|
|
1.IntroductionRaman spectroscopy is an optical measurement technique that enables the retrieval of sample molecular information, based on the inelastic scattering of light.1 As it is based purely on an optical process, it has the advantages of being noninvasive and not requiring the insertion of any dye for measurement, which makes it ideal for biological samples which can be measured nondestructively and without prior processing. Raman spectroscopy has, therefore, been extensively used as a characterization tool, where the spectral information acquired from locations in the sample can be employed to extract information, which then allows the mapping of the molecular content of cells and/or tissues, with extension to assess the viability of cell lines as models for primary cells2 or to measure molecular changes during cell death processes.3 It has also been employed to discriminate between species, such as different bone cell types,4 or to separate differentiated cells from embryonic ones,5 detect cancer cells6,7 or tissues,8 or analyze blood samples in situ.9 Since Raman spectroscopy extracts molecular signatures based on excitation in the visible or near-infrared light range, it shares the spatial resolution benefits of light microscopy so that label-free images based on molecular contrast can be obtained by scanning the excitation beam through the sample.10,11 The measurement becomes, in this case, a three-dimensional data set, containing both spectral and spatial (x, y) information from imaging. There is, thus, a duality in the treatment of the information. Strictly spectroscopic analysis, based on peak assignments or multivariate analysis, can be carried out with additional imaging analysis, based on the treatment of specific spectral band contrasts and/or the precise location of specific molecules. This approach has been applied, for instance, to monitor molecular changes during apoptosis,12 for the observation of malignant cells in tissue sections,13 or to discriminate cancerous cells at a subcellular level.14 However, the Raman process has the drawback of being fairly inefficient, so that the amount of signal retrieved from biological samples is rather low, significantly limiting the measurement speed that can be achieved. This can become critical, especially in imaging cases where the measurement requires scanning within a whole region, in contrast to pure spectroscopic point measurements. As experiments are usually limited by total laser exposure, the detector noise becomes significant when spreading the signals out over a large detector array as is done in Raman imaging.11 One way to circumvent this issue is to turn toward more efficient or enhanced processes, such as nonlinear optical responses15 [coherent anti-Stokes scattering16 (CARS) or stimulated Raman scattering17], but often at the cost of spectral range or resolution unless refined excitation schemes are employed.18 Another process is relying on surface-enhanced Raman scattering,19 which relies on the plasmonic resonance of metal nanoparticles, which can be employed in imaging, for instance, by tracking particle trajectories.20 Another approach consists of adding another imaging modality which can be employed to target specific regions, which can then be measured with Raman spectroscopy without needing the entire slow raster scan of the sample. This method has been employed in numerous applications where the selection of the region to be measured by the Raman channel are first identified, for example, with autofluorescence21 or CARS imaging22 for ex vivo section measurements, or with white light reflectance23 or optical coherence tomography24 for tissue observation, or two-photon autofluorescence25 or simultaneous quantitative phase microscopy (QPM)26 for cell imaging. Fluorescence has also been employed in this context, but the overlap between the fluorescence and Raman emission spectra often prevents the measurement of stained sample.27 If the additional mode provides enough information about the sample morphology compared to Raman spectral imaging, it may actually be advantageous in terms of useful sample information to forgo the full Raman imaging and instead collect the highest-quality Raman point spectra for chemical evaluation, while monitoring the sample morphology with the additional imaging mode. Therefore, we present here a hybrid approach where the Raman measurement is performed in point mode for optimum quality and higher throughput, coupled with QPM to provide detailed spatial imaging information. We also propose to exploit the spatial information from the phase imaging to define a target region in the observed sample, inside which the Raman excitation beam can be rapidly scanned, so that the resulting spectrum measures a whole region of the cell. By choosing the Raman scan region from the phase image, we include much of the ensemble of molecules which are key to discrimination and diagnosis of cellular reactions, avoiding the possibility that the point spectrum is emitted by specific molecules in the small measurement volume which are not necessarily representative of the cellular state. In other words, the best signal-to-noise ratio (SNR) Raman measurements come from single point measurements, but these may be unduly influenced by local conditions in the cell. By scanning a region of the cell chosen from the phase image and optically binning or descanning the signal beam so that all signal photons are aggregated at the detector, we gain the advantages of single point detection but retain imaging information through QPM. The simultaneous imaging information, the high SNR Raman measurements, and the low dependence on local variations in the sample, make this approach, denoted hybrid Raman imaging, ideal for robust spectroscopic analysis. To demonstrate this, we first assess the information content of spectra measured on a population of cells for the fixed-point and hybrid scanning approaches by employing estimators for the variation and robustness of the data. We then quantitatively compare three types of measurements in terms of their capability to spectrally discriminate between two different macrophage-like cell lines (J774A.1 and Raw264.7) using multivariate analysis. The analysis is based on (1) spectra extracted from Raman images, (2) spectra from point measurements with the beam parked in the cell, as is standard, and (3) spectra measured where our proposed hybrid imaging is used to select a measurement region and the signal is descanned before the detector. Measurements are performed on live cells, which are standard for both image and point-mode Raman spectral measurements in the literature. 2.Materials and Methods2.1.Cell CultureMacrophage-like cell lines J774A.1 and Raw264.7 are cultured on 5 cm culture dishes (BD Biosciences, San Jose, California) and immersed in Dulbecco’s modified Eagle’s medium (Nacalai Tesque, Kyoto, Japan) supplemented with 10% fetal bovine serum (Nacalai Tesque) and incubated at 37°C in a humidified atmosphere with 5% . One day before experiments, cells are trypsinized with a solution containing 0.25% trypsin and 1 mM ethylenediaminetetraacetic acid (Nacalai Tesque) for 5 to 10 min at 37°C to detach them from the plastic surface, and plated on 3.5 cm quartz-bottom dishes (Fine Plus International, Kyoto, Japan) containing cultured medium and incubated as above. Just before measurements, the culture medium is replaced with a phosphate buffer saline (Nacalai Tesque) solution supplemented with D-glucose (5 mM) and (2 mM) by washing the dish three to four times. Cells are then observed at room temperature. 2.2.Raman Acquisition and ImagingAll measurements are performed with a Raman imaging setup described in detail in Ref. 26. Briefly, cells are observed on an inverted microscope with a objective (NA 1.27, water immersion, Nikon, Tokyo, Japan). The excitation is performed with a continuous wave laser at 532 nm (Verdi V-6, Coherent, Santa Clara, California), with a power density of . A dual-axis galvano-mirror placed in the descanned region [GM2, see Fig. 2(a)] enables the selection of the location of measurement for point excitation. For imaging, the measurement is performed in slit-scanning conditions, i.e., the signal emitted by a whole line is measured during one frame acquisition [see Fig. 2(a)], enabling parallel detection. The excitation line is generated by rapidly scanning the laser spot (at 100 Hz) in the vertical direction with the galvano-mirror located outside the descanned region of the microscope (GM1) to form a line on both the sample and the detector, leading to an average power density of . The collected backscattered light is separated from the excitation laser with a long-pass dichroic mirror and sent into a 500 mm focal length Czerny-Turner spectrometer (Shamrock, Andor Technology, Belfast, Northern Ireland). The light dispersed by the grating () is then measured with a low-noise CMOS camera (Orca-Flash 4.0, Hamamatsu Photonics, Hamamatsu, Japan) with an exposure time of 3 s. Quantitative phase images are recorded with an interferometric setup based on off-axis digital holography.28 Acquisition and reconstruction of the holograms are performed according to the description given in Ref. 26. 2.3.Spectra PreprocessingBefore analysis, all spectra are baseline corrected by employing an estimated curve at large intervals based on low quantile values (i.e., it has a high probability of being smaller than the signal), which is then computed for all points with cubic spline interpolation.29 Cosmic ray removal is then applied by considering that points having a value outside a range of three times the standard deviation in a window of 20 pixels are cosmic rays, which are replaced by the median value within this interval. In the case of hyperspectral images, a background signal estimated by measuring an empty region of the substrate is also first subtracted from the spectra. The remaining baseline is then removed as described above. 2.4.Multivariate AnalysisAll multivariate analysis procedures are performed after spectra preprocessing and are implemented in the MATLAB® environment (The MathWorks, Natick, Massachusetts). In the case of imaging data, pixels containing cell spectra are separated from the background in the preprocessed Raman image by creating an image from the C-H stretching region (2910 to ). Cell spectra are then selected by applying a threshold on the image, as cells have a strong Raman response in this region. The pixels extracted from the three-dimensional data set are then concatenated into a two-dimensional matrix for multivariate analysis. Vertex component analysis (VCA) is a method that extracts the endmembers of a spectra data set as the vertices of a simplex, and is applied here based on the implementation described by Nascimento and Bioucas Dias.30 Principal component analysis (PCA) is a method that defines a new orthonormal basis representing the data which maximizes the variance of data. It is applied here by first mean-centering every variable in the data set and then factorizing the matrix through singular value decomposition, defined as where is a diagonal matrix, and represents the transpose operator. If represents the mean-centered data, then is the new vector basis of PCA.3.Results3.1.Raman Imaging and SpectroscopyRaman imaging is illustrated in Fig. 1(a), where a color-coded image of live J774A.1 cells can be extracted from the three-dimensional hyperspectral data by highlighting specific bands in the spectra [see Fig. 1(b)]. The general shape and content of the cells is highlighted in green with a C-H stretching band (2970 to ), while lipids (2850 to ) in red are essentially visible in the cytoplasm and in lipid droplets, making nuclei particularly visible by their absence. A specific protein, which provides a strong signal due to its resonance at 532 nm, cytochrome c, can also be seen in blue located in certain regions of the cytoplasm (1570 to ). The corresponding image acquired by QPM, containing a larger field of view, is shown in Fig. 1(c), with a full dynamic range corresponding to 5.1 rad. Fig. 1(a) Color-coded contrast of a Raman image of J774A.1 cells (scale bar ). Red: lipids (2850 to ). Green: C-H stretching (2970 to ). Blue: cytochrome c (1570 to ). (b) Raw Raman spectra extracted from pixels square regions of the image in (a), in various parts of the cells. Spectral bands used to make the image in (a) are also represented. (c) Corresponding QPM image with a larger field of view. 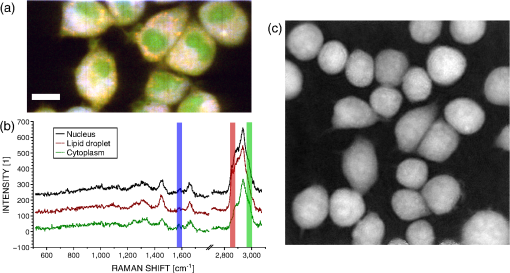 In the imaging case, the laser power is distributed along a whole region, effectively reducing the amount of signal retrieved per pixel. Furthermore, despite the parallel detection of the slit-scanning configuration,31 the 3-s exposure per line employed during measurement means that the pixels image of Fig. 1(a) still requires 10 min for acquisition. The limitation in the amount of signal can be partially overcome by averaging together detected signals, as shown in Fig. 1(b), where spectra extracted as spatial averages from square regions within specific locations of cells are shown. However, the spectra still exhibit a significant amount of noise despite the spatial averaging. It is also interesting to note that even though highly contrasted images can be extracted [see Fig. 1(a)], the spectral differences between different locations in cells are tenuous. As Raman imaging requires significant acquisition times and is technically challenging, a common measurement approach in single cell analysis is then to acquire spectra in point-mode acquisition, where a single spectrum from one point is measured with the detector acting as a line detector, as depicted in Fig. 2(a). This approach provides signals as shown in Fig. 3(a), where spectral features can be identified far more easily than in the case of specific pixels or averaged regions from within a Raman image, and are assumed to be representative of the molecular content of the whole cell. Fig. 2(a) Different approaches in scanning principles of the Raman microscope. Imaging is performed by slit-scanning with two scanning mirrors, with only one being in the descanned region. Fixed point measurement is done with no scanning, while hybrid scanning is performed with two descanned mirrors, resulting in a scan pattern at the sample but a nonscanned point at the slit plane, giving the same signal to noise as fixed point measurements. (b) Quantitative phase image of J774A.1 cells, with the square scanning region used for hybrid Raman imaging represented by the dashed square. 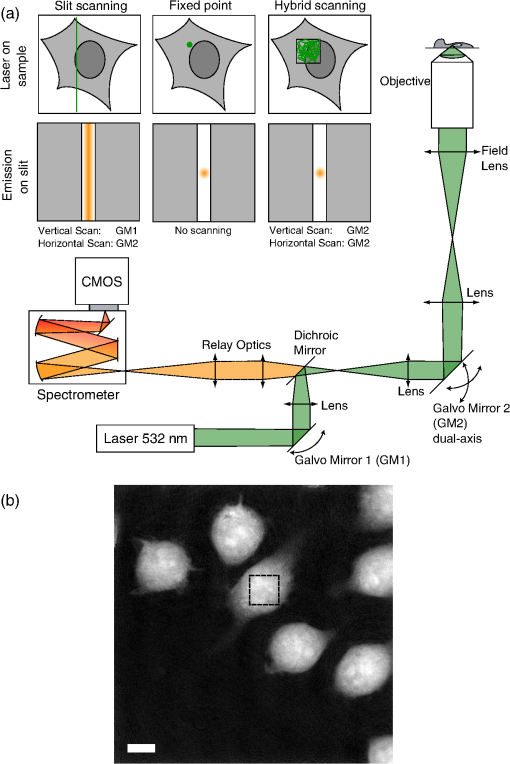 Fig. 3Spectra extracted from both fixed point (a) and hybrid scanning (b) excitation, where five endmembers obtained by vertex component analysis are displayed, showing the variability of spectral profiles for each scanning type. The fingerprint region (500 to ) has been scaled by a factor of 3 to improve visibility. 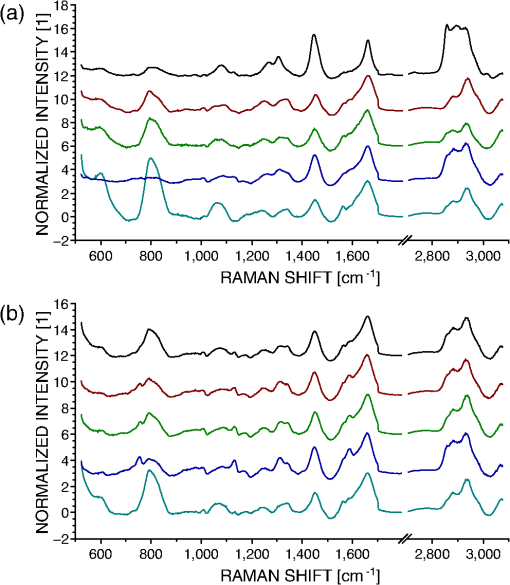 The important question then becomes how well a spectrum taken from a single location in the cell represents the overall cellular content. As illustrated in Fig. 1(a), different cell locations do possess different molecular contrasts. This is particularly important when considering, for example, the distribution of cytochrome c, which is sequestered in mitochondria and clearly not homogeneously distributed in the cytoplasm. The measured spectra may then vary significantly not only by the overall morphology of the cell (i.e., whether measured inside the cytoplasm, nucleus, etc.), but also by the local molecular content (for example, the mitochondrial distribution in the cytoplasm). These variations cannot easily be assessed unless a full Raman image is acquired, which is time-consuming, as noted. This implies that point-based measurement approaches may require a very large amount of samples in order to ensure that local variations within individual cells are not influencing the data analysis. To circumvent this local variation, we propose a hybrid spectral point measurement based on a random scanning pattern within a predefined region in the target cell. In this case, the excitation beam is rapidly scanned in a square region within the cell body, as represented in Fig. 2(a), which is selected from the additional imaging modality [see Fig. 2(b)]. Importantly, in contrast to the full Raman imaging case, the scanning is performed here through an x-y galvano-mirror located in the descanned region of the setup (GM2), so that the emitted signal stays located as a single point at the slit of the spectrometer, still yielding a strong signal as in the standard fixed-point case, but with a measurement derived from a larger region of the cell, which is expected to provide information more suitable for the analysis of cellular changes. This approach is also more efficient than averaging pixels from a Raman image, since by optically averaging the emission on one location for acquisition, the read-out noise (one of the most significant noise sources in Raman imaging) is significantly reduced. In order to reduce any systematic error in the measurement, the laser spot is scanned randomly within a square region of with a scanning frequency of 200 Hz in both directions, as represented by the dashed square in Fig. 2(b). The size of the square is first chosen according to the size of the sample to measure, so that the region should be large enough to enclose a region that can be considered significant of the whole sample (e.g., large enough to contain parts of the cytoplasm and the nucleus in case of cells in culture), while being small enough that it is contained within the cell area and does not sample significant regions outside the cell. The measured region shape is here arbitrarily chosen as a square, but could be adjusted to other shapes by simply changing the scanning pattern. The random scan is important so that, even for varying exposure times, we do not change the effective sampling patterns within the cell. To accurately set the location of the square region in the cell, an additional modality is necessary. In our case, we are using simultaneous QPM based on digital holography, which provides additional useful information, although the scanning approach, denoted here as hybrid Raman imaging, could be generalized to include other types of imaging. 3.2.Cell Molecular Content Represented with Point SpectraThe faster acquisition rates available using point rather than imaging measurements make it possible to measure a larger amount of samples, which is required for robust, statistically significant analysis of biological effects in cell populations. Measurements are performed on cells per data set to account for variations in cell condition. Furthermore, to also account for possible changes due to different substrates and culturing conditions, the measurements are spread over four dishes per data set, with measurements per dish. To ensure that the measurements are representative of the population, both procedures (point and hybrid) are measured in random locations in the cell. Since the measurement region of both hybrid and fixed-point measurements is small compared to the overall size of the cell, we position both square and point measurements randomly throughout the cell to avoid systematic errors in the measurements. Typical point spectra measured on J774A.1 cells are shown in Fig. 3 for both fixed-point [Fig. 3(a)] and hybrid scanning [Fig. 3(b)], with the fingerprint region scaled for the sake of visibility. In order to estimate the variability of spectra within a data set, the extreme ones are displayed here by retrieving five endmembers extracted by VCA. Spectra are then normalized by the intensity of the peak at , to account for overall changes in the signal intensity. The spectra in Fig. 3 clearly show that the fixed-point scanning approach provides more variability in the measurements, with some spectra emanating essentially from one molecular species, such as lipids (first curve), or with strong variations in the background signal, such as the last curve, where a significant contribution from the quartz substrate at is present. As expected, the spectra measured with the hybrid scanning approach seem far more similar to each other, as the measurement throughout a whole region reduces the influence of particular local content by optical averaging. It is worth remembering that in imaging mode, the spatial variations in the sample are part of the measured information, but single point Raman measurements carry the inherent assumption that variations are low so that they are representative of the whole sample. This implies that the two approaches have different assumptions on how molecular content is distributed in the cell. The large variability of fixed point measurements results from the spatial variation in the cell, whereas the hybrid scanning measurements are much more consistent, indicating that the hybrid scanning mode samples the molecular content more effectively. In order to quantitatively compare the information content of each data set, we also compute the covariance matrix defined as , where is the data matrix for measurements and data points in one spectrum. We then consider the rank of , which provides the number of linearly independent dimensions contributing to the variance of the data. The rank of then is an estimation of the amount of information present. The ranks of the data sets are shown in Fig. 4(a) for both scanning approaches (fixed and hybrid) and for two cell types, J774A.1 and Raw264.7. Each rank is the average of at least three experiments, with the error bars representing the standard deviation. Fig. 4The hybrid scanning approach generates significantly more information on cell content. (a) Ranks of covariance matrices for measurements performed with both fixed and hybrid scanning, and for different cells types (J774A.1 and Raw264.7), where 104 cells were measured for each experiment. (b) Cumulative variance described by principal component analysis (PCA) for both fixed and hybrid scanning. All bars and curves are the average of at least three experiments, with error bars representing standard deviations. 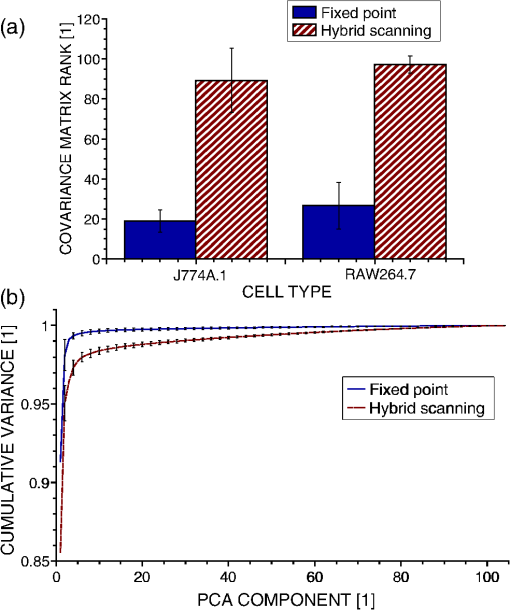 Surprisingly, although fixed-point measurements seem immediately more diverse than hybrid ones (see Fig. 3), the information content is actually lower as the mean rank of the fixed-point covariance matrices is 19 and 26.7 for J774A.1 and Raw264.7 cells, respectively, compared to 89.3 and 97.3 for hybrid scanning, for a total of 104 dimensions. This demonstrates that despite the fact that they appear similar, hybrid scanning measurements possess more linearly independent dimensions, denoting a larger information content compared to the fixed-point case. This aspect is also confirmed through another approach; when applying PCA to the J774A.1 data set, where by considering the contribution to the data variance of each principal component (PC) and plotting the cumulative variance [see Fig. 4(b)], it is possible to see that the data based on the fixed-point scanning can be represented with significantly fewer components. Three PCs already account for of the data variance, while 47 PCs are required in the hybrid case. Applying PCA on the Raw264.7 cell lines produces similar results. 3.3.Cell Line Classification Through Point SpectraIn order to test how the increased information available through hybrid scanning relates to an actual cellular analysis implementation, we investigate the possibility of distinguishing J774A.1 and Raw264.7 cell types based on their respective spectra by multivariate analysis. The cell line measurements, each composed of spectra from cells, are concatenated together to perform PCA on the whole set and estimate the separability of the two classes in the PCA space. Up to this point, there is no use of a priori knowledge of the cell types, and loading vectors are generated based on the inherent spectral variance in the data. Two loading vectors are then selected to visualize the scores of the data, as shown in Fig. 5. The best loading vectors for separating the cell lines are selected by choosing the ones that maximize the distance between the centroids of the two classes based on the a priori knowledge of the cell type. The two scanning methods (fixed point versus hybrid) are compared on the components best able to separate the cell lines since a one-to-one comparison on each PC is unsuitable since, as shown above, the number of relevant components is very different between the two approaches. The loading vectors used to compute the scores are shown in Figs. 5(a) and 5(c) for, respectively, the fixed-point and hybrid scanning. The corresponding scores are then displayed in Figs. 5(b) and 5(d), along with confidence intervals, shown as ellipses whose major axes contain 95% of the data points in their projection direction. Fig. 5Hybrid scanning improves the Raman-based separation between cell lines by multivariate analysis, as shown by PCA decomposition of data sets composed of J774A.1 and Raw264.7 cell lines. (a) and (c) Loading vectors used for displaying score plots, where centroid distances between the two classes are maximized. (b) and (d) Score plots showing the separation between the two classes, with 95% confidence regions, for fixed point [(a) and (b)] and hybrid scanning [(c) and (d)], respectively. Each score plot is representative of at least three experiments. 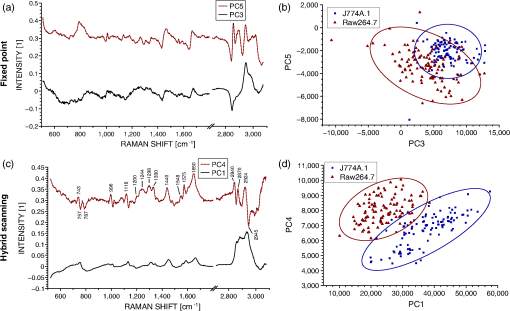 We can see that the hybrid scanning provides better separation, with the confidence regions being nearly fully separated, in contrast to the fixed-point case, where the whole distribution of J774A.1 cells is nearly fully enclosed within the confidence region of Raw264.7 data points. It is also possible to see that the data in the fixed-point case are less uniformly distributed within the confidence regions, as can be expected from their higher variability, identified in Fig. 3. The loading vectors extracted by PCA for both measurement methods also present rather different characteristics. In the case of hybrid scanning, PC1 is always one of the two components providing the best separation with a spectrum similar to the global features of cells which can be seen by comparing it with the spectra of Fig. 3. On the other hand, PCA applied to fixed-point data always extracts fewer significant vectors (here PC3 and PC5). This is likely due to the higher variability in fixed-point spectra, so that the low numbered PCs are dominated by the variation within the group rather than between the cell lines. A further point of note is the shape of the loading vectors, which indicate the spectral contrast providing the separation. PCA can produce loading vectors with both positive and negative features which can be difficult to interpret. The hybrid scanning loading vectors appear to have easily identified peaks and an overall shape that is more similar to raw spectra (for example, those shown in Fig. 1), which should make interpretation and later analysis simpler. 3.4.Imaging and ClassificationWhile the use of multivariate analysis for discrimination is common with point measurements in the literature, this analysis approach has also been used with imaging data.32 We can then extend our comparison of the separation power of Raman measurements to include imaging as well as fixed-point and hybrid scanning measurements. Prior to analysis, spectral data are first extracted separately from images of, respectively, J774A.1 and Raw264.7 cells (see Sec. 2.4), and then concatenated together, leading to a matrix containing spectra. The analysis is thereafter performed as in the previous section. The PCA of imaging spectra is shown in Fig. 6, which presents a separation distribution comparable to hybrid scanning in Fig. 5(d), while the fixed-point separation is different from both imaging and hybrid results. This similarity in separation happens despite the large differences in how the data are measured: in the case of hybrid spectra, each data point represents one cell, for a total of cells per class; on the other hand, PCA of image data involves one spectra from each pixel in the image, with around four to eight cells in the field of view and with far more data points ( per class). The data outside the 95% confidence region in the imaging case are also more spread than in the point measurements case, and the separation is less clear than for the hybrid scanning data. This can be explained by the fact that image pixels contain several types of spectra, leading, for example, to the image contrast in Fig. 1(a), which will induce more spread than optically averaged hybrid point spectra. Fig. 6Separation of cell lines by spectra extracted from pixels in Raman imaging by PCA decomposition. (a) Loading vectors that maximize the centroid distance, and the corresponding score plot. (b) The separation between the two classes with 95% confidence ellipses. For visibility, only 10% of the data points (randomly selected) are displayed on the graph. The figure represents three experiments. 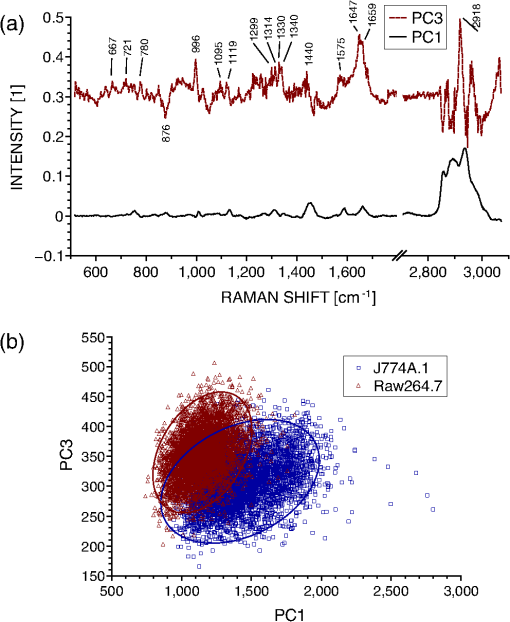 Despite these differences, it is interesting to note the similarities between the two types of measurements, where the score plots show almost identical trends between imaging data and hybrid scanning. The extracted loading vectors, which maximize the class centroid distance, are very similar (PC1 and PC3 or 4), and in both cases, PC1 is selected for all considered experiments. PC1 presents general cell features in both cases, with a nearly identical C-H stretching, and a fingerprint region presenting general features of lipids and proteins in the imaging case [ , 1650 to amide I)33,34 along with cytochrome c features (753, 1130, ),35 while the hybrid PC1 also possesses additional features along with the same peaks. Of special interest is the striking similarity of the second separating PC between the two measurement modes, where several identical spectral features can be identified, such as 1575 and as discussed above. Several identical features are also present as compensating factors of previously discussed features (asymmetric peaks), such as 1118 to 1134 and . While some features in the imaging case [see Fig. 6(a)] cannot be definitely identified as peaks due to noise levels, it is interesting to note that they correspond to peaks in the hybrid scanning case, such as several features in the 1200 to region. Both vectors also possess a strong asymmetric feature at 996 to , which can be attributed to phenylalanine (, aromatic ring breathing).36 By comparison, fixed-point decomposition leads to vectors that are far less characteristic, where PCs of lower order are selected for separation. Furthermore, there are far fewer visible spectral features in the fingerprint region, apart from the contribution at in PC5, which can be attributed to the quartz substrate and should not, in principle, significantly contribute to the separation between the two cell lines. The results indicate that fixed-point measurement is the weakest of the three scanning approaches in terms of the separation ability between groups, the richness of spectral information, and the resulting ease of analysis of the separation vectors. 4.DiscussionThe measurement approach proposed in this article is based on random scanning within a region of the cell body to retrieve a Raman spectrum accounting for the different molecular species contained in different locations of the cell. Since all the information gathered from the whole cell region is measured as one spectrum, the Raman spectra measured with the hybrid imaging are visibly far more consistent between cells compared to standard fixed-point measurement (see Fig. 3). However, spectra measured with the hybrid approach actually possess more linearly independent dimensions contributing to the variance of the data, as shown either by the rank of their covariance matrix [see Fig. 4(a)] or the amount of information carried by the first loading vectors of the PCA decomposition [see Fig. 4(b)]. This suggests that while fixed-point measurements provide information on the differences between individual locations in cells, the variability is such that the spectra can be represented as linear combinations of each other, while the hybrid measurements, instead, emphasize information related to cellular changes instead of pure local contributions. We should point out that if local molecular variations are of primary interest, then full Raman imaging should be considered. If there is not sufficient time and/or laser power to measure full Raman images, or if sample damage concerns preclude imaging, then nonimaging mode Raman measurements are quite capable of discrimination between samples while providing measurements on a larger population, and the hybrid scanning mode provides some unique advantages. The observed difference in the information content dramatically changes analysis results when considering a large population for statistical treatment, for instance, to classify two cell lines, such as macrophage-like cells J774A.1 and Raw264.7 with multivariate analysis (see Fig. 5). Clear separation is obtained with hybrid scanning, while the fixed-point approach does not discriminate the two classes. Interestingly, PCA provides similar results when hybrid scanning is compared with imaging data (see Fig. 6), which implies that the information content of a large cell population measured with hybrid scanning is comparable to a large number of data points (pixels) of a Raman image. This may be due to the fact that hybrid measurements account in a global way for the local differences that can be observed in images [see Fig. 1(a)]. Nevertheless, the separation is less clear in the case of imaging-based spectra, which can be expected from the local variations. If, during experiments, it becomes necessary to average together spectra acquired by Raman imaging to provide sufficient SNR for analysis, it appears more fruitful to average the data on the detector itself through a hybrid scan technique to avoid the increase in read-out noise. Furthermore, hybrid scan measurements are expected to be statistically more relevant due to their higher throughput providing a larger and more diverse population ( cells versus 4 to 8 cells in the imaging case), which also accounts for environmental variability by including measurements from several different dishes, while imaging data only contain cells located in a small area of one dish. This similarity in separation power indicates that the hybrid imaging approach, where spectral data are optically averaged and images of the sample are obtained with another modality, could be effectively employed as a substitute for full Raman imaging with much higher throughput. We should note that PCA is not an optimal technique for class discrimination applications, as it is a purely unsupervised decomposition. Several other methods, such as linear discriminant analysis or partial least square discrimination analysis, are usually more efficient at separating groups based on multivariate analysis, due to the incorporation of the a priori knowledge of measured classes into the decomposition. These approaches have been employed in several works classifying different cell types based on spectroscopic measurements.4,6,14 These limitations of PCA, however, serve to demonstrate the robustness of the separation comparisons. PCA is more suitable for assessing the intrinsic discriminating power of data since it is unsupervised. The point here is not to optimize the classification observed (for example, in Fig. 5) but to quantatively compare the amount of information available in each scan method from the variance characteristics of the data. An optimized separation algorithm could readily outperform PCA if classification alone was the goal. In addition to the similarity in the type of separation, PCA loading vectors from imaging and hybrid data are very similar [see Figs. 5(c) and 6(a)], with the second separating PC (respectively, PC4 and PC3) possessing identical spectral features, which can be assigned to specific molecular species, as discussed in Sec. 3.4. It is interesting that both measurement approaches lead to such similarities considering that they are based on very different principles (spatially resolved spectra at each pixel versus optically averaged signals), and yield different signals and SNR [see Figs. 1(b) and 3(b)]. This implies that the variance behavior is similar between imaging data on a small amount of cells and hybrid imaging on a relatively large population. It is likely that the high number of pixels in the imaging case then compensates for the lower SNR when statistical analysis is performed during multivariate analysis. These results show that the approach based on hybrid imaging accounts well for the spectral information contained in a cellular body in a global sense, while enabling a higher measurement throughput, which makes it possible to perform the analysis on populations that are statistically more relevant. Furthermore, it also provides a higher SNR, so that spectral features can be identified far more easily, as can be seen when comparing the PCs where some peaks are clearly identified in Fig. 5(c), while their presence in the imaging data [see Fig. 6(a)] is ambiguous. 5.ConclusionThe two main approaches that are usually employed in the case of Raman measurements, namely point acquisition at one location of the sample and imaging of a whole region of the sample to obtain spatial contrast, provide very different types of information and both have their own drawbacks. Imaging provides a very large amount of information where each pixel in the image contains spectral information, but it requires long acquisition times to ensure the collection of enough signal at each location, and is often sample damage limited. On the other hand, point measurements can be acquired fairly rapidly and provide a high SNR, thanks to the concentration of the excitation power at one location for measurement while the total laser exposure is still relatively low. However, the spectra measured in this fashion are very sensitive to the local content, so that repeated measurements on several cells are more sensitive to the changes in local content than to finer spectral changes originating from cell conditions. This can be problematic in cases where fine spectral changes are used to discriminate groups of cells, such as cancer versus noncancer. The proposed hybrid approach, which consists of rapidly scanning the excitation beam across a region of the sample while descanning and then detecting the signal as a point spectrum, provides an alternative to obtain more reliable measurements for statistical analysis while keeping the high throughput through point excitation. Furthermore, as this approach requires an additional imaging modality to position the scanning pattern within the region of interest, spatial information is still recorded to relate the spectral information to a known location. The comparison of the information content between the standard fixed-point and the hybrid approach showed that the latter provides more information despite the apparent stronger similarity of the measured spectra. This result is supported by the ability of hybrid scanning to effectively discriminate different cell lines, where it provides a far more reliable classification than the standard point approach. Extracted spectral features are also far easier to interpret and identify the molecular differences between the two cell lines, which is often of primary interest. Interestingly, even with very different types of data, the statistical treatment of a high number of low-SNR spectra from several cells (in imaging) and a low number of high-SNR spectra (hybrid) from a large number of cells lead to somewhat similar results, with the hybrid mode having additional advantages in classification power and clarity of discriminating spectra. The proposed hybrid imaging approach is, therefore, a viable alternative for noninvasive analysis of biological samples compared to more classical approaches (fixed-point or imaging measurements) by enabling the acquisition of signals which represent overall cellular molecular content, and maintaining a high throughput capability to measure representative populations. It could be employed not only for live cells as presented here as a proof of concept but also for other types of samples, such as histological sections or tissues. AcknowledgmentsThis work was funded by the Japan Society for the Promotion of Science (JSPS) through the Funding Program for World-Leading Innovative R&D on Science and Technology (FIRST Program), and by the JSPS World Premier International Research Center Initiative Funding Program, and Japan Science and Technology (JST) Agency PRESTO program. The authors would also like to thank Dr. Alison J. Hobro for fruitful discussion. ReferencesC. V. RamanK. S. Krishnan,
“A new type of secondary radiation,”
Nature, 121 501
–502
(1928). http://dx.doi.org/10.1038/121501c0 NATUAS 0028-0836 Google Scholar
R. J. Swainet al.,
“Assessment of cell line models of primary human cells by Raman spectral phenotyping,”
Biophys. J., 98
(8), 1703
–1711
(2010). http://dx.doi.org/10.1016/j.bpj.2009.12.4289 BIOJAU 0006-3495 Google Scholar
S. Verrieret al.,
“In situ monitoring of cell death using Raman microspectroscopy,”
Biopolymers, 74
(1–2), 157
–162
(2004). http://dx.doi.org/10.1002/(ISSN)1097-0282 BIPMAA 0006-3525 Google Scholar
I. Notingheret al.,
“In situ non-invasive spectral discrimination between bone cell phenotypes used in tissue engineering,”
J. Cell. Biochem., 92
(6), 1180
–1192
(2004). http://dx.doi.org/10.1002/(ISSN)1097-4644 JCEBD5 0730-2312 Google Scholar
I. Notingheret al.,
“In situ spectral monitoring of mRNA translation in embryonic stem cells during differentiation in vitro,”
Anal. Chem., 76
(11), 3185
–3193
(2004). http://dx.doi.org/10.1021/ac0498720 ANCHAM 0003-2700 Google Scholar
P. Crowet al.,
“The use of Raman spectroscopy to differentiate between different prostatic adenocarcinoma cell lines,”
Br. J. Cancer, 92
(12), 2166
–2170
(2005). http://dx.doi.org/10.1038/sj.bjc.6602638 BJCAAI 0007-0920 Google Scholar
Y. Oshimaet al.,
“Discrimination analysis of human lung cancer cells associated with histological type and malignancy using Raman spectroscopy,”
J. Biomed. Opt., 15
(1), 017009
(2010). http://dx.doi.org/10.1117/1.3316296 JBOPFO 1083-3668 Google Scholar
G. R. Lloydet al.,
“Discrimination between benign, primary and secondary malignancies in lymph nodes from the head and neck utilising Raman spectroscopy and multivariate analysis,”
Analyst, 138
(14), 3900
–3908
(2013). http://dx.doi.org/10.1039/c2an36579k ANLYAG 0365-4885 Google Scholar
I. Barmanet al.,
“Turbidity-corrected Raman spectroscopy for blood analyte detection,”
Anal. Chem., 81
(11), 4233
–4240
(2009). http://dx.doi.org/10.1021/ac8025509 ANCHAM 0003-2700 Google Scholar
C. Matthäuset al.,
“Label-free detection of mitochondrial distribution in cells by nonresonant Raman microspectroscopy,”
Biophys. J., 93
(2), 668
–673
(2007). http://dx.doi.org/10.1529/biophysj.106.102061 BIOJAU 0006-3495 Google Scholar
K. Hamadaet al.,
“Raman microscopy for dynamic molecular imaging of living cells,”
J. Biomed. Opt., 13
(4), 044027
(2008). http://dx.doi.org/10.1117/1.2952192 JBOPFO 1083-3668 Google Scholar
M. Okadaet al.,
“Label-free Raman observation of cytochrome c dynamics during apoptosis,”
Proc. Natl. Acad. Sci. USA, 109
(1), 28
–32
(2012). http://dx.doi.org/10.1073/pnas.1107524108 1091-6490 Google Scholar
C. Krafftet al.,
“Advances in optical biopsy—correlation of malignancy and cell density of primary brain tumors using Raman microspectroscopic imaging,”
Analyst, 137
(23), 5533
–5537
(2012). http://dx.doi.org/10.1039/c2an36083g ANLYAG 0365-4885 Google Scholar
M. Hedegaardet al.,
“Discriminating isogenic cancer cells and identifying altered unsaturated fatty acid content as associated with metastasis status, using K-means clustering and partial least squares-discriminant analysis of Raman maps,”
Anal. Chem., 82
(7), 2797
–2802
(2010). http://dx.doi.org/10.1021/ac902717d ANCHAM 0003-2700 Google Scholar
W. Minet al.,
“Coherent nonlinear optical imaging: beyond fluorescence microscopy,”
Annu. Rev. Phys. Chem., 62
(1), 507
–530
(2011). http://dx.doi.org/10.1146/annurev.physchem.012809.103512 ARPLAP 0066-426X Google Scholar
C.-Y. ChungJ. BoikE. O. Potma,
“Biomolecular imaging with coherent nonlinear vibrational microscopy,”
Annu. Rev. Phys. Chem., 64
(1), 77
–99
(2013). http://dx.doi.org/10.1146/annurev-physchem-040412-110103 ARPLAP 0066-426X Google Scholar
B. G. Saaret al.,
“Video-rate molecular imaging in vivo with stimulated Raman scattering,”
Science, 330
(6009), 1368
–1370
(2010). http://dx.doi.org/10.1126/science.1197236 SCIEAS 0036-8075 Google Scholar
C. H. J. Campet al.,
“High-speed coherent Raman fingerprint imaging of biological tissues,”
Nat. Photon., 8
(8), 627
–634
(2014). http://dx.doi.org/10.1038/nphoton.2014.145 1749-4885 Google Scholar
M. B. Mohamedet al.,
“The ‘lightning’ gold nanorods: fluorescence enhancement of over a million compared to the gold metal,”
Chem. Phys. Lett., 317
(6), 517
–523
(2000). http://dx.doi.org/10.1016/S0009-2614(99)01414-1 CHPLBC 0009-2614 Google Scholar
J. Andoet al.,
“Dynamic SERS imaging of cellular transport pathways with endocytosed gold nanoparticles,”
Nano Lett., 11
(12), 5344
–5348
(2011). http://dx.doi.org/10.1021/nl202877r NALEFD 1530-6984 Google Scholar
K. Konget al.,
“Diagnosis of tumors during tissue-conserving surgery with integrated autofluorescence and Raman scattering microscopy,”
Proc. Natl. Acad. Sci. U.S.A., 110
(38), 15189
–15194
(2013). http://dx.doi.org/10.1073/pnas.1311289110 PNASA6 0027-8424 Google Scholar
S. Yueet al.,
“Label-free analysis of breast tissue polarity by Raman imaging of lipid phase,”
Biophys. J., 102
(5), 1215
–1223
(2012). http://dx.doi.org/10.1016/j.bpj.2012.01.023 BIOJAU 0006-3495 Google Scholar
Z. Huanget al.,
“Integrated Raman spectroscopy and trimodal wide-field imaging techniques for real-time in vivo tissue Raman measurements at endoscopy,”
Opt. Lett., 34
(6), 758
–760
(2009). http://dx.doi.org/10.1364/OL.34.000758 OPLEDP 0146-9592 Google Scholar
C. A. Patilet al.,
“Integrated system for combined Raman spectroscopy—spectral domain optical coherence tomography,”
J. Biomed. Opt., 16
(1), 011007
(2011). http://dx.doi.org/10.1117/1.3520132 JBOPFO 1083-3668 Google Scholar
V. V. PullyA. LenferinkC. Otto,
“Hybrid Rayleigh, Raman and two-photon excited fluorescence spectral confocal microscopy of living cells,”
J. Raman Spectrosc., 41
(6), 599
–608
(2010). http://dx.doi.org/10.1002/jrs.v41:6 JRSPAF 0377-0486 Google Scholar
N. PavillonA. J. HobroN. I. Smith,
“Cell optical density and molecular composition revealed by simultaneous multimodal label-free imaging,”
Biophys. J., 105
(5), 1123
–1132
(2013). http://dx.doi.org/10.1016/j.bpj.2013.07.031 BIOJAU 0006-3495 Google Scholar
H.-J. van Manenet al.,
“Single-cell Raman and fluorescence microscopy reveal the association of lipid bodies with phagosomes in leukocytes,”
Proc. Natl. Acad. Sci. U.S.A., 102
(29), 10159
–10164
(2005). http://dx.doi.org/10.1073/pnas.0502746102 PNASA6 0027-8424 Google Scholar
P. Marquetet al.,
“Digital holographic microscopy: a noninvasive contrast imaging technique allowing quantitative visualization of living cells with subwavelength axial accuracy,”
Opt. Lett., 30
(5), 468
–470
(2005). http://dx.doi.org/10.1364/OL.30.000468 OPLEDP 0146-9592 Google Scholar
N. Pavillonet al.,
“Feature-based recognition of surface-enhanced Raman spectra for biological targets,”
J. Biophotonics, 6
(8), 587
–597
(2013). http://dx.doi.org/10.1002/jbio.v6.8 JBOIBX 1864-063X Google Scholar
J. M. P. NascimentoJ. M. Bioucas Dias,
“Vertex component analysis: a fast algorithm to unmix hyperspectral data,”
IEEE Trans. Geosci. Remote Sens., 43
(4), 898
–910
(2005). http://dx.doi.org/10.1109/TGRS.2005.844293 IGRSD2 0196-2892 Google Scholar
D. K. Veirset al.,
“Mapping materials properties with Raman spectroscopy utilizing a 2D detector,”
Appl. Opt., 29
(33), 4969
–4980
(1990). http://dx.doi.org/10.1364/AO.29.004969 APOPAI 0003-6935 Google Scholar
T. Ichimuraet al.,
“Visualizing cell state transition using Raman spectroscopy,”
PLoS ONE, 9
(1), e84478
(2014). http://dx.doi.org/10.1371/journal.pone.0084478 1932-6203 Google Scholar
S. P. VermaD. F. WallachR. Schmidt-Ullrich,
“The structure and thermotropism of thymocyte plasma membranes as revealed by laser-Raman spectroscopy,”
BBA-Biomembranes, 394
(4), 633
–645
(1975). http://dx.doi.org/10.1016/0005-2736(75)90148-0 0005-2736 Google Scholar
D. F. WallachS. P. VermaJ. Fookson,
“Application of laser Raman and infrared spectroscopy to the analysis of membrane structure,”
Biochim. Biophys. Acta, 559
(2–3), 153
–208
(1979). http://dx.doi.org/10.1016/0304-4157(79)90001-7 BBACAQ 0006-3002 Google Scholar
T. G. SpiroT. C. Strekas,
“Resonance Raman spectra of hemoglobin and cytochrome c: inverse polarization and vibronic scattering,”
Proc. Natl. Acad. Sci. USA, 69
(9), 2622
–2626
(1972). http://dx.doi.org/10.1073/pnas.69.9.2622 1091-6490 Google Scholar
J. Castroet al.,
“Vibrational spectra of 3-phenylpropionic acid and L-phenylalanine,”
J. Mol. Struct., 744–747 887
–891
(2005). http://dx.doi.org/10.1016/j.molstruc.2004.10.098 JMOSB4 0022-2860 Google Scholar
BiographyNicolas Pavillon obtained his BSc in microengineering in 2004 and his MSc in applied photonics in 2006 from the Ecole Polytechnique Fédérale de Lausanne (EPFL). He obtained his PhD in photonics from EPFL in 2011, during which he investigated phase imaging applications for cellular probing and quantitative three-dimensional imaging. He currently is a postdoctoral researcher at Osaka University, mainly focused on label-free imaging methods devoted to cellular dynamics investigation. Nicholas I. Smith studied undergraduate physics and mathematics at the University of Sydney and received his PhD from Osaka University with a focus on biophysics in 2003. He was an assistant professor in the Department of Frontier Biosciences, studying microscopy and laser irradiation effects on cell dynamics. He is currently an associate professor and principal investigator at the Biophotonics Laboratory at the Immunology Frontier Research Center, developing imaging-based techniques for immunology applications. |

