|
|
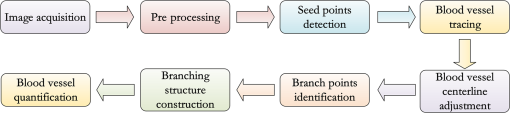
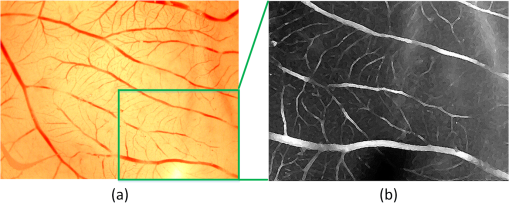
1.IntroductionAngiogenesis, the formation of new blood vessels, is an integral part of both normal developmental processes and numerous pathologies, ranging from tumor growth and metastasis to inflammation and ocular disease.1 Angiogenesis assays are used to test the efficacy of both pro- and anti-angiogenic agents.2,3 Among them, chick embryo chorioallantoic membrane (CAM) angiogenesis assay has been widely used for finding drugs targeting new blood vessel development in cancer research. The current CAM model gives us a way to quantify angiogenesis, which is used to evaluate the performance of blood vessel growth.4,5 This kind of assay uses the chick embryo as an animal model, and this model presents several advantages compared to mammal models, specifically, because the chick embryo is naturally immunocompetent, thus easily allowing mammal cell xenografts.6 The procedures are relatively simple, involving short experimental times and low costs. Meanwhile, the image analysis part, which contributed to improve the accuracy and efficiency of CAM assays,7,8 is still challenging because of the complexity of blood vessel branching structures. The traditional manual detection and measurement of blood vessels lacks efficiency, and is always limited to small regions of interest (ROIs) because of the high time consumption. To quantify angiogenesis more efficiently, especially in in vivo CAM assays, the automated quantification framework needs to be further improved to detect and remodel the blood vessel branching structures. Research on extracting linear structures in two-dimensional (2-D) images have already been applied for tracing similar branching structures such as neurons and retinal vessels, and can generally be divided into three categories according to the literature; the search-based approaches, the line–pixel detection approaches, and the direct exploratory tracing approaches. Search-based algorithms detect the line pixels using a live-wire segmentation paradigm.9–11 Starting from a user-specified initial point, these approaches search optimal paths from the initial point to all other pixels in the image, but these approaches need human interaction and are not fully automated. In line–pixel approaches, the local geometric properties of each pixel in the image is considered and the optimal pixels are linked to form the lines representing the linear structures.12–14 The heavy computational work of all pixel local geometric properties is the main drawback of these methods. In the last category, direct exploratory tracing algorithms automatically detect the initial points and extract the centerlines by iteratively sampling a short distance, then stop tracing until certain predefined conditions are satisfied.15,16 These approaches need fewer computations because only the geometric properties of the sampling points need to be calculated, but the tracing performance can be affected by low image quality or overly complicated branching structures. In order to provide an efficient scheme for the quantitative analysis of tree blood vessel structure in in vivo CAM assays, our framework made improvements based on some aspects of the direct exploratory tracing algorithm. First, to improve the image quality and enhance blood vessels for tracing, image preprocessing methods were applied to remove noises and enhance branching structures in the original images. Second, a direct exploratory tracing approach based on vector tracing was proposed to track blood vessels from automatically detected seed points, which are initial starting or ending points on or near the center line of a blood vessel segment. Third, to optimize the centerlines traced in the previous step, we proposed the blood vessel segmentation algorithm based the 2-D Rayburst method,17 which detects uneven diameters of blood vessels along centerlines from vector tracing. Fourth, “end” points of all detected blood vessel segments were classified into “branch” points and “terminal” points according to local neighborhoods, and were coded to construct an abstract model of the branching structure. Finally, statistical features extracted from labeled and segmented branching structures were provided as quantitative results of the image analysis. Experimental results showed that our algorithm has high accuracy in blood vessel detection and recognition validated by semimanual measurements, and also brings a fully automated method for CAM angiogenesis quantification with a high efficiency for in vivo applications. 2.Materials and MethodsDealing with the input microscopic images acquired from CAM assays, the processing and modeling parts were integrated into the automated computational framework. The workflow (Fig. 1) was then applied in quantification of the ICT inhibition effect on blood vessel growth, and a set of blood vessel growth indicators were extracted as outputs. 2.1.CAM Assay and Image AcquisitionICT (purity of 98.5%) was purchased from Shanghai Usea Biotech Company (Shanghai, China). The fertilized eggs were incubated at 37°C for eight days and divided into groups (eight eggs/group), then 100 μl of 0, 10, 20, or 40-mM ICT was added to the top of the CAM. After incubation for three more days, the CAM of each live egg was harvested and individually placed in six well plates. CAM blood vessels were captured using Motic Images Plus 2.0 (MediaCybernetics, Bethesda, Maryland) as 24-bits RGB images. The image resolution taken by the microscope was about . A set of blood vessel growth indicators including the blood vessel area were quantified using the proposed automated computational framework, such as a percentage of the total CAM area, blood vessel length, number of blood vessel segments, and other representative features. 2.2.Image PreprocessingInput microscopic images need to be preprocessed to remove noises in acquisition and to enhance branching structures. At first, the input RGB images were converted to a gray-scale format while retaining all information except the colors, which are redundant for the following pipeline processing. Usually, an image system generates three typical kinds of noises; shot noise, uneven illumination, and degradation. A median filter18 with a radius of 2 pixels erases shot noises efficiently by averaging the local areas of images. The uneven illumination is corrected by Gaussian smoothing with a sigma value set as 2 pixels, which is based on a low-pass filtering. We also employed the Lucy–Richardson deconvolution method19,20 to correct out of focus blurred images caused. Following the pipeline of image preprocessing, microscopy images with a low quality were enhanced for image analysis. Most of noises were erased and the edges of linear structures were sharpened compared to background (Fig. 2). Blood vessel structures from the original images are enhanced for the next tracing procedure. 2.3.Seed Points DetectionIn the proposed framework, the direct exploratory tracing algorithm automatically detects the initial points to start tracing blood vessels. One seed point detection method21 applied in neuron tracing was applied. We also used two steps including rough searching and screening to make improvements on removing redundant candidate points. In the rough search, an input image was divided into small blocks by grids, where horizontal lines and vertical lines in the image were considered. Ideally, the size of one block should be just a little larger than most of the blood vessels’ widths. In the pretests, we measured that the average width of blood vessels was about 12 pixels, and the thickest could be about 15 pixels, so one window formed by grids was normally set up as pixels in this case. By traversing each line horizontally and then vertically, local intensity maxima were detected along those grids. In practice, candidate points of local maxima would have a higher intensity than the median intensity of the image plus its standard deviation to ensure they are not parts of the background. Furthermore, candidate points must be screened to remove redundant detected points according to local signal-to-noise ratios (SNRs). As part of the blood vessel structure, candidate points would have a larger SNR than their neighborhood pixels in a local area. As the center of a block of pixels, candidate points with an SNR higher than a preset local threshold for the neighborhoods using Otsu’s algorithm22 would be selected as seed points. After finishing two procedures for seed point detection, blood vessel segments were traced and labeled from seed points in the following steps. 2.4.Blood Vessel Tracing and LabelingThe main purpose of tracing and labeling is to find the best track along a blood vessel centerline from a seed point. In order to make full use of the detected seed points and to get a balance between accuracy and speed in tracing for in vivo CAM angiogenesis assay, we proposed an exploratory algorithm based on a Hessian matrix23,24 in the tracing of blood vessels. The basic idea of the exploratory algorithm is to find an optimal path along the linear structure by following its natural trend in the image. Therefore, the key is to estimate local directions of the path at each small interval . Then, points on the track could be defined as Starting from a seed point, the centerline is traced following specific rules and stops when certain conditions are satisfied. Assuming a point is reached and the next point is going to be determined after a quite small interval, the direction from to is estimated as shown in Fig. 3. Fig. 3Illustration of centerline tracing. , , and are traced centerline points by interval distance , , , and are directions estimated at each points using Hessian matrix. 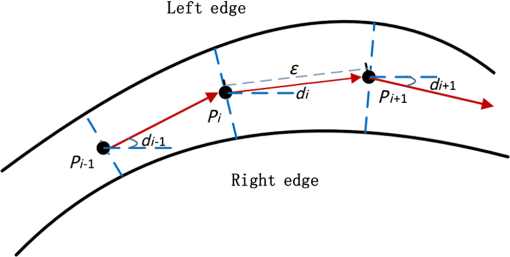 To proceed with the tracing at small intervals, we define the direction of each tracing point derived from the eigen-analysis of a Hessian matrix, i.e., the normalized direction of the eigenvector from the Hessian matrix. Based on Eq. (1), the next tracing point is estimated, and the tracing centerline of a blood vessel is completed by reaching the other seed point and linking all the estimated tracing points between the two seed points. 2.5.Blood Vessel Edge Detection and Centerline AdjustmentSince the main purpose vector tracing part of our proposed algorithm is to improve the tracing efficiency with a rapidly estimated centerline, in order to quantify the indicators of blood vessel growth more accurately, more precise blood vessels edges and centerlines need to be detected. A 2-D cast rays method was used to make an adjustment on the traced blood vessel path, and even more importantly, to estimate diameters along blood vessels for the area calculation. The casting rays method (Fig. 4) used in this paper is similar to Rayburst sampling, an algorithm for both 2-D and three-dimensional (3-D) automated shape analyses, which was first proposed by Rodriguez et al.17 The centerline extraction then gets an adjusted representation of the blood vessel with a centerline and radii in different parts. Since the blood vessels always have smooth boundaries except at the branch points, Rayburst performs well for getting accurate edges by casting rays. Fig. 4Centerline adjustment using the casting ray method. Blue circles represent the adjusted diameters with casting rays in different areas, whereas the green curve represents the adjusted centerline of the blood vessels. Circles are only shown in part of one blood vessel for a clear view. 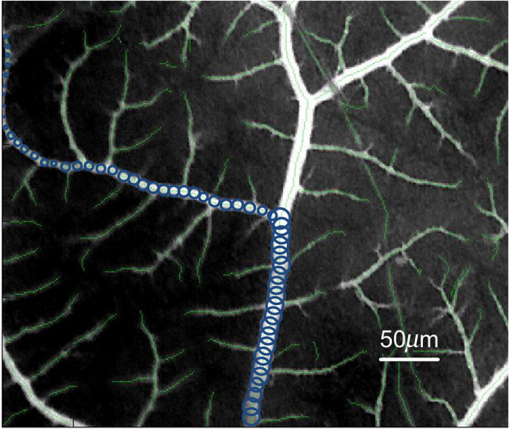 In addition to length, blood vessel width is an important feature of angiogenesis in two aspects. First, blood vessel width shows primary and secondary orders for identifying main and minor blood vessels, especially when only part of the vascular structure is shown in a small ROI, and is also helpful for building the branching structure in the next step. Second, the distribution of width along a single-blood vessel enables the drawing of a more detailed map of the vessel, and could be a useful feature for observing blood vessel growth in a timely manner. With the blood vessel length and width being measured, the area of a blood vessel could be estimated. Rayburst used the same small interval as the tracing as shown in Fig. 3. In each segment, the blood vessel in that section could be estimated as a trapezium which has two edges with the lengths as diameters of the two adjacent Rayburst circles, and the height as . Therefore, the total area of a single-blood vessel is estimated as and could be further simplified as where and are the area, length and a blood vessel, respectively. means the ’th segment of Rayburst starting from seed point 0 to end point . and are the diameter and radius of the Rayburst circle at the ’th segment. In practice, the adjustment process optimized the blood vessel branching structure for an in-depth analysis.2.6.Branch Points Identification and Branching Structure ModelingBased on the tracing results, the skeleton of a blood vessel tree made of centerline segments was extracted. Except for the structural measurements such as length, width, and other factors, research on CAM angiogenesis also concern structural changes during blood vessel growth, which is based on the in-depth analysis of the extracted branching structures. In such a structure, the “end” point is used to indicate points at the two ends of a blood vessel segment, which could be a seed point or the point where the centerline tracing stops. The “end” points can be further classified into “branch” points where blood vessels bifurcate, and “terminal” points where blood vessels end. Like a hierarchical tree, “children” branches protrude from a “parent” branch at branch points in the structure. Therefore, the branch points of each branch need to be identified to construct an abstract tree representing the complicated structure. Two steps were taken to automatically achieve the tree modeling, including branch and terminal point identification and coding following their natural hierarchical sequences. To accelerate the identification process, a set of well-defined connected domain models were adopted. A branch point is determined to be where the checked point has three neighbor points around in its eight-connected field, and an end point of a blood vessel segment with only one neighbor point in its connected domain is identified as a terminal point. There is never an obvious main blood vessel in CAM images because of the complex structure and the small focused region, so it is difficult to determine parents and children braches in the complex multilevel branching structures. We proposed a “root to leaves” identification strategy considering the width of the connected branches, where the “root” points are defined as the two terminal points located on the widest blood vessel which is also the main blood vessel. The main blood vessel is then determined by finding a way between both “root” points along other branch points. The detailed branch coding process (Fig. 5) is described as follows:
Fig. 5A hierarchical tree representing the abstract structure of an individual blood vessel in a CAM image. (a) Image in part of CAM region with a blood vessel tree labeled, where centerlines are represented as light green, branch points are illustrated as yellow dots, and terminal points as red dots. The abstract structure in (b) shows the hierarchical tree of main blood vessel between terminal points 0 to 1, where one of its children branches extending from point 10 is represented in detail. 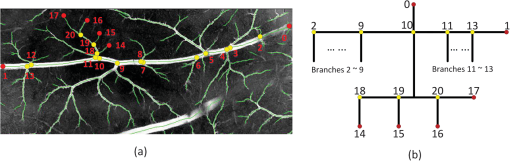 After finishing the modeling part, every end point of the blood vessel segments was coded and located according to its natural order. The hierarchical tree provided an insightful new way to look at the branching structure and the growth of blood vessels. 3.Results3.1.Comprehensive Measurements of CAM Angiogenesis in ICT Inhibition AssayTo evaluate the proposed framework in quantifying from CAM angiogenesis images, the automated quantification was applied in a set of CAM assays testing the ICT inhibition effects. The ICT used is a hydrolytic form of Icaritin purified from the genus Epimedium, a traditional Chinese herbal medicine,25 and has the possibility of multiple targets and multiple mechanisms of action through Epimedium preparations and their purified compounds.26 However, it is unknown whether ICT exerts inhibition effects on angiogenesis in any type of cancer. Following the protocols in Ref. 27, the proposed quantification framework was validated and applied to test the effects of ICT on CAM angiogenesis. To validate the proposed framework in a more comprehensive way, quantification of CAM angiogenesis images generated multiple features for in-depth analysis. We defined a series of criteria for feature calculations as shown in Table 1, which provide blood vessel growth indicators for more comprehensive understandings. Table 1Feature set of proposed chorioallantoic membrane (CAM) angiogenesis quantification framework.
Table 1 includes key indicators of blood vessel growth in CAM angiogenesis assay. Researchers may figure out the overall blood vessel growth under different conditions by taking all those features into consideration. Our computational framework automatically generates those features without manual labeling or measurement as required by popular image analysis software such as ImageJ. The outputs were further validated in quantification of the ICT inhibition effects of multiple concentrations. Except for the multiple outputs provided, the automated quantification also improved the analysis speed compared to semimanual measurements. Running in MATLAB, the computational work was performed using a desktop with a 2.4-GHz CPU and 4-Gbyte memory. Dealing with CAM microscopic images, it took about 55 s to complete the analysis of all blood vessels, which made real-time analysis possible for various in vivo experiments. 3.2.Validation of Quantification Results in ICT Inhibition Assay on CAM AngiogenesisTo knock-down the reproductive ability or kill the tumor, it is key to suppress the blood vessel growth close to the tumor area, which then blocks the blood supply. CAM assay was performed in 8-day fertilized eggs to quantify the suppression effect of ICT on the angiogenesis. Microscopic images were captured by Motic Images Plus 2.0, then quantified using the proposed automated framework. Furthermore, experts in our lab manually measured some blood vessels growth factors using Image Pro software (MediaCybernetics, Bethesda, Maryland) as the control data for validating the quantification performance. In Table 2, comparison between the results of the automated processing and the semimanual approach was conducted by simultaneously testing the ICT inhibition effects on angiogenesis. Table 2Average values and standard deviations of detailed CAM feature set.
The direct view of images under different conditions in Fig. 6 shows that blood vessel lengths gradually decrease when increasing the ICT dose, and blood vessels marked with green lines become shorter from left to right, which indicates a possible ICT inhibition on the angiogenesis took effect along with the concentrations. In order to validate our hypothesis, the features in Table 1 were extracted based on tracing and segmentation results. Detailed average values and standard deviations of the measurements are shown in Table 2. Fig. 6Validation of Icaritin inhibition effects on angiogenesis in the CAM assay. Panels (a) to (d) are images of the blood vessels of each CAM after treatment with 100 ml of 0, 10, 20, or 40 mM ICT added to the top of CAM. Panels (e) to (h) show blood vessels labeled with green centerlines under multiple conditions respectively. 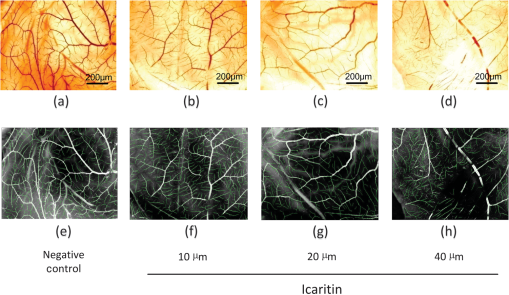 Among the feature values in Table 2, a comprehensive understanding of the relationship between the ICT inhibition effects and concentrations could be drawn. Four of the most popular features in related cancer research were taken for further evaluation. Statistical analysis of the indicated features including vessel area, vessel length, and the number of vessel segments is illustrated in Fig. 7. Fig. 7Statistical results of ICT inhibition effects in CAM assays, in which black bars are the results from semimanual measurements and gray bars are the results generated by the proposed computational framework. (a) Percentage of blood vessel area in the CAM region, (b) number of total blood vessel segments, (c) total blood vessel length, and (d) average blood vessel length in negative control group and ICT treated groups with concentrations of 10, 20, and 40 μm from left to right. 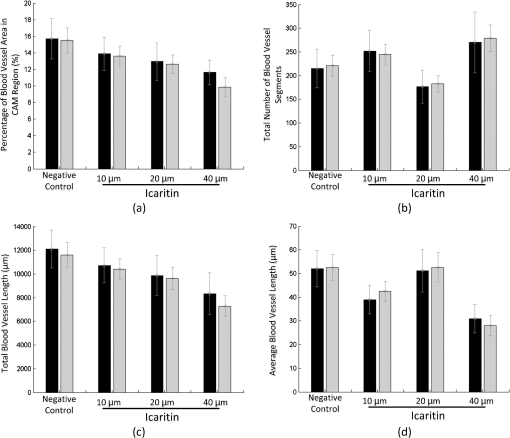 As shown in Fig. 7, blood vessel growth under different conditions was compared in multiple ways. First, the automated computational results were compared with results measured by human experts. In the validation part, the two bars of different colors under each of the conditions are quite close to each other in Fig. 7. Taking the semimanual measurements as control data, the error rates of the automated method are always less than 15%, which indicates the proposed framework has a high accuracy in CAM blood vessel quantification. Meanwhile, most of the error bars from the automated computational results are longer than their corresponding semimanual results, suggesting that the automated framework has a larger computational error but is still in a reasonable range for practical use. After validating the accuracy of the proposed method, it could be further applied in testing the inhibition effects of ICT on CAM angiogenesis. As the most important key indicators of blood vessel growth, the downtrend of “percentage of blood vessel area in CAM region” [Fig. 7(a)] and “total blood vessel length” [Fig. 7(c)] showed that the newly formed vessels were reduced in an ICT dose-dependent manner. Quantification results of those two features also demonstrated that ICT significantly decreased the vessel area and total vessel length with increasing concentrations, suggesting that ICT possesses the capacity for anti-angiogenesis, which is likely to help tumor shrinkage in vivo. There was an exception in that “total number of blood vessel segments” [Fig. 7(b)] does not decrease following the dose-dependent manner. Statistical results here showed that the ICT inhibition effect was too strong to suppress most of blood vessels’ growth, which caused a lot of poorly developed blood vessels to appear as shown in Fig. 6(d) and marked in Fig. 6(h). Meanwhile, the semimanual measurement results in Ref. 27 only showed branched points without counting the branches based on the tree modeling. In addition, more minor blood vessel segments were detected by application of the proposed preprocessing part in this paper. Therefore, the branch numbers of the ICT treated group are significantly high in Table 2 and Fig. 7, which were based on recalculating poorly developed branches with both the automated and semimanual ways. That caused the total number of segments to increase instead of decreasing under the condition of a ICT dosage. Another exception could be found in the “average blood vessel length” in Fig. 7(d), which is a feature derived by both “total blood vessel length” and “total number of blood vessel segments.” Although the total length of the ICT-treated group followed the dose-dependent manner, the total number of segments decreased much faster than the total length as shown in Figs. 6(c) and 6(g). Unlike the total segments number of the ICT-treated group, there were not so many poorly developed blood vessels in the ICT-treated group. Therefore, the division increased under this condition, which made the “average blood vessel length” treated by the ICT higher than that of other concentrations. Generally, statistical results proposed by automated calculation conformed to the semimanual quantification results in Ref. 27, and were much more efficient. With analysis of the CAM microscopic images, the most exciting finding proposed by our framework was that ICT has overall inhibition effects on angiogenesis in a dose-dependent manner. Since the angiogenesis plays an important role in the growth and progression of solid tumors,28 with the significant effect of anti-angiogenesis, the ICT anti-tumor effect would be further enhanced on its own or in combination with other techniques in vivo. 4.Conclusions and Discussions4.1.ConclusionsBlood vessel angiogenesis mainly occurs in embryo development, wound repair and inflammation, especially tumor growth or metastasis. The CAM assays, as some of the most widely used techniques for observing blood vessel angiogenesis, have many advantages, such as well-designed protocols and visible results. However, the quantification methods of CAM microscopic images developed far behind the application of CAM assays.29,30 Some software were employed to semimanually quantify blood vessel angiogenesis in small ROIs, and most of the automated linear structures extraction algorithms were applied for tracing similar structures like neurons and retinal vessels, but not for CAM blood vessels. Because of its complex branching structures, a fully automated and accurate system for the CAM blood vessel quantification is highly desired. To solve this problem, our algorithm focused on labeling centerlines, the segmentation of branch structures, and then modeling of the blood vessels from image analysis results. Dealing with the CAM microscopic images, a fully automated framework was proposed to quantify and analyze the blood vessel tree in-depth. The framework contains image preprocessing, centerline extraction and segmentation of the blood vessels, tree modeling, and branch order analysis. The centerlines of CAM blood vessels are extracted with exploratory approach and then adjusted by applying the Rayburst method, which considers the balance between speed and accuracy. Meanwhile, widths along each of the blood vessels were also recalculated along the centerlines, which was also an important improvement of our framework and provides more detailed information of blood vessel growth. By identifying “parents” and “children” branches by traversing all branch and terminal points in the image, the abstract tree of blood vessels was built. That gave a comprehensive view of the vascular network and its growth, which could be even more helpful for related research as well as for the quantification part. Based on the image processing and modeling results, morphological features including geometrical and topological properties are quantitatively measured. In the validation part, we employed the framework in the ICT inhibition assays in tumor research. Our automated labeling and analysis algorithms efficiently extracted multiple features for quantifying blood vessel growth. Experimental results showed that the newly formed vessels were reduced in an ICT dose-dependent manner, which pointed out a new possible way to suppress the blood vessel growth close to the tumor area and then kill the tumor. The proposed microscopic image processing framework is fully automated with no human interaction required, which avoids possible subjective errors. Except for the branching structure of the whole images, single or multiple blood vessels may also be selected for more accurate quantification or structural analysis. 4.2.DiscussionsIn addition to the advantages brought by the proposed algorithm, the proposed framework could be further improved in some aspects in the future. First, the image quality might be unstable in acquisition, and broken lines of blood vessels may occur, especially in the high ICT dose plates [see Fig. 6(d) and 6(h)]. An effective linking model is needed to treat such cases. The linking strategy in Ref. 31 might be adapted to low-quality CAM images, which considers the directions and distance between two breaking points. Meanwhile, in the CAM region, blood vessel widths and branch orders also need to be considered to link the correct breaking points together. Second, due to the limitation of 2-D microscopic acquisition, blood vessels in 3-D might be overlapped in the projection images, which may bring errors in branch point identification and affect the “average” features in quantification. It is quite difficult to identify projected blood vessels which are not in the surface plane. Most of the projected blood vessels on other planes may have lesser contrast as shown in figures with the original CAM images in this paper. A new model to erase those overlapped effects, based on contrast statistical differences, would be very useful. Third, both veins and arteries develop in chicken embryo, but it is still a problem to identify them respectively. Veins and arteries might be classified based on some observed and pathological criterions such as contrast and width differences. It might be another challenging work in CAM image analysis in the future. Hierarchical tree modeling is critical for analyzing CAM angiogenesis more comprehensively. The branching structure of CAM blood vessels has more branch layers than that of neurons or retinal vessels, which inherently brings more difficulty in hierarchical tree construction. The proposed framework uses “branch points” and “terminal points” to determine blood vessel segments and their hierarchical relations, which may be further improved in the modeling of other branching structures in biomedical applications. In summary, as a useful technique for research of in vivo vasodilatory effects, the presented computational framework proved the ICT anti-tumor effects after validation. Automated quantitative analysis of CAM is a useful tool for research on the pathogenetic mechanism and multiple factors of blood vessel angiogenesis. The application of the proposed framework will be helpful for both CAM-related pathological study and analysis of other complicated linear structures in microscopic images. AcknowledgmentsThe work is a joint advanced research project co-operated by Fujian Normal University and First Affiliated Hospital, Fujian Medical University. The authors also would like to thank every member in the laboratories of cooperation in Xiamen University and UF Shands Cancer Center. ReferencesL. M. KirchnerS. P. SchmidtB. S. Gruber,
“Quantitation of angiogenesis in the chick chorioallantoic membrane model using fractal analysis,”
Microvasc. Res., 51
(1), 2
–14
(1996). http://dx.doi.org/10.1006/mvre.1996.0002 MIVRA6 0026-2862 Google Scholar
R. Auerbachet al.,
“Angiogenesis assays: a critical overview,”
Clin. Chem., 49
(1), 32
–40
(2003). http://dx.doi.org/10.1373/49.1.32 CLCHAU 0009-9147 Google Scholar
R. Auerbachet al.,
“Angiogenesis assays: problems and pitfalls,”
Cancer Metastasis Rev., 19
(1–2), 167
–172
(2000). http://dx.doi.org/10.1023/A:1026574416001 CMRED4 0167-7659 Google Scholar
M. Bahramsoltaniet al.,
“Quantitation of angiogenesis and antiangiogenesis in vivo, ex vivo and in vitro - an overview,”
Altex, 26
(2), 95
–107
(2009). ALTCDD Google Scholar
C. N. Doukaset al.,
“Automated angiogenesis quantification through advanced image processing techniques,”
in Conf. Proc. IEEE Engineering in Medicine and Biology Society,
2345
–2348
(2006). Google Scholar
D. Ribatti,
“Chick embryo chorioallantoic membrane as a useful tool to study angiogenesis,”
Int. Rev. Cell Mol. Biol., 270 181
–224
(2008). http://dx.doi.org/10.1016/S1937-6448(08)01405-6 IRCMAH 1937-6448 Google Scholar
M. NguyenY. ShingJ. Folkman,
“Quantitation of angiogenesis and antiangiogenesis in the chick embryo chorioallantoic membrane,”
Microvasc. Res., 47
(1), 31
–40
(1994). http://dx.doi.org/10.1006/mvre.1994.1003 MIVRA6 0026-2862 Google Scholar
D. Ribattiet al.,
“New model for the study of angiogenesis and antiangiogenesis in the chick embryo chorioallantoic membrane: the gelatin sponge/ chorioallantoic membrane assay,”
J. Vasc. Res., 34
(6), 455
–463
(1997). http://dx.doi.org/10.1159/000159256 JVREE9 1018-1172 Google Scholar
W.A. BarrettE.N. Mortensen,
“Interactive live-wire boundary extraction,”
Med. Image Anal., 1
(4), 331
–341
(1997). http://dx.doi.org/10.1016/S1361-8415(97)85005-0 MIAECY 1361-8415 Google Scholar
A. X. FalcãoJ. K. UdupaF. K. Miyazawa,
“An ultra-fast user-steered image segmentation paradigm: livewire on the fly,”
IEEE Trans. Med. Imaging, 19
(1), 55
–62
(2000). http://dx.doi.org/10.1109/42.832960 ITMID4 0278-0062 Google Scholar
E. Meijeringet al.,
“Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images,”
Cytometry A, 58
(2), 167
–176
(2004). http://dx.doi.org/10.1002/(ISSN)1097-0320 1552-4922 Google Scholar
L. WangT. Pavlidis,
“Direct gray-scale extraction of features for character recognition,”
IEEE Trans. Pattern Anal. Mach. Intell., 15
(10), 1053
–1067
(1993). http://dx.doi.org/10.1109/34.254062 ITPIDJ 0162-8828 Google Scholar
S. Chaudhuriet al.,
“Detection of blood vessels in retinal images using two-dimensional matched filters,”
IEEE Trans. Med. Imaging, 8
(3), 263
–269
(1989). http://dx.doi.org/10.1109/42.34715 ITMID4 0278-0062 Google Scholar
I. S. KweonT. Kanade,
“Extracting topographic terrain features from elevation maps,”
Comput. Vis. Graph Image Process. Image Understanding, 59
(2), 171
–182
(1994). http://dx.doi.org/10.1006/cviu.1994.1013 0734-189X Google Scholar
R. PolliG. Valli,
“An algorithm for real-time vessel enhancement and detection,”
Comput. Methods Programs Biomed., 52
(1), 1
–22
(1997). http://dx.doi.org/10.1016/S0169-2607(96)01773-7 CMPBEK 0169-2607 Google Scholar
A. Canet al.,
“Rapid automated tracing and feature extraction from live high-resolution retinal fundus images using direct exploratory algorithms,”
IEEE Trans. Inf. Technol. Biomed., 3 125
–138
(1999). http://dx.doi.org/10.1109/4233.767088 ITIBFX 1089-7771 Google Scholar
A. Rodriguezet al.,
“Rayburst sampling, an algorithm for automated three-dimensional shape analysis from laser scanning microscopy images,”
Nat. Protoc., 1
(4), 2152
–2161
(2006). http://dx.doi.org/10.1038/nprot.2006.313 NPARDW 1750-2799 Google Scholar
T. N. SunY. Neurvo,
“Detail-preserving median based filters in image processing,”
Pattern Recognit. Lett., 15
(4), 341
–347
(1994). http://dx.doi.org/10.1016/0167-8655(94)90082-5 PRLEDG 0167-8655 Google Scholar
L. Lucy,
“An iterative technique for the rectification of observed distributions,”
Astron. J., 79
(6), 745
(1974). http://dx.doi.org/10.1086/111605 ANJOAA 0004-6256 Google Scholar
W. H. Richardson,
“Bayesian-based iterative method of image restoration,”
J. Opt. Soc. Am., 62
(1), 55
–59
(1972). http://dx.doi.org/10.1364/JOSA.62.000055 JOSAAH 0030-3941 Google Scholar
Y. Zhanget al.,
“Automated neurite extraction using dynamic programming for high-throughput screening of neuron-based assays,”
NeuroImage, 35
(4), 1502
–1515
(2007). http://dx.doi.org/10.1016/j.neuroimage.2007.01.014 NEIMEF 1053-8119 Google Scholar
N. Otsu,
“A threshold selection method from gray-level histograms,”
Automatica, 11
(285–286), 23
–27
(1975). Google Scholar
J. Lowellet al.,
“Measurement of retinal vessel widths from fundus images based on 2-D modeling,”
IEEE Trans. Med. Imaging, 23
(10), 1196
–1204
(2004). http://dx.doi.org/10.1109/TMI.2004.830524 ITMID4 0278-0062 Google Scholar
Y. Xuet al.,
“An improved algorithm for vessel centerline tracing in coronary angiograms,”
Comput. Methods Programs Biomed., 88
(2), 131
–143
(2007). http://dx.doi.org/10.1016/j.cmpb.2007.08.004 CMPBEK 0169-2607 Google Scholar
P. Shenet al.,
“Taxonomic, genetic, chemical and estrogenic characteristics of Epimedium species,”
Phytochemistry, 68
(10), 1448
–1458
(2007). PYTCAS Google Scholar
H. WuE. J. LienL. L. Lien,
“Chemical and pharmacological investigations of Epimedium species: a survey,”
Prog. Drug Res., 60 1
–57
(2003). http://dx.doi.org/10.1007/978-3-0348-8012-1 FAZMAE 0071-786X Google Scholar
J. Honget al.,
“Icaritin synergistically enhances the radiosensitivity of 4T1 breast cancer cells,”
PLoS One, 8
(8), e71347
(2013). http://dx.doi.org/10.1371/journal.pone.0071347 1932-6203 Google Scholar
D. HanahanJ. Folkman,
“Patterns and emerging mechanisms of the angiogenic switch during tumorigenesis,”
Cell, 86
(3), 353
–364
(1996). http://dx.doi.org/10.1016/S0092-8674(00)80108-7 CELLB5 0092-8674 Google Scholar
C. A. Statonet al.,
“Current methods for assaying angiogenesis in vitro and in vivo,”
Int. J. Exp. Pathol., 85
(5), 233
–248
(2004). http://dx.doi.org/10.1111/j.0959-9673.2004.00396.x IJEPEI 1365-2613 Google Scholar
C. A. StatonM. W. ReedN. J. Brown,
“A critical analysis of current in vitro and in vivo angiogenesis assays,”
Int. J. Exp. Pathol., 90
(3), 195
–221
(2009). http://dx.doi.org/10.1111/j.1365-2613.2008.00633.x IJEPEI 1365-2613 Google Scholar
G. Xionget al.,
“Automated neurite labeling and analysis in fluorescence microscopy images,”
Cytometry A, 69
(6), 494
–505
(2006). http://dx.doi.org/10.1002/(ISSN)1552-4930 1552-4922 Google Scholar
BiographyPeng Shi is an assistant professor at Fujian Normal University. He received his BS and MS degrees in computational fluid dynamics from Harbin Engineering University in 2001 and 2004, respectively, and his PhD degree in applied computer science from the Institute of Automation, Chinese Academy of Sciences, in 2008. His current research interests include biomedical image processing, microscopic image analysis, and pattern recognition. He is a member of IEEE. Jinsheng Hong is an associate professor at the First Affiliated Hospital, Fujian Medical University. He received his MS degrees in oncology from Fujian Medical University in 2005, and his PhD degree in medicine from Fujian Medical University in 2013. His current research interests include radiotherapy of head and neck cancer and lung cancer, radiation injury and radiation sensitivity. Yue Huang received a BS degree from the Department of Electrical Engineering, Xiamen University, and PhD degree from Department of Biomedical Engineering, Tsinghua University, Beijing, China, in 2005 and 2010, respectively. Since 2010, she has been an assistant professor with Xiamen University, Xiamen, China. Her main research interests include sparse signal representation, and machine learning. Zhenhuan Zhang is a research scientist at the University of Florida. He received his BS and MD degrees in Chongqing Medical University in 1993 and his PhD degree in endocrine oncology in Nagoya City University in 2006 in Japan. His current research interests include cancer radiation enhancers and mediators. He is member of RRS and AAAS. Mei Zhang is an assistant scientist in the Department of Radiation Oncology at the University of Florida. She graduated from Harbin Medical University and received her postdoctoral training in University of Rochester. Her primary research interest is the tumor angiogenesis and anti-tumor immunity. Lurong Zhang is a professor at the First Affiliated Hospital, Fujian Medical University, and the University of Florida. She received her MD degrees from Fujian Medical University in 1982, and her PhD degree from Second Medical College, Shanghai, China, in 1989. Her current research interests include immunotherapy of cancer, radiation injury and radiation sensitivity. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||