|
|
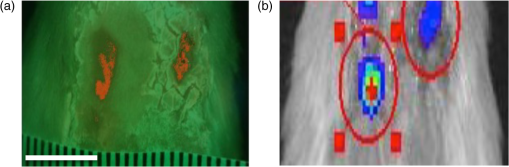
1.IntroductionBacterial infection of wounds is a major clinical problem and poses a significant burden to patients and healthcare systems worldwide.1 An infected wound can be polymicrobial and contain a larger number of microorganisms than a contaminated or colonized wound.2,3 Although a contaminated wound will heal (bacterial balance), an infected wound will not (bacterial imbalance). The key biological processes that occur during wound healing include tissue and cellular changes, such as inflammation and connective tissue remodeling in the dermis.4 Bacterial infection can significantly impede the healing process, extending it from days to months.5 The current clinical standard for diagnosing infected wounds involves the nonquantitative white light (WL) assessment of clinical signs and symptoms (CSS), such as pain, erythema, edema, lymphadenitis, or purulence.6 Microbiological swab cultures are occasionally used in conjunction with CSS to provide semiquantitative information about the relative amounts of pathogenic bacteria in a wound, as well as antibiotic susceptibility.7 However, the turnaround time is 3 to 5 days, which precludes real-time assessment. Therefore, there is a need for alternative approaches to wound diagnosis and treatment guidance. Real-time knowledge of wound bacterial load could improve sampling, treatment guidance (e.g., antibiotics, debridement), and treatment response assessment. We developed a new imaging device named Portable Real-time Optical Detection, Identification and Guidance for Intervention (PRODIGI) to help clinicians measure bacterial load over time in wounds. PRODIGI is a compact, hand-held, device for noncontact and noninvasive imaging. It captures both WL and autofluorescence (AF) signals produced by tissue components and bacteria without the use of contrast agents. Composite images are displayed by green AF, produced by endogenous connective tissues (e.g., collagen, elastin) in skin, and red AF, produced by endogenous porphyrins8,9 in clinically relevant bacteria such as Staphylococcus aureus. Siderophores/pyoverdins10,11 in other species such as Pseudomonas aeruginosa appear blue-green in color with in vivo PRODIGI fluorescence (FL) imaging. In the current preclinical study, we sought to demonstrate technical feasibility and validate the utility of PRODIGI as an imaging device for real-time detection and tracking of pathogenic bacteria in skin wounds. We used an established nude mouse skin wound model, previously reported for studying wound status and healing.7 Herein, we tested PRODIGI’s ability to accurately detect and measure bacterial load in wounds in real time, guide treatment decisions, and track wound healing over the course of antibacterial treatment. Additionally, we used bioluminescence imaging (BLI) to correlate absolute bacterial load with FL signals obtained using PRODIGI. The findings presented in this study have served to initiate on-going in-human validation and technical optimization studies of PRODIGI. 2.Methods2.1.PRODIGI Prototype Imaging DevicePRODIGI consists of a low-cost, consumer-grade, Super HAD™ charge-coupled device (CCD) sensor-based camera (Model DSC-T900, Sony Corp., Tokyo, Japan), with a 35 to 140 mm equivalent zoom lens housed in a plastic body and powered by rechargeable batteries (Fig. 1). It collects high-resolution 12.1 Mpixels color WL and AF images (or videos) in real time (), which are displayed in red-green-blue (RGB) format on a 3.5-in. touch-sensitive color liquid-crystal display (LCD) screen (Fig. 1). Broadband white light-emitting diodes (LEDs), electrically powered by a standard AC125V source, provide illumination during WL imaging, while two monochromatic violet-blue () LED arrays (Model LZ4, LedEngin, San Jose, California) provide 4-W excitation light power during FL imaging (bright, uniform illumination area at 10 cm distance from skin surface). WL and FL images are detected by a high-sensitivity CCD sensor mounted with a dual band FL filter ( to 550 and 590 to 690 nm) (Chroma Technologies Corp., Bellows Falls, Vermont) in front of the camera lens to block excitation light reflected from the skin surface. Tissue and bacteria AF are spectrally separated by the emission filter and displayed as a composite RGB image without image processing or color-correction, allowing the user to see the bacteria distribution within the anatomical context of the wound and body site. The CCD image sensor is sensitive across ultraviolet (), visible (400 to 700 nm), and near-infrared (700 to 900 nm) wavelengths to AF of tissues and bacteria, in the absence of exogenous contrast agents. A list of system components for the PRODIGI prototype device is provided in Table 1. Fig. 1The handheld prototype PRODIGI imaging device. (a) Front view of PRODIGI showing wound fluorescence (FL) image displayed in real time on the liquid-crystal display screen in high definition. (b) Back view of PRODIGI showing white light (WL) and 405-nm LED arrays providing illumination of the wound, while the FL emission filter is placed in front of the CCD sensor. 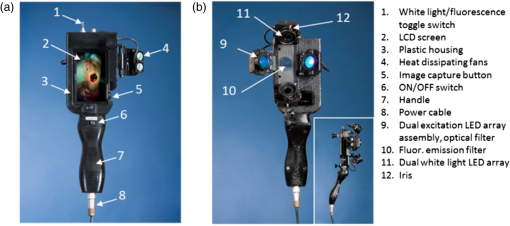 Table 1Main components of the PRODIGI prototype imaging device. The PRODIGI prototype imaging system consists of a Sony DSC-T900 camera, two 405 nm light-emitting diodes, two neutral white LEDs, two switches, four cooling fans, a male connector, a female connector, a DC/DC converter, two heat sinks, a spare Sony lithium-ion battery, an AC/DC power supply, two excitation light source filters, a detection dual bandpass filter, a system shell (built in house), and a carrying case.
Although the current study is a preclinical in vivo validation of PRODIGI in a mouse wound model, PRODIGI is approved by Health Canada as a Class II medical device for clinical investigational testing (Health Canada Investigational Testing Authorization #16114) as well as approvals from our institutional REB (UHN REB #09-0015-A and UHN REB #12-5003) and the Canadian Standards Association. A two-part Phase I, single center, nonrandomized clinical trial of PRODIGI for detecting pathogenic bacteria, guiding wound treatment, and assessing treatment response in chronic wound patients (clinicaltrials.gov identifiers: NCT01378728 and NCT01651845) is ongoing at the University Health Network. A separate paper detailing the results of this clinical trial is forthcoming. 2.2.PRODIGI Mobile Imaging DeviceThe PRODIGI mobile device (Fig. 2) is a modified version of the PRODIGI prototype device (Fig. 1), for easier and more convenient use in an animal colony. Although the PRODIGI prototype device is handheld and compact, its uses proved to be cumbersome during FL imaging of the mice within the confined space of the biosafety cabinet in the institution’s animal facility. As such, we developed an iPhone version of PRODIGI, which is a physically different representation of the same technology, operating based on the same optical principles and parameters of FL imaging, and providing the same imaging output as the prototype device described above. A technical comparison of the two devices is provided in Table 2. Fig. 2The PRODIGI mobile imaging device. (a) Front view showing the optical components and battery holder of the accessory adapter, which is mounted onto a standard iPhone 4S smart phone. (b) Back view of the device showing the on/off switch and the LCD viewing screen on which the WL and FL images are viewed by the user. 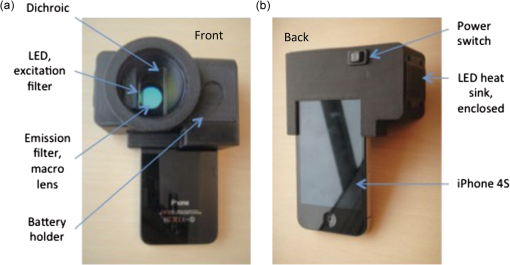 Table 2Comparison of the PRODIGI prototype versus PRODIGI mobile imaging systems. Both the PRODIGI prototype and PRODIGI mobile systems use the same excitation and emission filters, LED wavelength, and LCD screen. They differ in the image sensor type, physical weight, and dimensions and image spatial resolution. Technically, both devices operate similarly based on autofluorescence imaging achieved using different components.
2.3.Mouse Skin Wound ModelThe procedures described herein have been completed under a specific Animal Use Protocol approved by the University Health Network Animal Care Committee in compliance with all relevant regulatory and institutional agencies, regulations, and guidelines (UHN AUP# 2614.3). Female nude mice aged 8 to 12 weeks (NCRNU-F) were used. Nude mice were chosen for this model because their immune systems are compromised, thereby enabling a robust infection response following inoculation. On the day of surgery (day 0), mice were anesthetized using 2% isofluorane mixed with 100% oxygen. Dorsal skin areas were shaved and cleaned with 70% isopropyl ethanol and 10% povidone-iodine. Two circular full thickness wounds (6-mm diameter) were created on either side of the spine using a biopsy punch with the assistance of scissors and forceps. Mice were subcutaneously inoculated with Gram-positive bioluminescent S. aureus-Xen8.1 from the parental strain S. aureus 8325-4 (Caliper) () suspended in 30 μL of phosphate-buffered saline (PBS) in one wound and 30 μL PBS in the other wound as a control. Solutions were injected until bubbles formed on the wound surface, and the residual solution was spread on the surface of the infected wound. Gauze and Tegaderm™ were used to cover the wounds after the surgery (Fig. 3). All mice were monitored daily and both WL and FL images were taken of the wounds. Tegaderm™ was changed after each imaging procedure. Fig. 3Procedures for creating the mouse skin wound model based on the Guthrie mouse model.7 (a) Anesthetized mice receive two equal-sized wounds on both sides of the spine using a biopsy punch. (b) One wound was inoculated with phosphate-buffered saline (PBS) (control; left), the other with Staphylococcus aureus in PBS (right). (c) Tegaderm was applied across the wounds over sterile gauze 15 min after the inoculation. Scale bar: (a–c) 6 mm.  2.4.White Light and FL ImagingRegular room lighting was left on during WL imaging of wounds. During FL imaging, the lights were turned off to eliminate artifacts. WL and corresponding FL still images of wounds were collected in a noncontact manner with the device held within 10 to 15 cm from the wound. This distance was kept constant and the image was focused on the center of the wound. The camera’s built-in macromode was used, image acquisition took approximately 1 s, and the images taken were stored for future analysis on the camera’s standard digital memory card. Switching between WL and FL modes was instantaneous using a built in “toggle switch” on the PRODIGI prototype device and by manually turning on and off the FL light source mounted on the top of the iPhone for the PRODIGI mobile device (Fig. 2). Devices were decontaminated between uses with standard 70% ethyl alcohol as an antiseptic wipe. 2.5.Image AnalysisWL and FL images were transferred to a laptop for image processing. Two programs were used for image processing: MATLAB software (Version 7.9.0, The MathWorks, Natick, Massachusetts) using a custom-written program and ImageJ Software (Version 1.45n). In the MATLAB program, regions of interests (ROIs) were identified from individual pixel FL images of each wound. RGB images were separated into individual channels. Green (500 to 550 nm emission) and red AF () from tissue components and bacteria, respectively, detected by the CCD sensor were naturally aligned spectrally with the red and green filters on the Sony CCD image sensor. Thus, the green and red channels of the RGB image displayed on PRODIGI’s LCD screen were representative of the true tissue and bacterial AF signals detected in vivo. To quantify bacterial levels from individual FL images, the following image processing procedures were used. Briefly, individual green and red image channels from each RGB image were converted to gray scale (the blue channel was not used) and pixels whose gray scale intensity was above a given histogram threshold (selected to reduce the background noise of the raw image) were counted. A red color mask for red FL bacteria was created by finding the local maxima in the color range 100 to 255 gray scale. Then, an inverted green color mask was used to remove the green FL. All pixels with red FL (above the histogram threshold) were binarized and the sum of all “1” pixels was calculated. This was repeated for the green channel of each image. These data gave an estimate of the amount of red bacteria in each image. The number of FL pixels was converted into a more useful pixel area measure () by applying a ruler on the pixel image, thereby providing the total amount of fluorescent bacteria as an area measurement (). The sizes of the wounds were traced and measured similarly by converting pixel areas to of the circled wound area on the WL images. The resolution of the FL images was sufficient to localize bacteria based on regions of FL. ImageJ software was used to separate FL images into red, green, and blue channels using the built-in batch processing function “Split Channels” located within the image menu and color submenu. Each resulting channel was displayed and saved in gray scale. For further analysis, an ROI was identified in each corresponding red, green, and blue channel image. Under the built-in analysis menu, we used the “Set Measurement” function to select and measure the following measurement parameters for each color channel image: pixel area, min. and max. gray scale intensity values, and mean gray intensity values. The average red channel intensity value was determined as (bacterial) FL intensity per square pixel in each red channel image and then used for data analysis and comparison. 2.6.Tracking Bacterial Load over TimeThe mouse skin wound model described above was used to correlate wound status with the progression of bacterial infection (; 8 to 12 weeks; NCRNU-F). Daily WL and FL images were taken of the wounds as they became infected over time. Antibacterial treatment (topical Mupirocin three times daily, for a total of 1 day) was applied to the wound site when the red FL intensity peaked. The anti-microbial effect of the treatment was monitored over time using PRODIGI to acquire daily WL and FL images of the wound after treatment. The wounds were monitored for a total of 10 days, after which the mice were sacrificed. Bacterial amounts from FL images and wound size from WL images were quantified using the MATLAB program, and compared over time to determine the wound healing status. 2.7.Bioluminescence ImagingBLI was used in the current study to measure the absolute amount of bacteria in vivo, because it is one of the most sensitive and reliable screening tools for determining bacterial load.12,13 BLI collects the light emitted from the enzymatic reaction of luciferase and luciferin and therefore does not require excitation light.14 Dinjaski et al. recently showed that near-infrared FL imaging with a dye injection can be an alternative to BLI imaging.15 In the current study, FL imaging using PRODIGI (without any exogenous FL contrast agent administration) and BLI imaging of inoculated S. aureus bacteria were tracked over time and the FL and BLI intensities were compared (). The bacterial BLI signal did not contribute to the FL signal detected by PRODIGI’s consumer grade-CCD camera. Gram-positive bioluminescent S. aureus-Xen8.1 from the parental strain S. aureus 8325-4 (Caliper) was grown to midexponential phase the day before pathogen inoculation. Bacteria with the BLI cassette produce the luciferase enzyme and its substrate (luciferin), thereby emitting a 440 to 490 nm bioluminescent signal when metabolically active16,17 (Fig. 4). The bacteria () were suspended in 0.5 mL of PBS and injected into the wounds of female athymic nude mice (; 8 to 12 weeks; NCRNU-F Homozygous). To detect S. aureus bioluminescence, BLI images of the wound were acquired before, immediately after, and 1, 2, 3, 4, 5, 6, and 7 days postinoculation inside the dark chamber of the Xenogen IVIS Spectrum Imaging System 100 (Caliper, Massachusetts), using an exposure time of 10 s. The mice were kept under gas anesthesia consisting of 100% oxygen mixed with 2% isoflurane during imaging. BLI images were captured using Living Image In Vivo Imaging software (Caliper, Massachusetts). ROIs were digitally circumscribed over the wound and the total luminescence intensity counts were measured within the ROIs for each time point imaged. The absolute amount of bacteria measured from the BLI signals was tested for correlation with the corresponding FL signals on the FL images taken over time of the same wound using PRODIGI (as described above). 3.Results3.1.Tracking Bacterial Load over TimeThe red FL signal was first detected on day (range 2 to 4) and peaked on day (range 4 to 6). Two mice had serious cross-contamination of the adjacent wounds and were sacrificed. In the remaining mice (), red FL was observed in the wounds that were inoculated with S. aureus but not in the adjacent control wounds where only PBS was injected. Representative WL and FL images for a single mouse tracked over 10 days are shown in Fig. 5. Bacteria (red FL) were not visible on days 1 and 2 postinoculation, but as the bacteria proliferated and colonized the wound, red FL was visualized on day 3 and increased over days 4 to 6, by which time the infection could be visualized under WL. On day 7, the red FL intensity peaked and then began to drop. Mupirocin was applied to the wound on day 7 and the bacterial load, visualized as red FL, decreased significantly, to almost zero intensity on day 8. However, red FL reappeared on days 9 and 10. Fig. 5Representative WL and FL images for a single mouse tracked over 10 days. (a) PRODIGI prototype images showing the two equal-sized wounds on both sides of the spine. The right wound was inoculated with S. aureus in PBS and the left wound was inoculated with PBS only (control). The top row shows WL images, the middle row shows FL images, and the bottom row shows MATLAB quantified images, corresponding to bacterial areas and intensities. The FL imaging data demonstrated a significant increase in bacterial FL intensity in the wound inoculated with S. aureus, compared with the control wound, peaking on day 6. Mupirocin (day 7, red arrow) significantly decreased bacterial FL on day 8 to almost zero, indicating treatment effect. Bacteria increased again on days 9 and 10. (b) Graph showing quantitative changes in bacterial load from FL images obtained in panel (a). 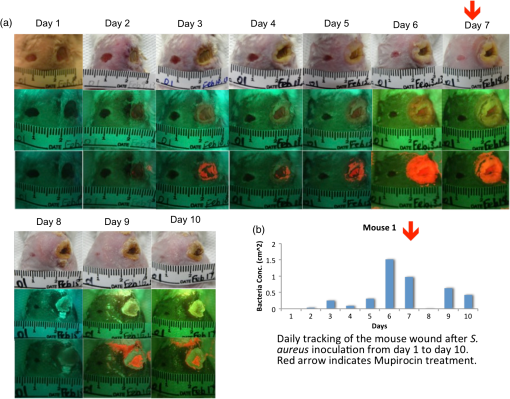 3.2.Correlating FL with BLIA significant positive correlation was observed between the bacterial FL captured by PRODIGI and the absolute bacterial load measured by the Xenogen imaging system (Pearson correlation coefficient 0.6889, -value 0.04; Fig. 6). Both imaging modalities showed an increase in bacterial load from day 0 to day 4, with a drop in bacterial load after day 4. Overall, the changes in FL intensity of the bacteria correlated with the bacterial BLI intensity (e.g., absolute bacterial load) as the bacterial amount increased and subsequently decreased in the wounds. A representative example of a mouse tracked over time using WL, FL, and BLI imaging is shown in Fig. 6. FL intensity was calculated from the red channel of RGB images using ImageJ software. Fig. 6Preclinical data show that pathogenic bacterial autofluorescence (AF) intensity correlates with bacterial load in vivo. (a) Time course PRODIGI mobile images of skin wounds in a mouse prior to and after inoculation with bioluminescent S. aureus-Xen8.1 ( in 30 μL PBS). Representative WL (top row), AF (middle row), and bioluminescence (bottom row) images are shown for each time point to 7 days after inoculation in a wounded mouse. BLI imaging gives absolute bacterial amount in vivo. Red arrows show when the tegaderm bandage was exchanged, causing some bacteria to be removed from the surface. (b) Average red FL from S. aureus-Xen8.1 ( nude mice) shown as a function of time demonstrating an increase in daily S. aureus bacterial FL (calculated from red channel of RGB images using ImageJ software). At days 2 and 7, tegaderm bandages were exchanged as per animal protocol. Average bacterial FL peaked at day 4 postinoculation. (c) Corresponding time course bioluminescence data (calculated from ROI) show similar increase and peaking at day 4 in total bacterial load in the wound. Data indicate strong positive correlation (Pearson correlation coefficient ) between total bacterial AF in a wound and the bacterial load in vivo. Standard errors are shown. Scale bars: (a) WL 1.5 cm and AF, BLI 1 cm. 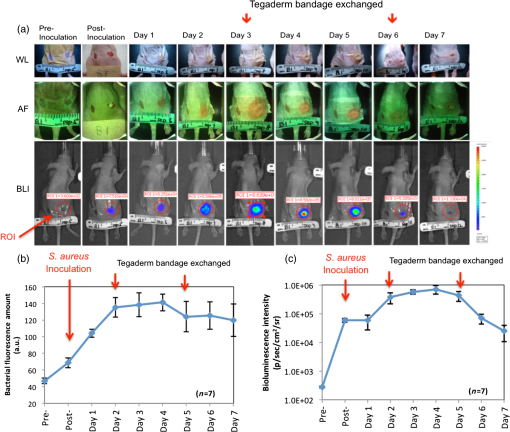 4.DiscussionAll wounds contain bacteria (e.g., Staphylococcus, Streptococcus, Pseudomonas species, and Coliform bacteria1 including aerobic and anaerobic types18), at levels ranging from contamination through critical colonization to infection.1 The role of bacteria in wounds depends on their concentration, species composition, and host response.19 Contamination and colonization by low concentrations of microbes are considered normal, and are not believed to inhibit healing.20 However, critical colonization and infection are associated with a significant delay in wound healing.21 The increased bacterial burden may be confined to the superficial wound bed or present in the deep compartment and surrounding tissue of the wound margins. Infections can delay wound healing by days, months, or years.2,4,5 Thus, early recognition of subclinical bacterial load is necessary if the early signs of infection are to be identified. The ongoing work of our group has aimed to create new technologies to diagnose wounds and guide clinical treatment. PRODIGI represents a novel point-of-care imaging device for real-time detection and tracking of pathogenic bacteria in wounds. In this study, preclinical in vivo validation of PRODIGI was conducted in a mouse wound model. Using PRODIGI, we were able to track bacterial load before and during antibiotic treatment. Moreover, we found FL measures obtained with PRODIGI were positively correlated with absolute bacterial load measured by BLI. AF imaging (without contrast agents) provides a powerful means for in situ visualization of bacteria and connective tissue that are invisible under normal WL.22 It has been used in other clinical applications, such as gastroenterology, to image both collagen and bacterial FL.23,24 PRODIGI spectrally distinguishes bacteria from connective tissues and blood in vivo.22 Using and to 550 nm, 590 to 690 nm, PRODIGI detects AF signals of S. aureus, Staphylococcus epidermidis, P. aeruginosa, Candida, Serratia marcescens, Viridans streptococci (α-hemolytic streptococci), Streptococcus pyogenes (-hemolytic streptococci), Corynebacterium diphtheriae, Enterobacter, Enterococcus, and methicillin-resistant S. aureus (MRSA), as verified by microbiological swab cultures (data from a human clinical trial by our group to be published in a forthcoming paper). This is a representative of the major types of pathogenic bacteria commonly found in infected wounds. Clinical microbiology tests confirmed that S. aureus, S. epidermidis, Candida, S. marcescens, Viridans streptococci, Corynebacterium diphtheriae, S. pyogenes, Enterobacter, and Enterococcus produced red FL (from porphyrin25–27) while P. aeruginosa produced a bluish-green FL (from pyoverdin10,11) detected by PRODIGI. These spectral characteristics differ significantly from connective tissues (collagen, elastin) and blood, which appear green and dark red, respectively. A representative image of these spectral characteristics is shown in Fig. 7. Fig. 7Representative spectral characteristics of green and red fluorescing bacteria. (a) WL image of a chronic wound on a patient’s right ankle. (b) Corresponding FL image distinguishing the red FL from the endogenous porphyrins in S. aureus (smaller circle) and the green FL from the siderophores/pyoverdins in P. aeruginosa (larger circle). (c) Corresponding point FL spectra confirming that, while both species emit green FL between 490 and 550 nm with 405-nm excitation, S. aureus emits red while P. aeruginosa emits bright bluish-green FL peaking at 480 nm. Scale bars (a and b) 1 cm. 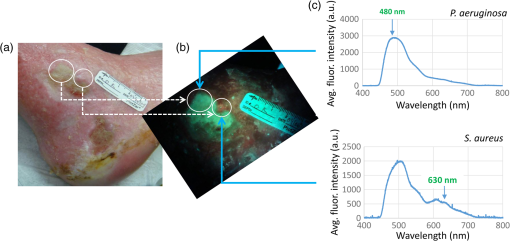 Microbiological data provide important wound management guidance on whether to treat a wound and, if so, on the choice of antibiotic treatment. However, most clinicians prescribe broad-spectrum antimicrobial agents before reviewing a microbiology report, and in many cases the treatment may be inappropriate or unnecessary. The misuse of broad-spectrum antibiotics can lead to the spread of antibiotic-resistant bacteria (e.g., MRSA, Clostridium difficile, vancomycin-resistant Enterococci).28 At present, PRODIGI can detect MRSA (an important class of antibiotic-resistant bacteria), but cannot differentiate between different strains of therapy-resistant bacteria. This may be possible in the future by improving in vivo FL classification of bacteria using alternate excitation/emission or optimizing FL filters. Nevertheless, PRODIGI’s current capability of AF image-guidance may enable earlier detection and appropriate antibiotic treatment of bacteria-infected wounds, thereby reducing the total bacterial load, preventing the emergence of antibiotic-resistance, and promoting wound healing. To the best of our knowledge, PRODIGI is the first FL imaging device to be used for wound care, providing biologically relevant information at the tissue and cellular levels in real time. To test PRODIGI’s capability of monitoring changes in bacterial load, we treated infected wounds with Mupirocin. Antibacterial treatment was applied to the wound site 7 days after the bacterial inoculation. As expected, Mupirocin decreased bacterial load, and with PRODIGI we observed nearly a complete absence of red FL 1 day after treatment. Similar reductions in bacterial load have been seen using BLI methods in wounds treated with Mupirocin in mice.29 By day 9, red FL intensity increased which indicated bacterial regrowth, likely from bacterial load in the underlying deeper layers of the wound (AF imaging accurately detects superficial bacterial load up to 1.5-mm deep). This was not unexpected as mice only received 1 day of Mupirocin treatment. Mupirocin requires of application for successful antimicrobial treatment.30 In conclusion, our data support that PRODIGI is capable of collecting FL images of wounds to detect the presence and relative amounts of bacteria within the wound based on endogenous bacterial FL, thus enabling quantification of “infection status.” This may significantly impact clinical wound care by allowing clinicians to monitor wound healing and treatment response over time. PRODIGI AF imaging may also be used to guide debridement of devitalized tissues with high bacterial load, directly visualize residual bacteria following debridement, and guide the targeted application of topical antimicrobial treatment. We further demonstrated the versatility of the PRODIGI device by developing a technically equivalent mobile version, producing the same WL and FL images as the original PRODIGI prototype, but with a smaller and more compact physical form for use in space-confined settings. Overall, PRODIGI could have significant impact in clinical wound care by (1) reducing the complications associated with missed detection of bacterial infection under conventional practice, (2) facilitating image-guided wound sampling by targeted swabbing, and (3) monitoring wound healing and treatment response over time. As such, results from this study have been used to optimize the design of an on-going multicenter clinical trial aiming to validate our FL imaging platform for clinical wound care. AcknowledgmentsThe authors wish to thank Yaxal Arenas (Princess Margaret Cancer Centre, UHN) for her technical support with microbiology procedures. Funding was provided by the Canadian Institutes for Health Research (CIHR) and Cancer Care Ontario (CCO) to R.S.D. ReferencesP. G. BowlerB. I. DuerdenD. G. Armstrong,
“Wound microbiology and associated approaches to wound management,”
Clin. Microbiol. Rev., 14
(2), 244
–269
(2001). http://dx.doi.org/10.1128/CMR.14.2.244-269.2001 CMIREX 0893-8512 Google Scholar
K. F. CuttingR. White,
“Defined and refined: criteria for identifying wound infection revisited,”
Br. J. Community Nurs., 9
(3), S6
–S15
(2004). http://dx.doi.org/10.12968/bjcn.2004.9.Sup1.12495 1462-4753 Google Scholar
R. J. WhiteK. F. Cutting,
“Critical colonization—the concept under scrutiny,”
Ostomy Wound Manage., 52
(11), 50
–56
(2006). 0889-5899 Google Scholar
K. F. CuttingR. J. White,
“Criteria for identifying wound infection—revisited,”
Ostomy Wound Manage., 51
(1), 28
–34
(2005). 0889-5899 Google Scholar
G. DowA. BrowneR. G. Sibbald,
“Infection in chronic wounds: controversies in diagnosis and treatment,”
Ostomy Wound Manage., 45
(8), 23
–27
(1999). 0889-5899 Google Scholar
M. McGuckinet al.,
“The clinical relevance of microbiology in acute and chronic wounds,”
Adv. Skin Wound Care, 16
(1), 12
–23
(2003). http://dx.doi.org/10.1097/00129334-200301000-00011 1527-7941 Google Scholar
K. M. Guthrieet al.,
“Antibacterial efficacy of silver-impregnated polyelectrolyte multilayers immobilized on a biological dressing in a murine wound infection model,”
Ann. Surg., 256
(2), 371
–377
(2012). http://dx.doi.org/10.1097/SLA.0b013e318256ff99 ANSUA5 0003-4932 Google Scholar
Y. Moriwakiet al.,
“Molecular basis of recognition of antibacterial porphyrins by heme-transporter IsdH-NEAT3 of Staphylococcus aureus,”
Biochemistry, 50
(34), 7311
–7320
(2011). http://dx.doi.org/10.1021/bi200493h BICHAW 0006-2960 Google Scholar
M. M. Goiset al.,
“Susceptibility of Staphylococcus aureus to porphyrin-mediated photodynamic antimicrobial chemotherapy: an in vitro study,”
Lasers Med. Sci., 25
(3), 391
–395
(2010). http://dx.doi.org/10.1007/s10103-009-0705-0 LMSCEZ 1435-604X Google Scholar
Y. S. CodyD. C. Gross,
“Characterization of pyoverdin(pss), the fluorescent siderophore produced by Pseudomonas syringae pv. syringae,”
Appl. Environ. Microbiol., 53
(5), 928
–934
(1987). AEMIDF 0099-2240 Google Scholar
C. D. CoxP. Adams,
“Siderophore activity of pyoverdin for Pseudomonas aeruginosa,”
Infect. Immun., 48
(1), 130
–138
(1985). INFIBR 1070-6313 Google Scholar
V. Ivancicet al.,
“Rapid antimicrobial susceptibility determination of uropathogens in clinical urine specimens by use of ATP bioluminescence,”
J. Clin. Microbiol., 46
(4), 1213
–1219
(2008). http://dx.doi.org/10.1128/JCM.02036-07 JCMIDW 1070-633X Google Scholar
C. M. Sedgleyet al.,
“Influence of irrigant needle depth in removing bioluminescent bacteria inoculated into instrumented root canals using real-time imaging in vitro,”
Int. Endod. J., 38
(2), 97
–104
(2005). http://dx.doi.org/10.1111/iej.2005.38.issue-2 IENJEA 1365-2591 Google Scholar
W. D. McElroyH. H. SeligerE. H. White,
“Mechanism of bioluminescence, chemiluminescence and enzyme function in the oxidation of firefly luciferin,”
Photochem. Photobiol., 10
(3), 153
–170
(1969). http://dx.doi.org/10.1111/php.1969.10.issue-3 PHCBAP 0031-8655 Google Scholar
N. Dinjaskiet al.,
“Near-infrared fluorescence imaging as an alternative to bioluminescent bacteria to monitor biomaterial-associated infections,”
Acta Biomater., 10
(7), 2935
–2944
(2014). ABCICB 1742-7061 Google Scholar
K. E. LukerG. D. Luker,
“Applications of bioluminescence imaging to antiviral research and therapy: multiple luciferase enzymes and quantitation,”
Antiviral Res., 78
(3), 179
–187
(2008). http://dx.doi.org/10.1016/j.antiviral.2008.01.158 ARSRDR 0166-3542 Google Scholar
A. SatoB. KlaunbergR. Tolwani,
“In vivo bioluminescence imaging,”
Comp. Med., 54
(6), 631
–634
(2004). COMEFT 1532-0820 Google Scholar
“Consensus development conference on diabetic foot wound care. 7–8 April 1999, Boston, Massachusetts. American Diabetes Association,”
J. Am. Podiatr. Med. Assoc., 89
(9), 475
–483
(1999). http://dx.doi.org/10.7547/87507315-89-9-475 JAPAEA 8750-7315 Google Scholar
F. Annoniet al.,
“The effects of a hydrocolloid dressing on bacterial growth and the healing process of leg ulcers,”
Int. Angiol., 8
(4), 224
–228
(1989). INANEK 0392-9590 Google Scholar
D. G. ArmstrongL. A. Lavery,
“Evidence-based options for off-loading diabetic wounds,”
Clin. Podiatr. Med. Surg., 15
(1), 95
–104
(1998). Google Scholar
D. G. ArmstrongL. A. LaveryT. R. Bushman,
“Peak foot pressures influence the healing time of diabetic foot ulcers treated with total contact casts,”
J. Rehabil. Res. Dev., 35
(1), 1
–5
(1998). JRRDEC 0748-7711 Google Scholar
R. S. DaCostaH. AnderssonB. C. Wilson,
“Molecular fluorescence excitation-emission matrices relevant to tissue spectroscopy,”
Photochem. Photobiol., 78
(4), 384
–392
(2003). http://dx.doi.org/10.1562/0031-8655(2003)078<0384:MFEMRT>2.0.CO;2 PHCBAP 0031-8655 Google Scholar
R. S. DaCostaet al.,
“Autofluorescence characterisation of isolated whole crypts and primary cultured human epithelial cells from normal, hyperplastic, and adenomatous colonic mucosa,”
J. Clin. Pathol., 58
(7), 766
–774
(2005). http://dx.doi.org/10.1136/jcp.2004.023804 JCPAAK 0021-9746 Google Scholar
R. S. DacostaB. C. WilsonN. E. Marcon,
“New optical technologies for earlier endoscopic diagnosis of premalignant gastrointestinal lesions,”
J. Gastroenterol. Hepatol., 17
(Suppl.), S85
–S104
(2002). http://dx.doi.org/10.1046/j.1440-1746.17.s1.8.x JGHEEO 1440-1746 Google Scholar
B. KjeldstadT. ChristensenA. Johnsson,
“Porphyrin photosensitization of bacteria,”
Adv. Exp. Med. Biol., 193 155
–159
(1985). http://dx.doi.org/10.1007/978-1-4613-2165-1 AEMBAP 0065-2598 Google Scholar
W. K. Philipp-DormstonM. Doss,
“Comparison of porphyrin and heme biosynthesis in various heterotrophic bacteria,”
Enzyme, 16
(1), 57
–64
(1973). ENZYBT 0013-9432 Google Scholar
F. M. StoneC. B. Coulter,
“Porphyrin compounds derived from bacteria,”
J. Gen. Physiol., 15
(6), 629
–639
(1932). http://dx.doi.org/10.1085/jgp.15.6.629 JGPLAD 0022-1295 Google Scholar
C. J. PapasianP. J. Kragel,
“The microbiology laboratory’s role in life-threatening infections,”
Crit. Care Nurs. Q., 20
(3), 44
–59
(1997). http://dx.doi.org/10.1097/00002727-199711000-00006 CCNQEJ 0887-9303 Google Scholar
Y. Guoet al.,
“In vivo bioluminescence imaging to evaluate systemic and topical antibiotics against community-acquired methicillin-resistant Staphylococcus aureus-infected skin wounds in mice,”
Antimicrob. Agents Chemother., 57
(2), 855
–863
(2013). http://dx.doi.org/10.1128/AAC.01003-12 0066-4804 Google Scholar
H. S. Ammerlaanet al.,
“Eradication of methicillin-resistant Staphylococcus aureus carriage: a systematic review,”
Clin. Infect. Dis., 48
(7), 922
–930
(2009). http://dx.doi.org/10.1086/598925 CIDIEL 1058-4838 Google Scholar
BiographyYichao Charilie Wu is a biomedical engineer (BSc) currently pursuing his MSc degree in clinical engineering. He served as a medic in the Canadian military for 6 years. Iris Kulbatski, PhD is a medical researcher and writer. She holds a PhD in medical science from the University of Toronto, Toronto, Canada, and has extensive research and science communication experience. Her interests include translational research and science communication. Philip J. Medeiros is a postdoctoral fellow at the Princess Margaret Cancer Center, University Health Network, Toronto, Ontario. He obtained his MSc and PhD degrees from the University of Western Ontario in the Department of Medical Biophysics, London, Ontario. Azusa Maeda is a graduate student pursuing a PhD degree in the Department of Medical Biophysics at the University of Toronto, Toronto, Canada. She obtained her BSc degree in microbiology and immunology from McGill University, Montreal, Canada. Jiachuan Bu holds an MSChE from the University of West Virginia (2009). His current research interests include the development and evaluation of high-affinity molecular imaging probes for the optical detection of cancer at its earliest premalignant stage, as well as clearly distinguishing tumor and normal tissue during oncological image-guided surgery. Lizhen Xu received her PhD in the Department of Statistics at the University of Toronto in 2012. She is a research biostatistician and postdoctoral fellow of Dalla Lana School of Public Health, University of Toronto. She is currently a CIHR STAGE trainee. She develops and applies statistical methods to biological and clinical topics. Her research interest focuses on statistical modeling for multilevel longitudinal data, mixed effect modeling, zero inflated data, and Bayesian design to genetic studies. |