|
|
1.Understanding Brain Machinery Requires Multilevel InvestigationThe brain encompasses vast numbers of interconnected neurons that constitute anatomical and functional networks. In order to understand how the specific wiring of neurons accounts for brain functions, neuroscientists need various types of data. First, they need to build three-dimensional (3-D) maps of neurons within their circuits on multiple scales, ranging from interhemispheric axonal connections to single synaptic contacts. Second, the flow of electrical signals through brain circuits needs to be computed. Third, since the circuitry undergoes continuous modifications, long-term monitoring of dynamic remodeling like structural reshaping and functional adaptation is required. A vast repertoire of experimental tools is currently available to map neuronal connectivity at multiple levels (Fig. 1). Functional and structural maps of (animal models and) human brain in their entirety can be obtained with low-resolution imaging techniques like functional magnetic resonance imaging (fMRI), electroencephalography (EEG), diffusion magnetic resonance imaging, and polarized light imaging.1–7 These methods achieve maximum spatial resolutions of hundreds of micrometers (fMRI) and temporal precision of the order of tens of milliseconds (EEG). These minimally invasive techniques play a key role in the exploration of the human functional and structural connectome.8,9 Moving toward finer details on smaller samples (human brain samples or entire mouse brain), light microscopy-based techniques like light sheet microscopy (LSM) are well suited to obtain mesoscopic maps of connectivity with micrometric resolution.10–12 Other techniques such as the knife-edge scanning microscopy,13 micro-optical sectioning tomography (MOST),14 fluorescence MOST (fMOST),15 and serial two-photon tomography16 (lateral resolution ) can achieve similar goals with slightly better contrast, though they do not preserve the sample and usually require a longer acquisition time. The subcellular resolution over the whole mouse brain is entangled by the need for sparse fluorescent labeling; brainbow techniques might overcome this limitation.17 Wide-field electron microscopy (EM) can achieve better spatial resolution (10 to 100 nm) than light microscopy, but cell types cannot be specifically labeled and the imaging process is orders of magnitude slower, making the reconstruction of the whole mouse brain really demanding. All of these techniques for whole brain reconstruction work ex vivo on mammalian samples and cannot reveal functional and/or dynamic features. The activity of microcircuits of neurons can be characterized with cellular and subcellular resolution over spatial scales of a few hundreds of micrometers by confocal microscopy and two-photon fluorescence microscopy (TPFM). With its unique ability to image with high resolution and high sensitivity inside scattering tissue,18 two-photon optical recordings combined with fluorescent reporters of cellular activity (e.g., calcium indicators or voltage sensitive probes) revealed crucial features of neural computation in vivo,19,20 in head-fixed awake animals21 and virtually22 or truly23 freely moving. Longitudinal studies (temporal scale of the order of weeks/months) of neuronal reshaping with in vivo TPFM revealed the structural correlation of neuronal functional adaptivity within its dynamics perspective. The structural features of medium-sized local circuits (e.g., a cortical column or the hippocampus) can be reconstructed in their entirety by EM-based techniques like serial block-face scanning electron microscopy (SBSEM)24 or Automatic Tape-Collecting Lathe UltraMicrotome (ATLUM)25 (voxel size ), which mechanically slice the sample. For smaller neural circuits (up to side) at higher resolution (near-isotropic voxels of ), focussed-ion beam scanning electron microscopy26 (FIBSEM) may be the most suitable. The identification of synaptic connections at the subcellular level and the molecular properties of individual synapses can be reached quite exclusively by electron microscopy and optical super-resolution techniques like photo-activated localization microscopy27 and stimulated emission depletion28 that break the diffraction barrier (nanometer resolution). Fig. 1Multiple spatial scales in the brain. On the top, different structures are depicted in proximity to their typical size, showing how relevant spatial scales in the brain span several orders of magnitude. On the bottom, typical working ranges of state-of-the-art imaging technologies. 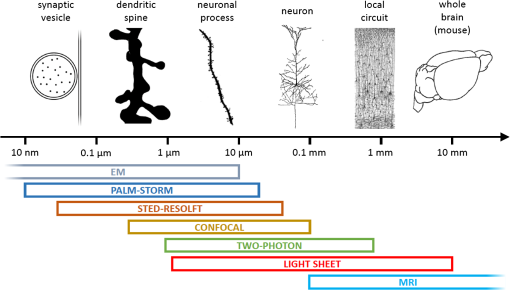 However, despite many decades of studies, we are still far from achieving comprehensive descriptions of brain machinery across all those levels. Recently, multiple attempts have addressed the multilevel complexity of this machinery by combining multiple imaging techniques providing different information. This integrated approach might overcome the intrinsic limit of spatial and temporal resolution and can provide multiple level information on the same sample. Here, we describe the state of the art of correlative approaches for investigating brain structure and functionality. 2.Correlative Imaging Overcomes the Limitation of Single TechniquesNovel approaches may provide new ways to bridge the gap between “postmortem” microscopic and in vivo macroscopic worlds through functional measures reflecting neural connectivity. Several orders of magnitudes can be crossed by combining the spatiotemporal resolution of complementary techniques. Here, we provide four examples of correlative approaches that linked different temporal/spatial scales, going from noninvasive whole brain functional MRI to subsynaptic structure imaging with FIBSEM. 2.1.Simultaneous fMRI and Optical Functional ImagingThe functional connectivity at macro- and mesoscopic scales can be inferred with different neuroimaging techniques. fMRI enables noninvasive monitoring of activity in healthy and diseased brains with submillimeter spatial resolution in humans and animals.2 Alterations in the blood oxygenation levels (BOLD contrast) arising from changes in cerebral blood flow, blood volume, and oxygenation are used to estimate brain activity.29,30 In order to understand the link between BOLD signals (macroscale) and the underlying neural activity (mesoscale), Helmchen et al. used a combination of BOLD fMRI and simultaneous recording of calcium activity31 [Fig. 2(a)]. The authors provided a proof of principle of the integration of fluorescence measures of brain functionality through an optical fiber with fMRI scanners. Further, they demonstrated the close relationship between the two signals by predicting BOLD signals from the fluorescence responses measured with the optical fiber. This study highlights the complexity of fMRI BOLD signals, involving both neuronal and glial activity. The hybrid method for simultaneous recording of BOLD fMRI and calcium transients imaged with TPFM could facilitate further understanding of cellular mechanisms of neurovascular coupling. Fig. 2Correlative microscopy in neuroscience. (a) Experimental approach for examining the relationship between single-cell activity and fMRI signals. Single-cell measured by two-photon microscopy were correlated with bulk responses in the same region measured by one-photon fiber-optic microscopy. Then the bulk one-photon responses were correlated with the BOLD fMRI signals. Figure modified with permission from Ref. 32. (b) On the left, maximum intensity projections of several stacks TPF stitched together in a single image. The red dashed line highlights blood vessel shadows. Red arrows highlight characteristic features of a dendritic arbor, to help finding it back in the CLSM images. On the right, CLSM imaging of the same neuron observed with TPF. Starting from the apical portion of the dendritic tree, the neuron has been segmented and is shown inside a maximum-projection 3-D rendering. The scale of the figure can be inferred from the red cube down on the right, which has side. Figure modified with permission from Ref. 33. (c) Functional characterization of direction-selective retinal ganglion cells (DSGCs) and their localization within SBSEM volume. On the left, polar tuning curves for 25 DSGCs sorted and color-coded by preferred direction. The corresponding soma locations superimposed onto a two-photon image from the recorded region of the ganglion cell layer and the acquired SBSEM volume (scale bars: 100 nm). On the right, skeleton reconstructions of DSGCs. DSGCs, color-coded by preferred direction (inset), normal to the plane of the retina (scale bars: ). Figure modified with permission from Ref. 34. (d) Correlative in vivo TPF and focused ion beam scanning electron microscopy of cortical neurons. In vivo TPF imaging of an axon showing two stable boutons (scale bar: ). Both boutons make multiple synaptic contacts, as visible in a single plane of the correspondent EM images, with multiple dendritic spines (scale bar: 500 nm). 3-D rendering of the same axon imaged in TFP microscopy. The cytoplasm of the axon is represented in light blue, mitochondria in green, synaptic vesicles in yellow and synapses in red. The postsynaptic spiny neurons are shown in gray. Figure modified with permission from Ref. 35. 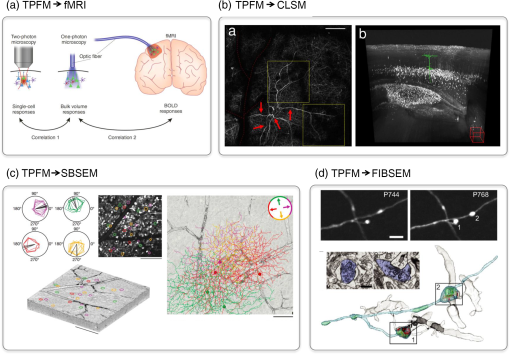 2.2.Correlative Light Sheet and In Vivo Two-Photon MicroscopyTPFM is a powerful tool for longitudinal studies of brain reshaping, both in terms of functional and structural plasticity. The structural plasticity of selected neuronal populations in vivo can be visualized by time-lapse imaging of fluorescently labeled neuronal structures like varicosities, spines (i.e., pre- and postsynaptic portions), axons and dendrites.36–40 The wide-ranging neuroanatomical tracing of the neuron previously observed in vivo allows associating univocally the dynamic fingerprint of a neuron to a specific neuronal type and to its partners of connectivity. A reliable correlative reconstruction of the neuron previously observed in vivo and of the surrounding network can be obtained by combining TPFM with other ex vivo imaging techniques that can explore large volumes. Unlike other large-scale neuroanatomical techniques, ultramicroscopy,11 confocal light sheet microscopy (CLSM),10 and CLARITY optimized light sheet microscopy (COLM)12 are the only ones in which the sample is preserved without slicing, allowing multiple imaging rounds and, therefore, providing a greater flexibility. By using the blood vessel pattern as an internal reference, Silvestri et al. were able to image the apical portion of a dendritic arbor in a living mouse using TPFM, to find the same neuronal process after tissue fixation, dehydration and clearing, and to trace the entire neuron from CLSM images33 [Fig. 2(b)]. The same correlative two-photon and LSM approach could allow reconstructing the distribution and the anatomy of neurons whose activity has been monitored through in vivo functional imaging. 2.3.Correlative In Vivo Two-Photon Calcium Imaging and Serial Block-Face Scanning Electron MicroscopyLinking functional TPFM with the structural connectivity obtained with large-scale electron microscopy allows answering otherwise intractable neurobiological questions. For example, though discovered 50 years ago,41 the computation of motion direction by direction-selective retinal ganglion cells (DSGCs) lacks a complete explanation. Though both light microscopy and EM studies independently attempted to study direction-selectivity circuit anatomy, the results were either contradictory42,43 or incomplete.44,45 Until the recent advancement in serial processing, EM was not capable of reconstructing large fractions of cells in the same piece of tissue, covering the spatial distances over which individual neurons project (hundreds of micrometers) with the resolution of single axonal projections (tens of nanometers). In addition, since DSGC’s preferred direction cannot be inferred from its dendritic morphology,46,47 functional optical imaging techniques are needed to complement structural reconstructions to determine these neural circuit diagrams. In a very accurate and in depth study, Briggman et al. combined in vivo calcium imaging in the intact retina and SBSEM-based reconstruction of the circuitry in the same piece of tissue to explain the behavior of retinal ganglion cells (DSGCs)34 [Fig. 2(c)]. They show that dendrites of mouse starburst amacrine cells make highly specific synapses with direction-selective ganglion cells depending on the ganglion cell’s preferred direction. This pattern provides the structural substrate for the functional asymmetry in the inhibitory input currents observed in DSGCs. This study directly correlated a structural (wiring) asymmetry with the functional properties of the cell, i.e., to the computation of direction selectivity. Accurate 3-D maps of connectivity between neurons will be essential for the implementation of the algorithms used in neural computations, such as the detection of directed motion by the retina. 2.4.Correlative In Vivo Two-Photon Imaging of Structural Plasticity and Focused-Ion Beam Electron MicroscopyThe synaptic connections between axons and dendrites can quite exclusively be visualized with EM. When combining high-resolution EM with in vivo light microscopy, the time lapse structural remodeling can be linked with the underlying ultrastructural morphology.48,49 In vivo-imaged dendrites and axons in adult mouse brains can subsequently be prepared and imaged with EM. The growth of dendritic spines in the adult mammalian brain, seen with two-photon in vivo microscopy, has been verified as new synaptic connections by using post hoc serial section transmission electron microscopy (TEM).50,51 The advent of FIBSEM greatly increased the level of automation, reliability, and speed of EM imaging.26 One of the most commonly used tricks to retrieve the position of the in vivo imaged structure is to burn fiducial marks with the pulsed laser next to it in the fixed tissue.52,53 Since this combination of techniques avoids the use of specific labels to identify the structures of interest in the electron microscope, optimal structural preservation for 3-D analysis is guaranteed [Fig. 2(d)]. In the last years, several studies provided proof of evidence of the added value of correlating in vivo TPFM and FIBSEM for targeting micro- and ultrastructural features of synaptic plasticity.35,54,55 Recently, two parallel works by Allegra Mascaro et al.56 and Canty et al.57 combined two-photon in vivo imaging with FIBSEM to study how laser axotomized and regenerated axonal branches interact with the surrounding neuropil and possible postsynaptic targets. These EM reconstructions allowed visualizing both the distribution of mitochondria and synaptic vesicles inside the axon and inferring the structural interplay between the axon and the dendrite. 3.Wider Methodological Frameworks Fusing Multiple Levels of Investigation might Boost Our Understanding of the BrainThe correlative methods presented in the previous section showed fundamental insight into different spatiotemporal scales of brain functioning. The small though solid bridge they provided between different types of data can be promisingly expanded toward a unified approach covering most perspectives. In detail, in vivo imaging by noninvasive human-targeted techniques like fMRI could be the starting point of a long pipeline that interrogates the long-term plasticity of small populations of neurons through in vivo TPFM functional and structural imaging. Actually, this translational step is the most critical and less explored, probably because fMRI remains a clinically oriented technique while TPFM is a basic science research tool. Nonetheless, understanding the cellular activation patterns underlying fMRI signals would be beneficial for diagnostic purposes, and it would boost the translational potential of light microscopy. Several studies on neurovascular coupling58–61 are now helping to fill this gap, and hopefully new correlative investigations combining simultaneous one- or two-photon fluorescence microendoscopy (e.g., Jung et al.)62 and MRI will come. Once functional and structural data are obtained in vivo at the small-circuitry level, the same sample shall be processed with LSM or analogous techniques for ex vivo long-range anatomical analysis. This contextualization into a wider framework is refined up to the synaptic scale when imaged through electron microscopy and/or super-resolution techniques. The need for sample processing procedures (e.g., clearing methods) compatible for all the techniques along the pipeline is one of the main limiting factor. In this view, the CLARITY technique, developed in Deisseroth’s lab to drastically reduce tissue scattering and perform optical imaging and immunolabeling through entire intact brains, preserves ultrastructural features and is thus EM compatible.63 Anyway, different degrees of tissue deformation throughout the pipeline are unavoidable and have to be considered when building multilevel maps of the brain. Informatics tools to align data at different scales within a common framework (i.e., stereotaxic atlases of murine and human brain) and big data storage facilities need to be further implemented and routinely used. Big transnational research partnerships like the Human Brain Project are working in this direction by “developing the integration and algorithmic reconstruction processes required for high fidelity reconstruction of the mouse brain across all levels of biological organization, from genes to cognition.”64 Since a completely integrated correlative approach implies that the same animal should be studied by multilevel analysis on several devices, the possibility of having this wide set of tools near to each other, e.g., in the same campus, is not a negligible issue. Multidisciplinary facilities provided with the above-described imaging devices are essential requirements for setting up this working strategy. In addition, considering the intrinsic difference between individuals (even between mice of the same strain), a multilevel investigation of the same sample would be extremely beneficial to reduce statistic variability and to cut the number of animals used in the experiments. We will try now to speculate on the information that can be gained on a neurological disease like stroke by following this pipeline. Stroke alters and triggers changes in intra- and inter-hemispheric connectivity; this rewiring aims at compensating for the loss of function.65 fMRI on (human and) mouse brain affected by stroke can tell the progression of the pathology over time, showing the plastic remapping of distant regions over the whole brain.66 MRI does not have enough resolution to infer what the cellular trigger of this remodeling is; simultaneously performed TPF imaging of labeled neuronal cells could reveal the structural and functional rewiring underlying fMRI signals in the newly activated cortical area of the same mouse with subcellular detail.67 Optical imaging on stroke animal models is capable of providing fundamental insight into dendritic remodeling, axonal rewiring, and spine plasticity67–69 while accurately depicting the functional remapping of the damaged cortex,70,71 and revealing angiogenesis and hemodynamic adaptation over time.72–74 Alterations in long-range projections underlying inter-hemispheric plasticity can be studied ex vivo on the same brain once cleared. Moreover, stroke-induced expression of several molecules and proteins, like growth-associated factors and inflammatory chemokines, can be addressed with multiround immunohistochemistry over the entire clarified brain by LSM or similar techniques. Once having the big picture in terms of long-term dynamics and wide-range remodeling, fine details like the presence of synaptic contacts on regenerated axons are available by electron microscopy on targeted regions of the same sample. In our opinion, this multidimensional hybrid strategy could be extremely useful in the investigation of complex brain diseases and would speed up the translation of neurobiology studies to clinical settings. Moreover, the setup of pharmacological treatments would crucially benefit from this multilevel investigation given the multitude of information that can be gained at once. We believe this kind of cross-disciplines multiscale studies is the missing tile to boost our knowledge of the brain. AcknowledgmentsThis research has received funding from LASERLAB-EUROPE (grant agreements no. 284464, EC’s Seventh Framework Programme) and has been supported by the Italian Ministry for Education, University and Research in the framework of the Flagship Project NANOMAX. This work has been supported by Regione Toscana in the framework of POR-CreO 2007-2013 action (SMAG project). This work has been supported by “Ente Cassa di Risparmio di Firenze.” This work is part of the activities of the European Flagship Human Brain Project (grant agreement no. 604102). Part of this work was performed in the frame of the Proof of Concept Studies for the ESFRI research infrastructure project Euro-BioImaging at the PCS facility LENS. ReferencesE. Bullmore and O. Sporns,
“Complex brain networks: graph theoretical analysis of structural and functional systems,”
Nature Rev. Neurosci., 10 186
–198
(2009). http://dx.doi.org/10.1038/nrn2575 NRNAAN 1471-0048 Google Scholar
N. K. Logothetis,
“What we can do and what we cannot do with fMRI?,”
Nature, 453 869
–878
(2008). http://dx.doi.org/10.1038/nature06976 NATUAS 0028-0836 Google Scholar
K. S. Saleem et al.,
“Magnetic resonance imaging of neuronal connections in the macaque monkey,”
Neuron, 34 685
–700
(2002). http://dx.doi.org/10.1016/S0896-6273(02)00718-3 NERNET 0896-6273 Google Scholar
A. Van der Linden et al.,
“In vivo manganese-enhanced magnetic resonance imaging reveals connections and functional properties of the songbird vocal control system,”
Neuroscience, 112 467
–474
(2002). http://dx.doi.org/10.1016/S0306-4522(02)00070-2 NERSD9 0735-2743 Google Scholar
M. Massimini et al.,
“Breakdown of cortical effective connectivity during sleep,”
Science, 309 2228
–2232
(2005). http://dx.doi.org/10.1126/science.1117256 SCIEAS 0036-8075 Google Scholar
M. Axer et al.,
“A novel approach to the human connectome: Ultra-high resolution mapping of fiber tracts in the brain,”
NeuroImage, 54 1091
–1101
(2011). http://dx.doi.org/10.1016/j.neuroimage.2010.08.075 NEIMEF 1053-8119 Google Scholar
P. Hagmann et al.,
“MR connectomics: principles and challenges,”
J. Neurosci. Methods, 194 34
–45
(2010). http://dx.doi.org/10.1016/j.jneumeth.2010.01.014 JNMEDT 0165-0270 Google Scholar
O. Sporns,
“The human connectome: origins and challenges,”
Neuroimage, 80 53
–61
(2013). http://dx.doi.org/10.1016/j.neuroimage.2013.03.023 NEIMEF 1053-8119 Google Scholar
T. B. Leergaard, C. C. Hilgetag and O. Sporns,
“Mapping the connectome: multi-level analysis of brain connectivity,”
Front. Neuroinf., 6 1
–6
(2012). http://dx.doi.org/10.3389/fninf.2012.00014 1662-5196 Google Scholar
L. Silvestri et al.,
“Confocal light sheet microscopy: micron-scale neuroanatomy of the entire mouse brain,”
Opt. Express, 20 20582
–20598
(2012). http://dx.doi.org/10.1364/OE.20.020582 OPEXFF 1094-4087 Google Scholar
H. U. Dodt et al.,
“Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain,”
Nat. Methods, 4 331
–336
(2007). http://dx.doi.org/10.1038/nmeth1036 1548-7091 Google Scholar
R. Tomer et al.,
“Advanced CLARITY for rapid and high-resolution imaging of intact tissues,”
Nature Protoc., 9 1682
–1697
(2014). http://dx.doi.org/10.1038/nprot.2014.123 NPARDW 1754-2189 Google Scholar
D. Mayerich, L. Abbott and B. McCormick,
“Knife-edge scanning microscopy for imaging and reconstruction of three-dimensional anatomical structures of the mouse brain,”
J. Microsc., 231 134
–143
(2008). http://dx.doi.org/10.1111/jmi.2008.231.issue-1 JMICAR 0022-2720 Google Scholar
A. Li et al.,
“Micro-optical sectioning tomography to obtain a high-resolution atlas of the mouse brain,”
Science, 330 1404
–1408
(2010). http://dx.doi.org/10.1126/science.1191776 SCIEAS 0036-8075 Google Scholar
H. Gong et al.,
“Continuously tracing brain-wide long-distance axonal projections in mice at a one-micron voxel resolution,”
Neuroimage, 74 87
–98
(2013). http://dx.doi.org/10.1016/j.neuroimage.2013.02.005 NEIMEF 1053-8119 Google Scholar
T. Ragan et al.,
“Serial two-photon tomography for automated ex vivo mouse brain imaging,”
Nat. Methods, 9 255
–258
(2012). http://dx.doi.org/10.1038/nmeth.1854 1548-7091 Google Scholar
J. W. Lichtman, J. Livet and J. R. Sanes,
“A technicolour approach to the connectome,”
Nat. Rev. Neurosci., 9 417
–422
(2008). http://dx.doi.org/10.1038/nrn2391 NRNAAN 1471-0048 Google Scholar
F. Helmchen and W. Denk,
“Deep tissue two-photon microscopy,”
Nat. Methods, 2 932
–940
(2005). http://dx.doi.org/10.1038/nmeth818 1548-7091 Google Scholar
K. Svoboda et al.,
“In vivo dendritic calcium dynamics in neocortical pyramidal neurons,”
Nature, 385 161
–165
(1997). http://dx.doi.org/10.1038/385161a0 NATUAS 0028-0836 Google Scholar
K. Ohki et al.,
“Functional imaging with cellular resolution reveals precise micro-architecture in visual cortex,”
Nature, 433 597
–603
(2005). http://dx.doi.org/10.1038/nature03274 NATUAS 0028-0836 Google Scholar
D. A. Dombeck et al.,
“Imaging large-scale neural activity with cellular resolution in awake, mobile mice,”
Neuron, 56 43
–57
(2007). http://dx.doi.org/10.1016/j.neuron.2007.08.003 NERNET 0896-6273 Google Scholar
D. A. Dombeck et al.,
“Functional imaging of hippocampal place cells at cellular resolution during virtual navigation,”
Nat. Neurosci., 13 1433
–1440
(2010). http://dx.doi.org/10.1038/nn.2648 NANEFN 1097-6256 Google Scholar
J. Sawinski et al.,
“Visually evoked activity in cortical cells imaged in freely moving animals,”
Proc. Natl. Acad. Sci. U. S. A., 106 19557
–19562
(2009). http://dx.doi.org/10.1073/pnas.0903680106 PNASA6 0027-8424 Google Scholar
W. Denk and H. Horstmann,
“Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure,”
PLoS Biol., 2 e329
(2004). http://dx.doi.org/10.1371/journal.pbio.0020329 1544-9173 Google Scholar
R. Schalek et al.,
“Development of high-throughput, high-resolution 3D reconstruction of large-volume biological tissue using automated tape collection ultramicrotomy and scanning electron microscopy,”
Microsc. Microanal., 17 966
–967
(2011). http://dx.doi.org/10.1017/S1431927611005708 MIMIF7 1431-9276 Google Scholar
G. Knott et al.,
“Serial section scanning electron microscopy of adult brain tissue using focused ion beam milling,”
J. Neurosci., 28 2959
–2964
(2008). http://dx.doi.org/10.1523/JNEUROSCI.3189-07.2008 JNRSDS 0270-6474 Google Scholar
E. Betzig et al.,
“Imaging intracellular fluorescent proteins at nanometer resolution,”
Science, 313 1642
–1645
(2006). http://dx.doi.org/10.1126/science.1127344 SCIEAS 0036-8075 Google Scholar
S. W. Hell and J. Wichmann,
“Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy,”
Opt. Lett., 19 780
–782
(1994). http://dx.doi.org/10.1364/OL.19.000780 OPLEDP 0146-9592 Google Scholar
D. J. Heeger and D. Ress,
“What does fMRI tell us about neuronal activity?,”
Nat. Rev. Neuroscience, 3 142
–151
(2002). http://dx.doi.org/10.1038/nrn730 NRNAAN 1471-0048 Google Scholar
K. E. Stephan et al.,
“Comparing hemodynamic models with DCM,”
Neuroimage, 38 387
–401
(2007). http://dx.doi.org/10.1016/j.neuroimage.2007.07.040 NEIMEF 1053-8119 Google Scholar
K. Schulz et al.,
“Simultaneous BOLD fMRI and fiber-optic calcium recording in rat neocortex,”
Nat. Methods, 9 597
–602
(2012). http://dx.doi.org/10.1038/nmeth.2013 1548-7091 Google Scholar
S. Charpak and B. Stefanovic,
“Shedding light on the BOLD fMRI response,”
Nat. Methods, 9 547
–549
(2012). http://dx.doi.org/10.1038/nmeth.2039 1548-7091 Google Scholar
L. Silvestri et al.,
“Correlative two-photon and light sheet microscopy,”
Methods, 66 268
–272
(2014). http://dx.doi.org/10.1016/j.ymeth.2013.06.013 MTHDE9 1046-2023 Google Scholar
K. L. Briggman, M. Helmstaedter and W. Denk,
“Wiring specificity in the direction-selectivity circuit of the retina,”
Nature, 471 183
–188
(2011). http://dx.doi.org/10.1038/nature09818 NATUAS 0028-0836 Google Scholar
F. W. Grillo et al.,
“Increased axonal bouton dynamics in the aging mouse cortex,”
Proc. Natl. Acad. Sci., 110 E1514
–E1523
(2013). http://dx.doi.org/10.1073/pnas.1218731110 PMASAX 0096-9206 Google Scholar
K. Svoboda and R. Yasuda,
“Principles of two-photon excitation microscopy and its applications to neuroscience,”
Neuron, 50 823
–839
(2006). http://dx.doi.org/10.1016/j.neuron.2006.05.019 NERNET 0896-6273 Google Scholar
A. Holtmaat and K. Svoboda,
“Experience-dependent structural synaptic plasticity in the mammalian brain,”
Nat. Rev. Neurosci., 10 647
–658
(2009). http://dx.doi.org/10.1038/nrn2699 NRNAAN 1471-0048 Google Scholar
D. Stettler et al.,
“Axons and synaptic boutons are highly dynamic in adult visual cortex,”
Neuron, 49 877
–887
(2006). http://dx.doi.org/10.1016/j.neuron.2006.02.018 NERNET 0896-6273 Google Scholar
V. De Paola et al.,
“Cell type-specific structural plasticity of axonal branches and boutons in the adult neocortex,”
Neuron, 49 861
–875
(2006). http://dx.doi.org/10.1016/j.neuron.2006.02.017 NERNET 0896-6273 Google Scholar
D. J. Margolis et al.,
“Reorganization of cortical population activity imaged throughout long-term sensory deprivation,”
Nat. Neurosci., 15 1539
–1546
(2012). http://dx.doi.org/10.1038/nn.3240 NANEFN 1097-6256 Google Scholar
H. B. Barlow, R. M. Hill and W. R. Levick,
“Retinal ganglion cells responding selectively to direction and speed of image motion in the rabbit,”
J. Physiol., 173 377
–407
(1964). http://dx.doi.org/10.1113/jphysiol.1964.sp007463 JPHYA7 0022-3751 Google Scholar
S. I. Fried, T. A. Munch and F. S. Werblin,
“Mechanisms and circuitry underlying directional selectivity in the retina,”
Nature, 420 411
–414
(2002). http://dx.doi.org/10.1038/nature01179 NATUAS 0028-0836 Google Scholar
E. V. A. Famiglietti,
“A structural basis for omnidirectional connections between starburst amacrine cells and directionally selective ganglion cells in rabbit retina, with associated bipolar cells,”
Vis. Neurosci., 19 145
–162
(2002). http://dx.doi.org/10.1017/S0952523802191139 VNEUEY 0952-5238 Google Scholar
E. V. Famiglietti,
“Synaptic organization of starburst amacrine cells in rabbit retina: analysis of serial thin sections by electron microscopy and graphic reconstruction,”
J. Comp. Neurol., 309 40
–70
(1991). http://dx.doi.org/10.1002/(ISSN)1096-9861 JCNEAM 0021-9967 Google Scholar
R. F. Dacheux, M. F. Chimento and F. R. Amthor,
“Synaptic input to the on-off directionally selective ganglion cell in the rabbit retina,”
J. Comparative Neurol., 456 267
–278
(2003). http://dx.doi.org/10.1002/(ISSN)1096-9861 JCNEAM 0021-9967 Google Scholar
C. W. Oyster, F. R. Amthor and E. S. Takahashi,
“Dendritic architecture of ON-OFF direction-selective ganglion cells in the rabbit retina,”
Vision Res., 33 579
–608
(1993). http://dx.doi.org/10.1016/0042-6989(93)90181-U VISRAM 0042-6989 Google Scholar
G. Yang and R. H. Masland,
“Receptive fields and dendritic structure of directionally selective retinal ganglion cells,”
J. Neurosci., 14 5267
–5280
(1994). JNRSDS 0270-6474 Google Scholar
A. Holtmaat et al.,
“Long-term, high-resolution imaging in the mouse neocortex through a chronic cranial window,”
Nat. Protoc., 4 1128
–1144
(2009). http://dx.doi.org/10.1038/nprot.2009.89 NPARDW 1754-2189 Google Scholar
A. Holtmaat et al.,
“Experience-dependent and cell-type-specific spine growth in the neocortex,”
Nature, 441 979
–983
(2006). http://dx.doi.org/10.1038/nature04783 NATUAS 0028-0836 Google Scholar
G. W. Knott et al.,
“Spine growth precedes synapse formation in the adult neocortex in vivo,”
Nat. Neurosci., 9 1117
–1124
(2006). http://dx.doi.org/10.1038/nn1747 NANEFN 1097-6256 Google Scholar
J. T. Trachtenberg et al.,
“Long-term in vivo imaging of experience-dependent synaptic plasticity in adult cortex,”
Nature, 420 788
–794
(2002). http://dx.doi.org/10.1038/nature01273 NATUAS 0028-0836 Google Scholar
D. Bishop et al.,
“Near-infrared branding efficiently correlates light and electron microscopy,”
Nat. Methods, 8 568
–570
(2011). http://dx.doi.org/10.1038/nmeth.1622 1548-7091 Google Scholar
B. Maco et al.,
“Correlative in vivo 2 photon and focused ion beam scanning electron microscopy of cortical neurons,”
PloS One, 8 e57405
(2013). http://dx.doi.org/10.1371/journal.pone.0057405 1932-6203 Google Scholar
M. Cane et al.,
“The relationship between PSD-95 clustering and spine stability in vivo,”
J. Neurosci., 34 2075
–2086
(2014). http://dx.doi.org/10.1523/JNEUROSCI.3353-13.2014 JNRSDS 0270-6474 Google Scholar
R. Mostany et al.,
“Altered synaptic dynamics during normal brain aging,”
J. Neurosci., 33 4094
–4104
(2013). http://dx.doi.org/10.1523/JNEUROSCI.4825-12.2013 JNRSDS 0270-6474 Google Scholar
A. L. Allegra Mascaro et al.,
“In vivo single branch axotomy induces GAP-43-dependent sprouting and synaptic remodeling in cerebellar cortex,”
Proc. Natl. Acad. Sci. U. S. A., 110 10824
–10829
(2013). http://dx.doi.org/10.1073/pnas.1219256110 PNASA6 0027-8424 Google Scholar
A. J. Canty et al.,
“In-vivo single neuron axotomy triggers axon regeneration to restore synaptic density in specific cortical circuits,”
Nat. Commun., 4 2038
(2013). http://dx.doi.org/10.1038/ncomms3038 NCAOBW 2041-1723 Google Scholar
E. M. C. Hillman,
“Coupling mechanism and significance of the BOLD signal: a status report,”
Annu. Rev. Neurosci., 37 161
–181
(2014). http://dx.doi.org/10.1146/annurev-neuro-071013-014111 ARNSD5 0147-006X Google Scholar
A. Devor et al.,
“Neuronal basis of non-invasive functional imaging: from microscopic neurovascular dynamics to BOLD fMRI,”
Neural Metabolism In Vivo, 433
–500 Springer, USA
(2012). Google Scholar
A. Devor et al.,
“Frontiers in optical imaging of cerebral blood flow and metabolism,”
J. Cereb. Blood Flow Metab., 32 1259
–1276
(2012). http://dx.doi.org/10.1038/jcbfm.2011.195 JCBMDN 0271-678X Google Scholar
C. Du et al.,
“Low-frequency calcium oscillations accompany deoxyhemoglobin oscillations in rat somatosensory cortex,”
Proc. Natl. Acad. Sci., 111 E4677
–E4686
(2014). http://dx.doi.org/10.1073/pnas.1410800111 PMASAX 0096-9206 Google Scholar
J. C. Jung et al.,
“In vivo mammalian brain imaging using one- and two-photon fluorescence microendoscopy,”
J. Neurophysiol., 92 3121
–3133
(2004). http://dx.doi.org/10.1152/jn.00234.2004 JONEA4 0022-3077 Google Scholar
K. Chung et al.,
“Structural and molecular interrogation of intact biological systems,”
Nature, 497 332
–337
(2013). http://dx.doi.org/10.1038/nature12107 NATUAS 0028-0836 Google Scholar
T. H. Murphy and D. Corbett,
“Plasticity during stroke recovery: from synapse to behaviour,”
Nat. Rev. Neurosci., 10 861
–872
(2009). http://dx.doi.org/10.1038/nrn2735 NRNAAN 1471-0048 Google Scholar
N. S. Ward et al.,
“Neural correlates of motor recovery after stroke: a longitudinal fMRI study,”
Brain, 126 2476
–2496
(2003). http://dx.doi.org/10.1093/brain/awg245 BRAIAK 0006-8950 Google Scholar
C. E. Brown et al.,
“Extensive turnover of dendritic spines and vascular remodeling in cortical tissues recovering from stroke,”
J. Neurosci., 27 4101
–4109
(2007). http://dx.doi.org/10.1523/JNEUROSCI.4295-06.2007 JNRSDS 0270-6474 Google Scholar
R. Mostany et al.,
“Local hemodynamics dictate long-term dendritic plasticity in peri-infarct cortex,”
J. Neurosci., 30 14116
–14126
(2010). http://dx.doi.org/10.1523/JNEUROSCI.3908-10.2010 JNRSDS 0270-6474 Google Scholar
C. E. Brown and T. H. Murphy,
“Livin’ on the edge: imaging dendritic spine turnover in the peri-infarct zone during ischemic stroke and recovery,”
Neuroscientist, 14 139
–146
(2008). http://dx.doi.org/10.1177/1073858407309854 NROSFJ 1073-8584 Google Scholar
A. Sigler, M. H. Mohajerani and T. H. Murphy,
“Imaging rapid redistribution of sensory-evoked depolarization through existing cortical pathways after targeted stroke in mice,”
Proc. Natl. Acad. Sci. U. S. A., 106 11759
–11764
(2009). http://dx.doi.org/10.1073/pnas.0812695106 PNASA6 0027-8424 Google Scholar
T. C. Harrison et al.,
“Displacement of sensory maps and disorganization of motor cortex after targeted stroke in mice,”
Stroke, 44 2300
–2306
(2013). http://dx.doi.org/10.1161/STROKEAHA.113.001272 SJCCA7 0039-2499 Google Scholar
A. Y. Shih et al.,
“Active dilation of penetrating arterioles restores red blood cell flux to penumbral neocortex after focal stroke,”
J. Cereb. Blood Flow Metab., 29 738
–751
(2009). http://dx.doi.org/10.1038/jcbfm.2008.166 JCBMDN 0271-678X Google Scholar
J. Lee et al.,
“Quantitative imaging of cerebral blood flow velocity and intracellular motility using dynamic light scattering-optical coherence tomography,”
J. Cereb. Blood Flow Metab., 33 819
–825
(2013). http://dx.doi.org/10.1038/jcbfm.2013.20 JCBMDN 0271-678X Google Scholar
A. Y. Shih et al.,
“Two-photon microscopy as a tool to study blood flow and neurovascular coupling in the rodent brain,”
J. Cereb. Blood Flow Metab., 32 1277
–1309
(2012). http://dx.doi.org/10.1038/jcbfm.2011.196 JCBMDN 0271-678X Google Scholar
BiographyAnna Letizia Allegra Mascaro obtained a master’s degree in chemistry at the University of Florence in 2007. She obtained a PhD degree in atomic and molecular spectroscopy in 2011. Her research as a postdoctoral fellow (2011 to 2014) at the European Laboratory for Non-Linear Spectroscopy concerned the study of neuronal plasticity and regeneration with two-photon microscopy in vivo, which still represents her main research interest. She currently is a researcher at the National Institute of Optics in Florence. Ludovico Silvestri received a master’s degree in physics at the University of Pisa in 2008. He then moved to the University of Florence, where he obtained a PhD degree in atomic and molecular spectroscopy in 2012. From 2012 to 2014, he was a postdoctoral at the European Laboratory for Non-Linear Spectroscopy. Currently, he is a researcher at the National Institute of Optics in Florence. His research activity is focused on light sheet microscopy and its applications. Leonardo Sacconi received a master’s degree in physics at the University of Florence in 2001. He then moved to Trento, where he obtained a PhD degree in physics. He is a visiting scientist at Cornell University, Ithaca, New York, USA, in the group of Watt Webb. From 2005 to 2011, he started new research lines on functional imaging of neuronal networks in vivo. He is now a researcher at the National Institute of Optics in Florence. Francesco S. Pavone received a master’s degree in physics at the University of Florence in 1989. In 1993 he obtained a PhD degree in optics at the National Institute for Optics. In 1997, he spent a year and a half as Maitre de Conférences Associe au College de France, Paris. In 1998, he became an associate professor of physics at the University of Perugia. In 2001, he moved as an associate professor to the University of Florence. In 2004, he became a full professor. |

