models are used to reconstruct changes in oxy- and deoxyhemoglobin concentration in a three-dimensional (3-D) volume and can localize activity to the brain.4 This method for brain activity reconstruction requires segmentation of the tissue types in the whole head, not just the brain, as the optical probes are placed on the scalp so photons traverse the head tissue surrounding the brain. It is especially important to have the correct cerebrospinal fluid (CSF) layer thickness because CSF has much lower optical absorbance and higher electrical conductivity than other tissues. EEG forward models also require modeling all tissues in the head. Scalp and skull segmentation are important for creating accurate models of the head with the correct conductivity. Physical property values can differ by an order of magnitude between extracerebral head tissue types, as shown in Table 1, underscoring the necessity of accurate tissue segmentation for realistic head models. Table 1Extracerebral tissue physical properties. Optical properties are from Ref. 5 and conductivity is from Ref. 6.
Segmenting the skull from MRI is important for both DOT and EEG forward modeling. Unfortunately, this task is complicated by the fact that the standard high-resolution T1 image collected for fMRI localization is optimized for gray matter/white matter tissue contrast, not skull segmentation. Other types of MRI weighted images such as T2-weighted and proton density images are often used to segment these other tissue types.6,7 Methods for creating high-quality head meshes from MRI have been described,8 but require at least one T1 and one T2 scan, and optimally require four MRI scans. However, from a practical perspective it would be preferable to have an accurate head segmentation using a single MRI. T1 images are commonly available and would be the most cost-effective and fastest pulse sequence use to create accurate head segmentations. There has been a fair amount of attention paid to the accuracy of gray matter/white matter segmentation9 and skull stripping.10 Far less attention has been paid to accurate skull and scalp segmentation, despite studies which have shown that errors in modeling tissue thickness can lead to source localization errors in EEG11,12 and incorrect estimates of light reaching the cortex in optical forward modeling.13,14 A few software packages have been developed to address the problem of creating head segmentations from T1 structural MRIs. We identified four software packages with the capability to segment head tissue types from T1 MRIs only. The required inputs and general methodology are shown in Table 2. The comparison software packages we used are SkullFinder,15 the FMRIB Software Library (FSL) tools,16 Freesurfer’s mri_watershed,17 and the Statistical Parameter Mapping 8 (SPM8) MATLAB toolbox.18 In this work, we present a novel hybrid method to accurately segment all head tissues and compare this novel method to the publicly available packages. Table 2Evaluated segmentation methods.
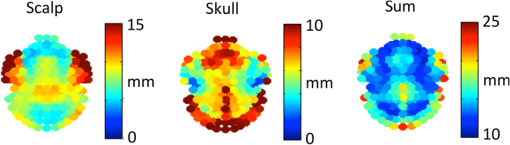
For multimodal neuroimaging, it is necessary to have accurate segmentation of scalp, skull, CSF, gray matter, and white matter. Multimodal neuroimaging with DOT is particularly sensitive to skull and scalp thickness accuracy. For imaging studies including EEG or DOT, it is necessary to have the appropriate CSF layer thickness. A fully automated pathway for creating the segmentations is also preferred, both to save researcher time and avoid researcher bias in the segmentations. Evaluation of segmentation algorithms can be complicated by the fact that algorithms are often validated against manual segmentation, which is time-consuming and may not be entirely consistent between studies. To test algorithms of interest in this work, we employed the BrainWeb Simulated Brain Database.7 This database includes a set of 20 realistic head segmentations which were created from real MRI scans, and simulated T1 images that were generated from these segmentations. Therefore, it is possible to test the accuracy of segmentation algorithms by comparing the segmentations of the simulated T1s created by each algorithm to the segmentation that generated the T1s. The goal of this work is to present a novel algorithm for whole head segmentation from T1 MRI scans. We quantitatively compare this new method against currently available methods using both traditional voxel-based segmentation accuracy metrics and a novel thickness metric. This new layer-based thickness metric is particularly useful for creating head models for noninvasive DOT forward modeling, and also allows for better comparisons with theoretical work that models the head as slabs with layers of tissues. The novel segmentation method employs a hybrid approach based on the aspects of the previously published algorithms that performed well as determined by our segmentation evaluation metric, and we show that this hybrid method faithfully reconstructs head tissue thicknesses in regions of the head that are relevant for noninvasive brain imaging. 2.Methods2.1.Segmentation Algorithms2.1.1.Ground truth segmentationsThe MRIs used to test the algorithms in this work were from the normal anatomical models section of the BrainWeb Simulated Brain Database.7 The segmentation models were created from multisequence MRI scans of subjects, and then these digital phantoms were used to create companion simulated T1 images. The methodology means that the anatomical segmentation models serve as “ground truth” for the T1s and allow for comparison of the segmentation algorithms of interest. The BrainWeb segmentations contain more tissue types than necessary for optical and M/EEG forward models, so the “fat,” “around fat,” “muscle,” and “muscle/skin” tissues were mapped to the scalp class, and the “dura,” and “bone marrow” classes were mapped to the skull class. The “vessel” tissue class was mapped to the second-most likely tissue class. Head models, therefore, contained scalp, skull, CSF, gray matter, white matter, and vessel tissue classes. 2.1.2.Standard segmentation methodsSegmentations of the BrainWeb-generated T1s were performed with the Freesurfer Watershed, BrainSuite Skullfinder, FSL BET, and SPM algorithms. The input T1 images were simulated to approximate a spoiled fast low angle shot (SFLASH) sequence (, , flip , and 1 mm isotropic voxel size). All 20 of the simulated T1 subjects in the normal anatomical database were used. These segmentations were used to extract the scalp, skull, CSF, and brain tissue types. The Freesurfer algorithm provides a “fat” classification, which was mapped to scalp. The BrainSuite algorithm does not differentiate between gray matter and white matter, so to compare the BrainSuite results with the BrainWeb segmentation both gray matter and white matter were combined to form a “brain” segmentation. The FSL method used brain extraction tool (BET) and the brain surface extraction tool BETSURF to extract the scalp, skull, and CSF, and FMRIB’s Automated Segmentation Tool (FAST) to segment gray and white matters and refine the CSF segmentation. The general strategies employed by each segmentation method are highlighted in Table 2. 2.1.3.Novel hybrid segmentation algorithm to accurately recover skull, scalp, and CSF layer thicknessA novel hybrid algorithm was developed for multimodal neuroimaging forward models with a focus on accurately reconstructing skull, scalp, and CSF layer thickness from T1 images. This algorithm was inspired by the Skullfinder algorithm as described in Dogdas et al.15 and the FSL BET algorithm. In general, this hybrid segmentation algorithm used morphological operations such as thickening and thinning implemented in the iso2mesh tools. Anatomically, relevant information was also used as a prior to constrain the skull and scalp thicknesses. The algorithm was used to create surfaces that delineate each tissue boundary, and for the purposes of comparing the surface-based segmentation with the original generating segmentations, the surfaces were combined to create a voxelized segmentation volume. The code was developed in MATLAB and is available from the authors. The process of extracting the scalp, outer skull, inner skull, and brain surfaces is described visually in Fig. 1, which shows how the boundary surfaces of each tissue type were calculated and combined to form the total head segmentation. Fig. 1Segmentation algorithm. B1: T1 image. B2: Gray matter segmentation from Freesurfer. B3: White matter segmentation from Freesurfer. B4: Combined white matter and gray matter brain mask. S1: T1 image. S2: Thresholded T1 image. S3: Largest connected region in the thresholded segment. S4. Active shapes algorithm performed on all two-dimensional slices. White indicates that the voxel was classified as scalp in all three slice orientations, and was therefore included in the final scalp segmentation. O1: T1 image, with the brain masked out and scalp mask applied. O2: Thresholded image A, also with two voxels of the scalp eroded. O3: Largest connected region in the thresholded segment. O4. Surface-smoothed and filled outer skull segmentation. I1: T1 image, with eroded outer skull mask applied. I2: Thresholded image I1. I3: Maximum skull thickness of 1.5 cm enforced. I4: Largest connected region in the thresholded segment. I5: Surface-smoothed and filled outer skull segmentation. I6: Final closing. 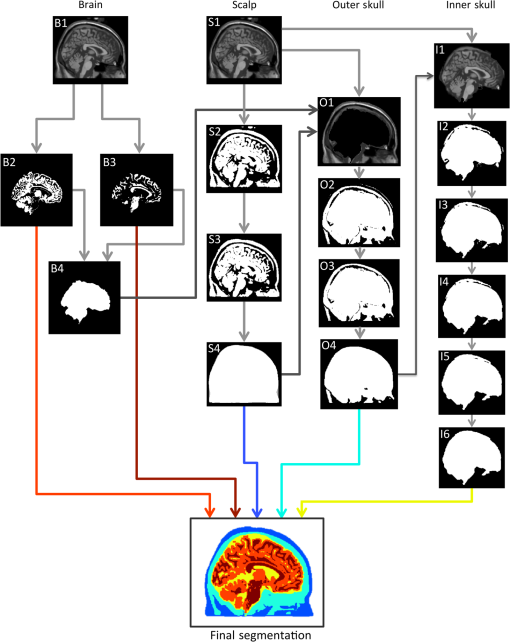 The first step in the algorithm after the standard Freesurfer gray matter and white matter segmentation was to segment the scalp. This process is described visually in Fig. 1 in the scalp column. First, a threshold was applied at 15% of the total range of intensity values in the T1 to binarize the volume. Voxels above this threshold include the scalp, brain tissue, and often external markers that might have been scanned with the subject. This threshold was suggested in the FSL segmentation implementation. Next, the largest contiguous set of voxels was selected as region of interest. Finally, an active shapes algorithm was applied to each two-dimensional slice of the volume, in all three directions, and only the voxels that were contained in all three slice orientations were kept. The active shapes algorithm used a probe radius . Next, the outer skull layer was segmented. This process is described visually in Fig. 1 in the outer skull column. First, the scalp was thinned by 2 voxels in all directions using the iso2mesh toolbox (thinbinvol) and used as a mask to ensure that the scalp was at least 2 mm thick. The thinning algorithm works by repeatedly removing the outermost layer of voxels until the specified number of thinning steps have been completed. Voxels in a brain mask as defined by Freesurfer were also set to zero. Next, a threshold at 25% of the total range of values in the T1 was applied, where voxels below this intensity were kept. This step allowed us to keep the low-intensity skull and exclude the scalp. The largest connected region of this image was kept to get rid of residual scalp voxels. Finally, surface smoothing (smoothbinvol) and 3-D volume filling (fillholes3d) algorithms from the iso2mesh toolbox were applied. The smooth surface algorithm had a radius bound of 4 mm and a distance bound of 1 mm. The last step was to extract the inner skull surface. This process is described visually in Fig. 1 in the inner skull column. A mask that was eroded 1 mm from the calculated outer skull surface was applied to the T1 image in order to ensure that the skull was at least 1 mm thick in all locations. The 25% threshold was applied, and this time voxels that were above the threshold became the region of interest. A second mask was applied that was the inner skull surface eroded by 15 mm, which enforced a maximum skull thickness of 15 mm. The largest connected region was again found, and surface smoothing and 3-D volume filling was again applied to get the final inner skull surface. The smoothed surface had a radius bound of 2 mm and a distance bound of 1 mm. 2.2.Segmentation Evaluation2.2.1.Voxel-based similarity metricSimilarity between two segmentations can be quantified in voxelated volumes or by using meshes that are generated from the segmentations. One common way of comparing voxelated segmentations is to calculate the Dice coefficient19 where is the Dice coefficient and and are the two binary images. The Dice coefficient is on the scale of 0 to 1, where 0 indicates that the segmentations are completely disjoint, and 1 indicates the segmentations are the same. The Dice coefficient does not assume that either the segmentation is correct, it merely describes the similarity between two segmentations. The Dice coefficient is only defined for binary-valued images, so each Dice coefficient for tissue type must be computed separately. Dice coefficients for each tissue type were averaged over subjects to get a metric describing the volumetric performance of each segmentation algorithm.2.2.2.Thickness-based similarity metricFor head models used in DOT and EEG forward model generation, accurate scalp, skull, and CSF layer thicknesses are particularly important. Although high Dice coefficients indicate overall similarity of segmented volumes, this does not necessarily indicate accurate layer thicknesses. Certain regions of the head are also more relevant to brain imaging, namely superior to the nasion, inion, and preauricular points. In order to quantify the mesh accuracy, we quantified the scalp and skull thickness at the locations on the scalp, which informs us if the segmentations are accurate in the most relevant regions of the head. Using these locations also allows us to compare between segmentation algorithms. Head meshes were created using the voxelated segmentations using the iso2mesh20 MATLAB toolbox. The positions were mapped on each head model using the National Food Research Institute (NFRI) tools21 as described in Ref. 22. Tissue layer thickness was calculated at 286 locations from the EEG positioning systems.23 For each segmentation and each subject, surfaces were created of the scalp, outer skull, and inner skull using iso2mesh. At each of the locations, all the nodes within 0.5 cm were projected toward the center of mass of the inner skull surface. The center of mass projection was found to be generally more robust than a surface normal projection from the scalp. The projection distance from the scalp to the inner skull and outer skull surfaces was recorded and averaged for all of the surface nodes within 0.5 cm of the node closest to the estimated locations. The distance to the outer skull is reported as the scalp thickness at the designated locations, and the difference between the inner skull distance and outer skull distance is reported as the skull thickness. The skull plus scalp thickness is also reported, as changes from the true values in this sum thickness generally indicate an incorrect CSF layer thickness. 3.Results and Discussion3.1.True Skull and Scalp ThicknessThe mean thicknesses of the skull, scalp, and skull plus scalp for the 20 subjects are shown in Fig. 2. The scalp is thin over the forehead and in the posterior region, and thick in the region in front of the ears. The skull is thin near the ears and thick in the posterior regions and the forehead region. The sum of skull and scalp thickness shows that the overall thinnest region is located in a ring around the head roughly just above the ears. Thicker regions are located along the medial section of the skull and along the inferior edges. These results are similar to those reported elsewhere.24,25 3.2.Qualitative Comparison Between Segmentation AlgorithmsExample sagittal slices of the volumetric segmentation images are shown in Fig. 3 for qualitative comparison. In this image, it is possible to see that the segmentation provided by the BrainSuite algorithm is not very accurate for the skull and CSF thickness. The Freesurfer segmentation is also not very accurate, particularly for the CSF layer, that appears to have extended thickness. The Freesurfer algorithm also separates the skull into two layers. Gray matter/white matter segmentation could also be done with the Freesurfer algorithm, but it is not shown in Fig. 3 due to the general poor head tissue segmentation. The FSL segmentation appears to be much more accurate than the segmentations resulting from the BrainSuite and FreeSurfer watershed algorithms. However, it is still possible to see that there is an extended CSF layer and corresponding thin skull layer near the top of the head. The SPM segmentation result also appears to be generally accurate, although it also has a thin skull layer near the top of the head. The SPM result also shows a thin layer of voxels classified as skull on the outer edge of the scalp. The segmentation results produced by the hybrid algorithm introduced in this work appears to be more accurate at avoiding the extended CSF layer/thin skull problems of the SPM/FSL algorithms. The comparative performance of these algorithms is next examined from a quantitative perspective. Fig. 3Example slice segmented with five different methods along with the generating segmentation. Color indicates tissue type. True is the provided generating segmentation, BS is the BrainSuite skullfinder segmentation, FS is the Freesurfer segmentation, FSL is the FSL/BET segmentation, SPM is the SPM method, and hybrid is the method described in this paper. 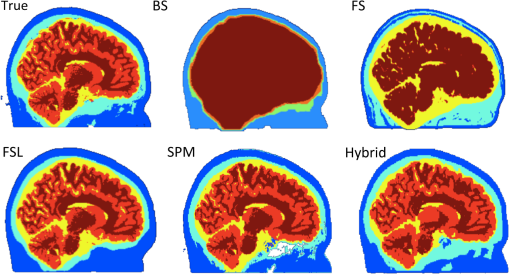 3.3.Quantitative Comparisons Between Segmentation AlgorithmsThe differences in the segmentations can be described by the Dice coefficients, reported in Table 3. The Dice coefficients indicate that when the entire head volume is accounted for, the SPM algorithm is the most accurate at generating scalp, CSF, and GM/WM segmentations, and the hybrid algorithm is most accurate at generating the skull segmentation. However, the Dice coefficients are not able to determine which algorithm is the best for the purposes of DOT forward models, as the metric is not specific to the regions of the scalp and skull near the brain. The values reported here for the Dice coefficients of the top-performing methods are similar to those reported elsewhere.15 Table 3Dice coefficient results between true and reconstructed segmentations. GM/WM indicates gray matter and white matter separately if allowed by the metric.
The top three segmentation methods (SPM, FSL, and hybrid) were compared using ray-based thickness methods to assess differences in scalp, skull, and scalp plus skull layer thickness. A histogram of errors for each tissue type at each locations for all subjects is shown in Fig. 4. The error distributions do not appear to be normally distributed. The differences in error distributions can be better appreciated in Fig. 5, which shows the fraction of scalp locations versus absolute error. In this representation, segmentation algorithms with low error will have a sharp initial slope and a flattened slope for large error values. The fraction of scalp locations with an error less than a particular value of interest is therefore shown for each tissue type and segmentation method. Fig. 4Histograms of scalp, skull, and summed scalp and skull thickness difference from truth for the BET, SPM, and hybrid segmentation methods. The horizontal indicates the amount of difference from the true segmentation thickness, while the vertical indicates the number of locations that have the thickness difference. 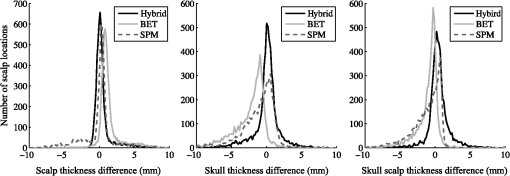 Fig. 5Thickness difference from generating segmentation. The horizontal indicates the absolute value of the segmentation thickness error, while the vertical indicates the fraction of locations that have the absolute value of that thickness difference or less. More accurate segmentation algorithms have a steeper initial slope. 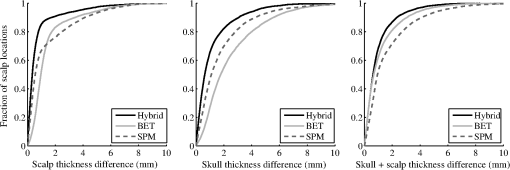 The spatial distribution of errors is displayed in Fig. 6. The errors tend to vary smoothly over the locations. Larger errors are often seen on the outer edge of the diagram, indicating regions that are on the inferior portion of the map. Generally, the scalp segmentations in the algorithms tested tend to be too thick, and the skull segmentations tend to be too thin. Larger errors are present along the middle of the maps for the skull thickness, indicating difficulties segmenting the sagittal sinus. Fig. 6Map of thickness differences from the true thickness displayed by scalp location for three segmentation methods, averaged over subjects. Positive numbers indicate that the tested segmentation was thicker than the generating segmentation, and negative numbers indicate that the tested segmentation was thinner than the generating segmentation in that region. 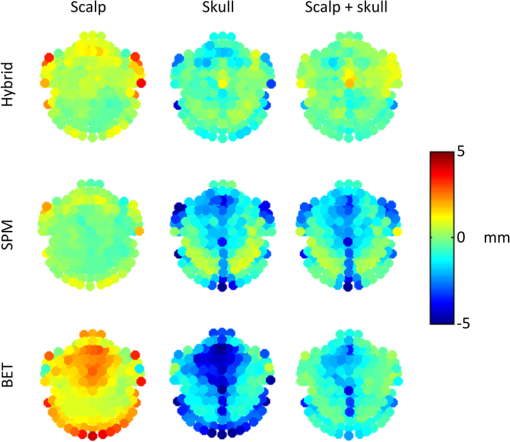 A summary root mean square (RMS) metric describing the overall accuracy of each segmentation method for each tissue type was calculated. The values in Table 4 are the RMS layer thickness differences over all scalp locations and all 20 tested cases. The RMS thickness metric does not allow cancellation of positive and negative errors, so we recommend its use for further studies evaluating segmentation layer thicknesses. The reported errors in Table 4 show that the hybrid method has the smallest error for each tissue type. Table 4RMS thickness difference in mm, full error distributions in Figs. 4 and 5.
3.4.Further ConsiderationsSegmentation errors in extracerebral tissues can cause inaccuracy in forward models, leading to source localization errors and errors in spatial resolution metrics. The SPM and BET/FSL algorithms both underestimate the summed thickness of the scalp and skull, indicating an increased CSF layer thickness compared to the generating segmentation. This extended CSF layer is a particular problem for DOT modeling, as it effectively introduces a light pipe that is not anatomically correct into the forward model. Inaccuracies in skull thicknesses are a problem with EEG forward models, as the skull is highly resistive and cannot reasonably be lumped with the scalp. Variations in tissue thickness have been shown to cause localization errors during EEG source reconstruction.11,12 The quantitative impact of incorrect tissue segmentation on brain source reconstruction has been investigated in other studies. For DOT, a tissue slab model and optical forward model showed that the estimated light reaching the gray matter decreases by approximately 50% if the skull thickness is increased from 5 to 10 mm.13 In our work, we show that underestimation of the skull thickness is a common error in head segmentation algorithms, as shown in Fig. 6, indicating that forward models created with the segmentations would overestimate the photons that would reach the gray matter. Similarly, if the sum total of the skull and scalp thickness is too thin, it indicates a thicker CSF layer. A CSF layer that increases from 1 to 3 mm has been shown to correspond to an increase of approximately 50% estimated light to the cortex.13 Together, these two effects mean that the amount of light to the cortex can be substantially under- or overestimated if inaccurate anatomical segmentations are used. For EEG, it has been estimated that localized skull thickness errors that are will not cause a large impact on source localization, although they can cause local magnitude estimation errors.26 In all of the algorithms tested, the treatment of the sagittal sinus is somewhat problematic. On T1 images, these voxels have a medium intensity similar to gray matter or scalp. The BrainWeb database segmentations have a vessel classification; however, most head models for brain imaging do not have a separate vessel tissue segmentation category. In order to compare the BrainWeb segmentations to head models with the usual tissue classifications, vessel voxels were mapped to the next-most-probable tissue type. Many of these voxels were mapped to the skull segmentation category following this rule. In a prior study,22 we showed that accurately modeling the vessel tissue class for DOT does not have a large impact on DOT sensitivity estimates, indicating that at least for DOT, inaccurate classification of the sagittal sinus may not be a significant concern. There has been increasing interest in using atlases to localize DOT activity instead of performing MRI scans and creating a subject-specific head model.27,28 Particular care should be taken in choosing the segmentation algorithms to create these atlases, as more accurate segmentation algorithms enable better atlas creation. The thickness metrics described here could also be used to create a simple mean head for a particular experiment based on subject skull and scalp thickness, and photon propagation could only be run once on the average head, saving the time usually spent running photon propagation code on multiple subjects. 4.ConclusionThe goal of this work was to introduce a novel hybrid algorithm for the purpose of generating accurate head models for EEG and DOT. For the purposes of creating accurate head models for noninvasive neuroimaging, the Dice coefficient metric for comparing segmentations is not sufficient, as all voxels are given the same weight regardless of location in the head. When generating head forward models, especially for DOT, the accuracy of scalp and skull layer thickness is critically important in the region covered by the scalp positioning systems. Small errors in the thickness of these regions may have a large impact on the amplitude of reconstructed brain activity, while segmentation errors in the jaw region, for example, would have no effect. Overall thickness accuracy for functional neuroimaging modeling applications is best summarized by RMS of the thickness difference at the scalp locations, as errors in the tissue layer thickness tend to be skewed. Overall, the quantitative evaluation of segmentation algorithms allows investigators to make informed decisions about processing steps for the creation of neuroimaging forward models. AcknowledgmentsFunding was provided by NSF (Graduate Research Fellowship to K.L.P.), NIH (5R01EB006385), a Director’s DISCOVERY grant from the Institute for Quantitative Biomedical Sciences (iQBS), and the Thayer School of Engineering at Dartmouth. ReferencesD. A. Boaset al.,
“Three dimensional Monte Carlo code for photon migration through complex heterogeneous media including the adult human head,”
Opt. Express, 10
(3), 159
–170
(2002). http://dx.doi.org/10.1364/OE.10.000159 OPEXFF 1094-4087 Google Scholar
J. C. MosherR. M. LeahyP. S. Lewis,
“EEG and MEG: forward solutions for inverse methods,”
IEEE Trans. Biomed. Eng., 46
(3), 245
–259
(1999). http://dx.doi.org/10.1109/10.748978 IEBEAX 0018-9294 Google Scholar
J. P. MuglerJ. R. Brookeman,
“Three-dimensional magnetization-prepared rapid gradient-echo imaging (3D MP RAGE),”
Magn. Reson. Med., 15
(1), 152
–157
(1990). http://dx.doi.org/10.1002/(ISSN)1522-2594 MRMEEN 0740-3194 Google Scholar
J. P. Culveret al.,
“Volumetric diffuse optical tomography of brain activity,”
Opt. Lett., 28
(21), 2061
–2063
(2003). http://dx.doi.org/10.1364/OL.28.002061 OPLEDP 0146-9592 Google Scholar
G. StrangmanM. A. FranceschiniD. A. Boas,
“Factors affecting the accuracy of near-infrared spectroscopy concentration calculations for focal changes in oxygenation parameters,”
NeuroImage, 18
(4), 865
–879
(2003). http://dx.doi.org/10.1016/S1053-8119(03)00021-1 NEIMEF 1053-8119 Google Scholar
C. H. Wolterset al.,
“Influence of tissue conductivity anisotropy on EEG/MEG field and return current computation in a realistic head model: a simulation and visualization study using high-resolution finite element modeling,”
NeuroImage, 30
(3), 813
–826
(2006). http://dx.doi.org/10.1016/j.neuroimage.2005.10.014 NEIMEF 1053-8119 Google Scholar
B. Aubert-Brocheet al.,
“Twenty new digital brain phantoms for creation of validation image data bases,”
IEEE Trans. Med. Imaging, 25
(11), 1410
–1416
(2006). http://dx.doi.org/10.1109/TMI.2006.883453 ITMID4 0278-0062 Google Scholar
M. WindhoffA. OpitzA. Thielscher,
“Electric field calculations in brain stimulation based on finite elements: an optimized processing pipeline for the generation and usage of accurate individual head models,”
Hum. Brain Mapp., 34
(4), 923
–935
(2013). http://dx.doi.org/ 10.1002/hbm.21479 HBRME7 1065-9471 Google Scholar
F. Klauschenet al.,
“Evaluation of automated brain MR image segmentation and volumetry methods,”
Hum. Brain Mapp., 30
(4), 1310
–1327
(2009). http://dx.doi.org/10.1002/hbm.v30:4 HBRME7 1065-9471 Google Scholar
D. W. Shattucket al.,
“Online resource for validation of brain segmentation methods,”
NeuroImage, 45
(2), 431
–439
(2009). http://dx.doi.org/10.1016/j.neuroimage.2008.10.066 NEIMEF 1053-8119 Google Scholar
B. Cuffin,
“Effects of local variations in skull and scalp thickness on EEG’s and MEG’s,”
IEEE Trans. Biomed. Eng., 40
(1), 42
–48
(1993). http://dx.doi.org/10.1109/10.204770 IEBEAX 0018-9294 Google Scholar
N. Chauveauet al.,
“Effects of skull thickness, anisotropy, and inhomogeneity on forward EEG/ERP computations using a spherical three-dimensional resistor mesh model,”
Hum. Brain Mapp., 21
(2), 86
–97
(2004). http://dx.doi.org/10.1002/(ISSN)1097-0193 HBRME7 1065-9471 Google Scholar
S. Wanget al.,
“Effects of spatial variation of skull and cerebrospinal fluid layers on optical mapping of brain activities,”
Opt. Rev., 17
(4), 410
–420
(2010). http://dx.doi.org/10.1007/s10043-010-0076-6 1340-6000 Google Scholar
G. E. StrangmanQ. ZhangZ. Li,
“Scalp and skull influence on near infrared photon propagation in the colin27 brain template,”
NeuroImage, 85
(1), 136
–149
(2014). http://dx.doi.org/10.1016/j.neuroimage.2013.04.090 NEIMEF 1053-8119 Google Scholar
B. DogdasD. W. ShattuckR. M. Leahy,
“Segmentation of skull and scalp in 3-D human MRI using mathematical morphology,”
Hum. Brain Mapp., 26
(4), 273
–285
(2005). http://dx.doi.org/10.1002/(ISSN)1097-0193 HBRME7 1065-9471 Google Scholar
S. M. Smithet al.,
“Advances in functional and structural MR image analysis and implementation as FSL,”
NeuroImage, 23
(Suppl 1), S208
–S219
(2004). http://dx.doi.org/10.1016/j.neuroimage.2004.07.051 NEIMEF 1053-8119 Google Scholar
F. Ségonneet al.,
“A hybrid approach to the skull stripping problem in MRI,”
NeuroImage, 22
(3), 1060
–1075
(2004). http://dx.doi.org/10.1016/j.neuroimage.2004.03.032 NEIMEF 1053-8119 Google Scholar
K. J. Fristonet al.,
“Statistical parametric maps in functional imaging: a general linear approach,”
Hum. Brain Mapp., 2
(4), 189
–210
(1994). http://dx.doi.org/10.1002/hbm.v2:4 HBRME7 1065-9471 Google Scholar
L. R. Dice,
“Measures of the amount of ecologic association between species,”
Ecology, 26
(3), 297
(1945). http://dx.doi.org/10.2307/1932409 ECOLAR 0012-9658 Google Scholar
Q. FangD. A. Boas,
“Tetrahedral mesh generation from volumetric binary and grayscale images,”
in Proc. of IEEE Int. Symposium on Biomedical Imaging 2009,
1142
–1145
(2009). Google Scholar
V. JurcakD. TsuzukiI. Dan,
“, , and systems revisited: their validity as relative head-surface-based positioning systems,”
NeuroImage, 34
(4), 1600
–1611
(2007). http://dx.doi.org/10.1016/j.neuroimage.2006.09.024 NEIMEF 1053-8119 Google Scholar
K. L. PerdueQ. FangS. G. Diamond,
“Quantitative assessment of diffuse optical tomography sensitivity to the cerebral cortex using a whole-head probe,”
Phys. Med. Biol., 57
(10), 2857
–2872
(2012). http://dx.doi.org/10.1088/0031-9155/57/10/2857 PHMBA7 0031-9155 Google Scholar
R. OostenveldP. Praamstra,
“The five percent electrode system for high-resolution EEG and ERP measurements,”
Clin. Neurophysiol., 112
(4), 713
–719
(2001). http://dx.doi.org/10.1016/S1388-2457(00)00527-7 CNEUFU 1388-2457 Google Scholar
M. S. Beauchampet al.,
“The developmental trajectory of brain-scalp distance from birth through childhood: implications for functional neuroimaging,”
PLoS One, 6
(9), e24981
(2011). http://dx.doi.org/10.1371/journal.pone.0024981 1932-6203 Google Scholar
M. Okamotoet al.,
“Three-dimensional probabilistic anatomical cranio-cerebral correlation via the international 10–20 system oriented for transcranial functional brain mapping,”
NeuroImage, 21
(1), 99
–111
(2004). http://dx.doi.org/10.1016/j.neuroimage.2003.08.026 NEIMEF 1053-8119 Google Scholar
B. Lanferet al.,
“Influences of skull segmentation inaccuracies on EEG source analysis,”
NeuroImage, 62
(1), 418
–431
(2012). http://dx.doi.org/10.1016/j.neuroimage.2012.05.006 NEIMEF 1053-8119 Google Scholar
A. Custoet al.,
“Anatomical atlas-guided diffuse optical tomography of brain activation,”
NeuroImage, 49
(1), 561
–567
(2010). http://dx.doi.org/10.1016/j.neuroimage.2009.07.033 NEIMEF 1053-8119 Google Scholar
R. J. Cooperet al.,
“Validating atlas-guided DOT: a comparison of diffuse optical tomography informed by atlas and subject-specific anatomies,”
NeuroImage, 62
(3), 1999
–2006
(2012). http://dx.doi.org/10.1016/j.neuroimage.2012.05.031 NEIMEF 1053-8119 Google Scholar
BiographyKatherine L. Perdue received a BS in physics from Harvey Mudd College in 2005 and a PhD in engineering sciences from the Thayer School of Engineering at Dartmouth in 2012. She has also worked as a research assistant at the Martinos Center for Biomedical Imaging at Massachusetts General Hospital. Currently, she is a research fellow at Boston Children’s Hospital. Solomon G. Diamond received an AB degree in engineering sciences from Dartmouth College in 1997, a BE degree from the Thayer School of Engineering at Dartmouth in 1998, and a PhD in engineering sciences from Harvard University in 2004. He conducted post-doctoral training at the Martinos Center for Biomedical Imaging at Massachusetts General Hospital. He is currently an assistant professor at Dartmouth where he teaches courses in neuroengineering, computer-aided design, and solid mechanics. |