|
|
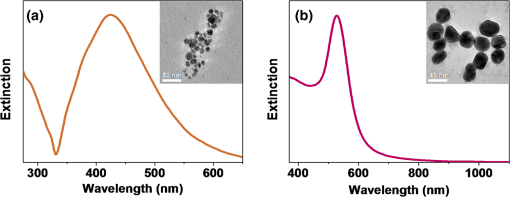
1.IntroductionColocalizations of nanoparticles and biomolecules in cellular environments are important for the development of efficient nano-biosensors and theranostics.1–4 Detection of theranostics can be based on their plasmonic signals or surface enhanced Raman scattering (SERS) signals obtained from the molecules in the vicinity of particles. To assess the intracellular specificity of theranostics, it is important to verify the exact size, shape, and state (single or dimer or aggregate) of nanoparticles along with the identification of molecules adsorbed on their surfaces.5,6 Various techniques have been developed for such observations at the single particle level.7–9 Among the available techniques, dark field scattering microspectroscopy (DFSMS) is a well-established technique for the observations of single plasmonic nanoparticles (particularly silver and gold).10,11 On the other side, confocal Raman scattering microspectroscopy (RSMS) has been considered as an efficient tool for the detection of single molecules, and where Raman signals from biomolecules are too weak to be detected in a noisy environment, noble metal nanoparticles (NMNPs) can be of help.12,13 Due to their SERS activity and thermal properties, NMNPs have been used as theranostics.12,13 In a complex environment, nanoparticles of different sizes and shapes may exhibit similar signals or SERS activities.14 Hence, confirmation of the signals obtained using multiple techniques is essential for improving detection and sensing capabilities.15 In such cases, the exact nature of particles can be determined by methodologies based on polarization and excitation wavelength-dependent measurements of their SERS activity or their plasmonic spectra.16–19 To precisely monitor specificity, correlated measurements using combined techniques and setups are required. These techniques generally involve correlation between two different signals obtained from the same nanoparticle15 or precise tracking of single particles in real time.20 In this context, if it is necessary to understand the exact state of nanoparticles and biomolecules responsible for the state change,4,21 DFSMS and RSMS are the important techniques which allow such identifications. A few demonstrations of such combined techniques have been done in the past. Lee et al. have demonstrated such correlated measurements for dimeric and trimeric forms of Au–Ag core shell nanoparticles tethered on DNA.22 In this study, Rayleigh scattering, SERS, and atomic force microscopy were used for correlated measurements. These results were used to determine relationships between particle structure, SERS, and Rayleigh scattering with the help of scattering intensity ratio and the incident polarization. In another study, such correlated measurements between Rayleigh scattering and SERS were performed on AgNP aggregates and the data obtained were used to determine the optimized conditions required for maximum SERS.23 Apart from such physicochemical applications and developments, correlated measurements have major applications in biology. A study from Henry et al. has shown the application of such a technique to collect SERS signals from multiple nanotags at the same time.24 In other studies performed by El-Sayed et al., combined Rayleigh and Raman spectroscopy was used to identify phases of cell cycle or other applications such as the determination of drug efficacy from single cell imaging.21,25 In a recent work, we have performed single cell investigations of silver nanoparticles within bacteria using separate DFSMS and RSMS measurements.10 With this background, development of such techniques is under progress and improvements in terms of resolution, ease of operation, various data analysis, and interpretation techniques are the criteria under consideration.21,25 Here, we discuss issues related to the specificity of detection techniques. In sensing and detection of intracellular nanoparticles, while using plasmonic/fluorescence imaging, changes in the scattering/fluorescence signals due to dimerization or aggregation of nanostructures are considered as signals.26–28 But due to the complexity of a biological environment, apart from the molecule of interest, many other moieties in the cellular environment can affect the system under investigation. For example, proteins in the cell culture medium itself cause aggregation of nanoparticles.29 Moreover, interactions among the nanoparticles themselves lead to their aggregation.5 Also, due to various chemical changes in the surroundings of nanoparticles, changes in the intrinsic properties of nanoparticles may also affect the confidence level of the diagnostic data.3 Hence, in such cases, it is necessary to identify the molecules in the vicinity of the nanoparticles as well as state of the particle such as dimer or aggregate. Similarly, while using SERS-based detection of intracellular nanoparticles, changes in the intensity of Raman signals are considered for the localization of nanoparticles.30,31 In such cases, for better understanding of the results, it is important to understand the state of the nanoparticle as well as the mechanisms involved in the SERS activity of the same. To solve the aforementioned issues, in this work, we have demonstrated a combination of high-resolution DFSMS with RSMS. A system of AgNP-treated bacteria was investigated as an example. We have shown successful colocalization between plasmonic signatures of nanoparticles along with the Raman signature of biomolecules in the vicinity of bacteria. Applicability of this setup for the measurements on mammalian cells was demonstrated with the system of HEK293 cells treated with gold nanoparticles. Further, we discuss the mechanism of SERS of nanostructures within cells. To the best of our knowledge, this is the first time such measurements have been used for intracellular colocalization of plasmonic nanoparticles with biomolecules and to investigate the mechanisms of SERS inside cells. 2.Materials and Methods2.1.MaterialsTetrachloroauric acid trihydrate () (99.9%), trisodium citrate (), poly-l-lysine solution (0.1% in ), and trypsin-Ethylenediaminetetraacetic acid (0.25% in ) were purchased from Sigma Chemicals. Silver nitrate (99.9%) was purchased from RANKEM, India. Dulbecco’s modified Eagle medium (DMEM) with high glucose, GlutaMAX™ Supplement and pyruvate, Antibiotic (, penicillin–streptomycin–glutamine), and fetal bovine serum were purchased from Invitrogen. Plasticware for cell culture was purchased from Tarson, India. Millipore deionized water (DI) () was used throughout the experiments. 2.2.Synthesis of Citrate Capped Silver NanoparticlesAgNPs were synthesized by the Turkevich method.32 Briefly, 17 mg of AgNO3 was dissolved in 100 mL of DI water. This solution was heated to 100°C and then 40 mg of trisodium citrate was added. Once the color of the solution started changing to pale yellow, it was cooled under tap water. As synthesized, AgNPs were characterized using UV–Vis spectroscopy and transmission electron microscopy (TEM) imaging. A broad surface plasmon resonance (SPR) distribution with a maximum at was observed [Fig. 1(a)]. TEM suggests nonuniformity in the size and shape of AgNPs. 2.3.Synthesis of Citrate Capped Gold NanoparticlesGNPs were synthesized by the Turkevich method.32 Briefly, 20 mL of 0.3 mM solution in DI water was heated in a synthesizer (400 rpm). Upon boiling, of 100 mM trisodium citrate solution was added. After 20 min of boiling, the color of the solution changed to wine red. Then the solution was cooled at room temperature. As synthesized, AuNPs were characterized using UV–Vis spectroscopy and TEM imaging. AuNPs of size in diameter with SPR around 529 nm were observed [Fig. 1(b)]. 2.4.Bacterial Culture and Sample PreparationEscherichia coli 739 bacterial culture was grown to an (optical density at 600 nm) value of 1. This culture (5 mL) was washed twice by centrifugation (3000 rpm, 10 min) with DI water. The final pellet was resuspended in the as-prepared AgNP solution and incubated at room temperature for 1 h. After that, this E. coli-AgNP suspension was washed twice with DI water and spotted on a poly-l-lysine-coated ultrasonically cleaned glass slide (1-mm thickness, SCHOTT). After 5 min of incubation, the slide was flushed with DI water to remove unbound bacteria and covered with 0.145-mm thick coverslips (SCHOTT) for imaging purposes. 2.5.Mammalian Cell Culture and Sample PreparationHEK293 cells were cultured on poly-l-lysine-coated glass coverslips in six-well plates. After reaching confluency, cells were washed twice with phosphate-buffered saline (PBS) and 2 mL DMEM containing citrate capped GNPs was added. These cells were incubated overnight inside the incubator at 37°C. After this, the cells were washed thrice with PBS and incubated with paraformaldehyde solution (4% in PBS) for 8 min. Then the cells were washed thrice with PBS and mounted on 1-mm thick glass slides (SCHOTT). 2.6.UV–Visible Extinction SpectroscopyEnsemble UV–Vis extinction spectroscopic measurements were performed using PerkinElmer Lambda 25 spectrophotometer in the range of 200 to 1100 nm. 2.7.Transmission Electron MicroscopyHigh resolution transmission electron microscopic measurements were performed using a JEOL 3010, 300 kV instrument. Carbon-coated copper grids were used for measurements. Samples were spotted by drop casting followed by ambient air drying. 2.8.Confocal Raman Scattering MicrospectroscopyRaman microspectroscopic measurements were performed using WiTec GmbH equipment. Frequency doubled Nd:YAG dye laser (532 nm) with a maximum output power of 40 mW was used for excitation of the sample. The laser was focused onto the sample using a oil immersion objective (UPLFLN, Olympus). The signal, after passing through a 532 nm super-notch filter, was dispersed using a grating () onto a charge-coupled device (CCD). Spectral images were scanned using the sample mounted on a piezo stage. 2.9.Plasmonic and Raman Scattering MicrospectroscopyFor plasmonic and Raman scattering microspectroscopy (PRSMS) measurements, an attachment was designed to use a Cytoviva™ high-resolution dark field condenser (oil immersion) and oil immersion objective (UPLFLN, Olympus) in the above-mentioned confocal Raman microspectroscope. For white light illumination (400 to 1000 nm), a L1090-Halogen Lamp from International Light Technologies Inc. was used. The other details of the instrument for Raman microspectroscopic measurements are the same as above. 3.Results and Discussion3.1.Setup for Combined Plasmonic and Raman Scattering MicrospectroscopyTo accomplish correlated DFSMS and RSMS measurements of biological samples using the same setup, we attached a high-resolution dark field condenser with a conventional point-scan RSMS setup. Schematic and photographs of the actual setup are shown in Fig. 2. In the PRSMS setup, a dark field condenser was used to illuminate samples in the transmitted dark-field configuration. For the DFSMS of samples, a visible–near-infrared (VNIR) light source with a wavelength range from 400 to 1000 nm was used to excite the plasmonic modes of NMNPs. For RSMS, a 532-nm laser was used to excite the Raman spectra and measurements were performed in the reflection mode. Samples sandwiched between a coverslip and glass slide were placed on a piezoelectric scanner and the scattered light was collected using a oil immersion objective. Substantial signal strength in conjunction with confocality was achieved using a optical fiber. This fiber was connected to a VNIR spectrometer equipped with appropriate gratings to resolve the spectra. Since for DFSMS a wide range of wavelengths has to be scanned, a grating with was used. For RSMS, a shorter wavelength range but better resolution is necessary. Hence, to resolve RSMS spectra, a grating with was used. It is important to mention that despite the use of wide field illumination in DFSMS, the method of signal collection was confocal due to the configuration of the RSMS setup. Such a signal collection in DFSMS has its pros and cons, which are discussed below. Fig. 2Schematic of the setup for correlated plasmonic and Raman scattering microspectroscopy (PRSMS) measurements. Briefly, it consists of a high-resolution dark field condenser attached with conventional point-scan confocal RSMS setup. Inset shows a dark field condenser (red arrow, top left) along with the designed attachment (blue arrow, down right). 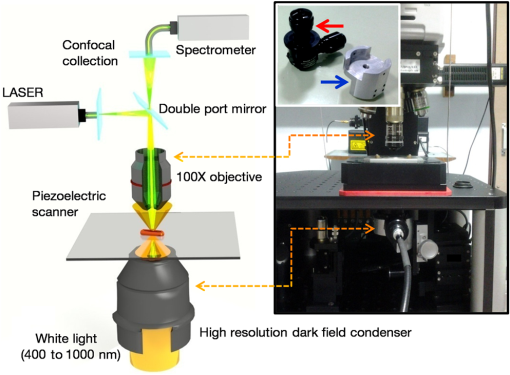 3.2.Images of AgNP Treated Bacteria Collected with Different ConfigurationsFigure 3(a) shows a dark field image of bacteria (E. coli 739) treated with citrate capped silver nanoparticles. This image was captured using a CCD camera of the PRSMS setup. In such an image, it appears that only borders (cell wall) of the bacteria scatter substantial light, whereas the central portion appears transparent. This is because the wide field image strongly reflects the relative intensity variations which occur due to light collection from all focal planes. But when this sample was measured for DFSMS using the PRSMS setup, the spectral image was captured pixel by pixel using an optical fiber which introduces confocality. Such a confocal signal collection produces an image constructed from the light collected only from a particular focal plane. This image has a diffused appearance as shown in Fig. 3(b). To eliminate the possibility of defocused imaging in the PRSMS setup, care was taken to focus the desired plane of imaging by using a laser spot for confocal RSMS. Figure 3(c) shows a line profile of the scattering intensity along the yellow line in Fig. 3(b). Line profile shows the maximum at the center similar to the airy disc pattern and confirms that the particle was properly focused. Fig. 3(a) Dark field image captured using CCD camera of the PRSMS setup. (b) Pseudo-colored dark field scattering microspectroscopy (DFSMS) image collected using the PRSMS setup for the same area. Red arrows indicate the presence of AgNPs which exhibit sharp SPRs. A dotted yellow line shows pixels for which scattering intensity profile is shown in (c). Scale bar is for images (a) and (b). 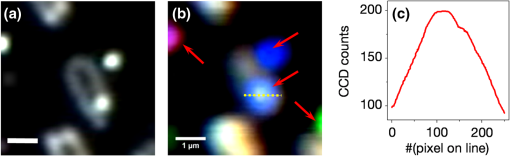 We also observed that when DFSMS images are presented as intensity scaled images, it does not reveal information about locating the NMNPs in complex environments. However, the sharp and nearly monochromatic scattering features of NMNPs allow them to be distinguished as bright and distinct colored spots. This can be achieved by presenting DFSMS images as pseudo-RGB color coded images [Fig. 3(b)]. In pseudo-colored DFSMS images, we have assigned 552-, 615-, and 688-nm wavelengths to blue, green, and red colors, respectively. With this, particles can be clearly located as shown in Fig. 3(b) by red arrows. On this background, we have performed PRSMS measurements on two different systems. These measurements combine our ability to locate NMNPs using DFSMS and colocalizing the same with biomolecules using RSMS. 3.3.Colocalization of AgNPs and Biomolecules in E. ColiA previous study10 by our group using RSMS has shown that AgNPs interact specifically with the bacterial DNA. It was observed that adenine (A), guanine (G), and cytosine (C) exhibit substantial SERS intensities while thymine (T) shows only limited SERS enhancement. This is because the interactions between AgNPs and A, G, and C are stronger than those with T. This specificity of interaction is due to the interaction between AgNPs and the exocyclic nitrogen present in A, G, and C. In the aforementioned study, DFSMS was used separately to show the attachment of AgNPs to the bacteria.10 Here, in the present study, we use PRSMS to colocalize the position of AgNPs with the Raman signatures of biomolecules. Figure 4 shows the consolidated data of these measurements. Fig. 4Images of AgNP treated bacteria collected using PRSMS setup in different configurations. (a) Dark field image captured using CCD camera. (b) Pseudo-colored DFSMS image. (c) Intensity scaled confocal RSMS image and corresponding color scale are shown. (d) Overlay of the clustered RSMS image with dark field image. (e) Scattering spectra of AgNPs 1 to 5 are shown along with the scattering spectrum of bacteria. (f) Averaged cluster spectra corresponding to image (d) are shown in matching colors. Scale bar is in all the images. 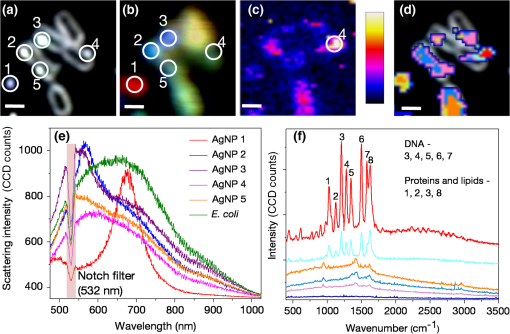 Figure 4(a) shows the dark field image captured using a CCD camera with the PRSMS setup. In the region of interest (ROI) selected for imaging, we can see AgNPs labeled 1, 2, 3, and 4 are attached on the surface of bacteria and particle 5 is on the glass slide. The next image shows a pseudo-colored DFSMS image [Fig. 4(b)] for the same ROI. Scattering spectra collected from particle positions are shown in Fig. 4(e). Based on the scattering spectra, it can be observed that particle 1 is possibly a rod or prism-shaped silver nanoparticle;33 whereas the other particles are relatively smaller and scatter light at lower wavelengths. Among the marked AgNPs, particles 2 and 3 scatter more light as compared to particles 4 and 5. This suggests that AgNPs 2 and 3 are relatively larger particles. Also as compared to AgNP 1, other particles show a broadening in their scattering peak, implying adsorption of biomolecules on their surfaces. Figure 4(c) shows the intensity scaled RSMS image of the same ROI shown in Fig. 4(a). It suggests a maximum intensity enhancement in the vicinity of AgNPs 2 and 4. To understand this data better, we have done cluster analysis of the RSMS image. Cluster analysis helped in grouping and color coding the Raman spectra on the basis of their similarities. Then this color coded image was overlaid with the dark field image as shown in Fig. 4(d). Averages of the spectral clusters are shown in Fig. 4(f) where it can be seen that although some intensity enhancement was observed on AgNP 2 and bacteria at the bottom, actual SERS activity was observed only in the vicinity of AgNP 4 (red colored Raman spectrum). Considering the scattering spectrum of AgNP 4, it can be concluded that in this case, SERS activity is due more to the electronic resonance-charge transfer between the AgNP and the molecule in its vicinity than the electric near-field enhancement. These observations suggest that although many AgNPs attach to the bacteria, all particles do not exhibit SERS activity. This is due to the dependence of SERS activity on the site of molecular adsorption on the surface of the nanoparticle as well as the size and shape of the nanoparticle. Assignments of Raman bands corresponding to the molecule in the vicinity of AgNP 4 are given in Table 1. Most of these bands were found to be due to DNA. These results are in good agreement with our previous observations on the interaction between AgNPs and bacteria.10 For a further demonstration of the applicability of PRSMS setup with mammalian cells, we have performed measurements on HEK293 cells treated with citrate capped AuNPs. These observations are discussed in Sec. 3.4. Table 1Assignments of Raman bands of AgNP treated Escherichia coli sample. 3.4.Colocalization of AuNPs and Biomolecules Inside HEK293 CellsFigure 5(a) shows the dark field image of HEK293 cells treated with citrate capped AuNPs of in diameter. The Z scanning (focus stacking) measurements performed on these cells have shown that the AuNPs under investigation were within the cells. To prove this, a movie compiled of a Z scanned image stack is shown in Video 1. Figure 5(b) shows a pseudo-colored DFSMS image of the ROI shown in Fig. 5(a). Corresponding scattering spectra shown in Fig. 5(d) suggest that particle 1 has a sharp plasmon resonance and relatively less FWHM, which implies that it is a single AuNP. Other particles (2 and 3) show scattering signatures of aggregates (multiple broad but sharp SPRs). The scattering signal from cellular components exhibits a broad spectrum as well. When the ROI shown in Fig. 5(a) was scanned for RSMS, particle 1 exhibited the most SERS activity [Figs. 5(c) and 5(e)] whereas an aggregate (particle 2) exhibits only a moderate enhancement in the Raman signal as compared to the background. This observation suggests that, although due to the plasmonic coupling effect aggregates are expected to exhibit better SERS activity than single nanoparticles, SERS activity is highly specific to the site of molecule adsorption. Hence, such PRSMS studies are necessary for additional confirmation about the nature of the nanoparticle under investigation. When diagnostic or sensing techniques are solely based on SERS activity or the expected hybridization of NMNPs by specific biomolecules, colocalization using PRSMS is necessary to prove the presence of moieties and nanostructures of interest. In this particular measurement, assignments for Raman bands are given in Table 2. A major band was assigned to protein molecules (1600 to ) and another band between 1050 and was assigned to DNA and RNA O-P-O stretching. It has been reported by previous studies that such an appearance of the Raman band is commonly found for mammalian cells undergoing apoptosis.25 Fig. 5Images of AuNP-treated HEK293 cells collected using PRSMS setup at different configurations. (a) Dark field image captured using CCD camera. Region of interest (ROI) scanned for PRSMS is shown by the square. Image is taken at such focus that nanoparticles can be seen clearly. Cells can be seen clearly in a movie made of Z stacking of images (Video 1). (b) Pseudo-colored DFSMS image of the ROI shown in (a). Particles are numbered as 1, 2, and 3. Scale bar is 1 micron. (c) Intensity scaled RSMS image. (d) Scattering spectra of AuNPs 1 to 3 are shown along with the scattering spectrum from cellular components. (e) Inset of this image shows RSMS image of particle 1 when it was rescanned. Scale bar is . Raman spectra corresponding to particles 1 (red) and 2 (blue) from RSMS image (c) are shown along with the Raman spectrum of particle 1 after rescanning (black) (Video 1, MPEG, 0.4 MB).[DOI: http://dx.doi.org/10.1117/1.JBO.20.4.046011.1]. 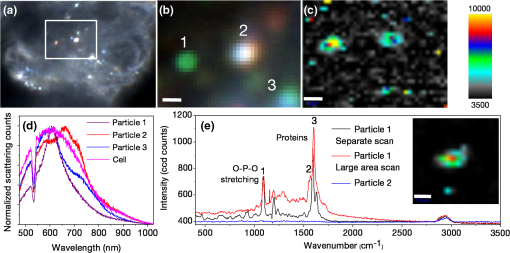 Table 2Assignments of Raman bands of AuNP treated HEK293 sample.
Further, from our observations, it can be seen that when the same ROI containing particle 1 was scanned separately for RSMS [Fig. 5(e), black trace], the Raman spectrum did not repeat with high fidelity upon rescanning [Fig. 5(e), red trace], although in both these measurements, the instrumental parameters were kept exactly the same. This may be due to the complexity of the biological samples and sensitivity of the measurements to the surrounding environment. Hence, we suggest that in biosensing measurements, it is first necessary to optimize experimental parameters using standard samples of similar types and then perform measurements with the experimental samples. 4.ConclusionsWe have developed a methodology for colocalizing intracellular biomolecules and plasmonic NMNPs by plasmonic and Raman scattering microspectroscopy. We demonstrate the applicability of PRSMS setup by imaging AgNP treated E. coli bacteria. SERS signals of biomolecules were detected from the same place where plasmonic scattering signals for nanoparticles were detected. These observations suggest that nanoparticles and observed biomolecules are in such close vicinity that SERS could occur and help us to colocalize the same. In the case of mammalian cells, considering the biocompatibility of AuNPs, an example of AuNPs treated HEK293 cells was demonstrated. Observations support the fact that the SERS activity of nanoparticles is specific to shape, size, and site of adsorption. Data further suggest that in the case of the samples comprised of multiple complex components, localization of molecules only on the basis of intensity scaled RSMS images or indirectly through the plasmonic features of NMNPs may be risky in sensing applications. In such cases, dual verification of the sensing signals is necessary. We believe that PRSMS provides a solution to such problems and can be efficiently used for molecule and particle colocalization in biological environments which will be of prime importance in improving the specificity of theranostics. AcknowledgmentsWe thank the Department of Science and Technology, Government of India for constantly supporting our research program on nanomaterials. We thank Hitesh Mamgain, application scientist, WITec GmbH, Bangalore, India, for discussions. ReferencesW. E. Moerner,
“A dozen years of single-molecule spectroscopy in physics, chemistry, and biophysics,”
J. Phys. Chem. B, 106
(5), 910
–927
(2002). http://dx.doi.org/10.1021/jp012992g JPCBFK 1520-6106 Google Scholar
Y. Gu et al.,
“Single particle orientation and rotational tracking (SPORT) in biophysical studies,”
Nanoscale, 5
(22), 10753
–10764
(2013). http://dx.doi.org/10.1039/c3nr02254d 1556-276X Google Scholar
M. Xu et al.,
“Formation of nano-bio-complex as nanomaterials dispersed in a biological solution for understanding nanobiological interactions,”
Sci. Rep., 2 406
(2012). http://dx.doi.org/10.1038/srep00406 SRCEC3 2045-2322 Google Scholar
P. Nativo, I. A. Prior and M. Brust,
“Uptake and intracellular fate of surface-modified gold nanoparticles,”
ACS Nano, 2
(8), 1639
–1644
(2008). http://dx.doi.org/10.1021/nn800330a 1936-0851 Google Scholar
A. Albanese and W. C. W. Chan,
“Effect of gold nanoparticle aggregation on cell uptake and toxicity,”
ACS Nano, 5
(7), 5478
–5489
(2011). http://dx.doi.org/10.1021/nn2007496 1936-0851 Google Scholar
K. Lee et al.,
“Quantitative imaging of single mRNA splice variants in living cells,”
Nat. Nano, 9 474
–480
(2014). http://dx.doi.org/10.1038/nnano.2014.73 1748-3387 Google Scholar
J. Chen and J. Irudayaraj,
“Quantitative investigation of compartmentalized dynamics of ErbB2 targeting gold nanorods in live cells by single molecule spectroscopy,”
ACS Nano, 3
(12), 4071
–4079
(2009). http://dx.doi.org/10.1021/nn900743v 1936-0851 Google Scholar
Y.-W. Jun et al.,
“Continuous imaging of plasmon rulers in live cells reveals early-stage caspase-3 activation at the single-molecule level,”
Proc. Natl. Acad. Sci. U. S. A., 106
(42), 17735
–17740
(2009). http://dx.doi.org/10.1073/pnas.0907367106 PNASA6 0027-8424 Google Scholar
C. Leduc et al.,
“A highly specific gold nanoprobe for live-cell single-molecule imaging,”
Nano Lett., 13
(4), 1489
–1494
(2013). http://dx.doi.org/10.1021/nl304561g NALEFD 1530-6984 Google Scholar
S. Vishnupriya et al.,
“Single-cell investigations of silver nanoparticle–bacteria interactions,”
Part. Part. Syst. Charact., 30
(12), 1056
–1062
(2013). http://dx.doi.org/10.1002/ppsc.201300165 PPCHEZ 0934-0866 Google Scholar
W. Qian et al.,
“Dark-field light scattering imaging of living cancer cell component from birth through division using bioconjugated gold nanoprobes,”
J. Biomed. Opt., 15
(4), 046025
(2010). http://dx.doi.org/10.1117/1.3477179 JBOPFO 1083-3668 Google Scholar
S. Jung et al.,
“Theragnostic pH-sensitive gold nanoparticles for the selective surface enhanced Raman scattering and photothermal cancer therapy,”
Anal. Chem., 85
(16), 7674
–7681
(2013). http://dx.doi.org/10.1021/ac401390m ANCHAM 0003-2700 Google Scholar
C. E. Talley et al.,
“Intracellular pH sensors based on surface-enhanced Raman scattering,”
Anal. Chem., 76
(23), 7064
–7068
(2004). http://dx.doi.org/10.1021/ac049093j ANCHAM 0003-2700 Google Scholar
E. Ringe et al.,
“Plasmon length: a universal parameter to describe size effects in gold nanoparticles,”
J. Phys. Chem. Lett., 3
(11), 1479
–1483
(2012). http://dx.doi.org/10.1021/jz300426p 1948-7185 Google Scholar
H. Deschout et al.,
“Correlation of dual colour single particle trajectories for improved detection and analysis of interactions in living cells,”
Int. J. Mol. Sci., 14
(8), 16485
–16514
(2013). http://dx.doi.org/10.3390/ijms140816485 IJMCFK 1661-6596 Google Scholar
C. J. Orendorff et al.,
“Aspect ratio dependence on surface enhanced Raman scattering using silver and gold nanorod substrates,”
Phys. Chem. Chem. Phys., 8
(1), 165
–170
(2006). http://dx.doi.org/10.1039/B512573A PPCPFQ 1463-9076 Google Scholar
J. Jiao et al.,
“Polarization-dependent SERS at differently oriented single gold nanorods,”
Chemphyschem, 13
(4), 952
–958
(2012). http://dx.doi.org/10.1002/cphc.201100718 CPCHFT 1439-4235 Google Scholar
K. Munechika et al.,
“Plasmon line widths of single silver nanoprisms as a function of particle size and plasmon peak position,”
J. Phys. Chem. C, 111
(51), 18906
–18911
(2007). http://dx.doi.org/10.1021/jp076099e 1932-7447 Google Scholar
T. Itoh, K. Hashimoto and Y. Ozaki,
“Polarization dependences of surface plasmon bands and surface-enhanced Raman bands of single Ag nanoparticles,”
Appl. Phys. Lett., 83
(11), 2274
–2276
(2003). http://dx.doi.org/10.1063/1.1604188 APPLAB 0003-6951 Google Scholar
C. J. Szymanski, W. H. I. V. Humphries and C. K. Payne,
“Single particle tracking as a method to resolve differences in highly colocalized proteins,”
Analyst, 136
(17), 3527
–3533
(2011). http://dx.doi.org/10.1039/c0an00855a ANLYAG 0365-4885 Google Scholar
B. Kang, L. A. Austin and M. A. El-Sayed,
“Real-time molecular imaging throughout the entire cell cycle by targeted plasmonic-enhanced Rayleigh/Raman spectroscopy,”
Nano Lett., 12
(10), 5369
–5375
(2012). http://dx.doi.org/10.1021/nl3027586 NALEFD 1530-6984 Google Scholar
H. M. Lee et al.,
“High-precision measurement-based correlation studies among atomic force microscopy, Rayleigh scattering, and surface-enhanced Raman scattering at the single-molecule level,”
Phys. Chem. Chem. Phys., 15
(12), 4243
–4249
(2013). http://dx.doi.org/10.1039/c3cp43817a PPCPFQ 1463-9076 Google Scholar
T. Itoh et al.,
“Correlated measurements of plasmon resonance Rayleigh scattering and surface-enhanced resonance Raman scattering using a dark-field microspectroscopic system,”
J. Photochem. Photobiol. A, 183
(3), 322
–328
(2006). http://dx.doi.org/10.1016/j.jphotochem.2006.06.031 JPPCEJ 1010-6030 Google Scholar
A. I. Henry, B. Sharma and R. P. Van Duyne,
“Continuous sensing of blood by dark-field microscopy and surface-enhanced Raman spectroscopy,”
Nanotechnology 2012: Bio Sensors, Instruments, Medical, Environment and Energy, 3 40
–43 Taylor & Francis, Boca Raton
(2012). Google Scholar
L. A. Austin, B. Kang and M. A. El-Sayed,
“A new nanotechnology technique for determining drug efficacy using targeted plasmonically enhanced single cell imaging spectroscopy,”
J. Am. Chem. Soc., 135
(12), 4688
–4691
(2013). http://dx.doi.org/10.1021/ja4011145 JACSAT 0002-7863 Google Scholar
M. Liu et al.,
“Dark-field microscopy in imaging of plasmon resonant nanoparticles,”
Colloids Surf. B, 124
(0), 111
–117
(2014). http://dx.doi.org/10.1016/j.colsurfb.2014.06.001 CSBBEQ 0927-7765 Google Scholar
N. Kawashima et al.,
“Reversible dimerization of EGFR revealed by single-molecule fluorescence imaging using quantum dots,”
Chem. Eur. J., 16
(4), 1186
–1192
(2010). http://dx.doi.org/10.1002/chem.v16:4 CEUJED 0947-6539 Google Scholar
L. Zhang et al.,
“Single gold nanoparticles as real-time optical probes for the detection of NADH-dependent intracellular metabolic enzymatic pathways,”
Angew. Chem., 123
(30), 6921
–6924
(2011). http://dx.doi.org/10.1002/ange.201102151 ANCEAD 0044-8249 Google Scholar
C. Rosman et al.,
“A new approach to assess gold nanoparticle uptake by mammalian cells: combining optical dark-field and transmission electron microscopy,”
Small, 8
(23), 3683
–3690
(2012). http://dx.doi.org/10.1002/smll.v8.23 1613-6829 Google Scholar
X. M. Qian and S. M. Nie,
“Single-molecule and single-nanoparticle SERS: from fundamental mechanisms to biomedical applications,”
Chem. Soc. Rev., 37
(5), 912
–920
(2008). http://dx.doi.org/10.1039/b708839f CSRVBR 0306-0012 Google Scholar
X. Qian et al.,
“In vivo tumor targeting and spectroscopic detection with surface-enhanced Raman nanoparticle tags,”
Nat. Biotechnol., 26
(1), 83
–90
(2008). http://dx.doi.org/10.1038/nbt1377 NABIF9 1087-0156 Google Scholar
J. Turkevich, P. C. Stevenson and J. Hillier,
“The nucleation and growth processes in the synthesis of colloidal gold,”
Discuss. Faraday Soc., 11 55
–75
(1951). http://dx.doi.org/10.1039/df9511100055 DFSOAW 0014-7664 Google Scholar
J. N. Anker et al.,
“Biosensing with plasmonic nanosensors,”
Nat. Mater., 7
(6), 442
–453
(2008). http://dx.doi.org/10.1038/nmat2162 NMAACR 1476-1122 Google Scholar
B. Kang, L. A. Austin and M. A. El-Sayed,
“Real-time molecular imaging throughout the entire cell cycle by targeted plasmonic-enhanced Rayleigh/Raman spectroscopy,”
Nano Lett., 12
(10), 5369
–5375
(2012). http://dx.doi.org/10.1021/nl3027586 NALEFD 1530-6984 Google Scholar
F. Draux et al.,
“Raman spectral imaging of single cancer cells: probing the impact of sample fixation methods,”
Anal. Bioanal. Chem., 397
(7), 2727
–2737
(2010). http://dx.doi.org/10.1007/s00216-010-3759-8 ABCNBP 1618-2642 Google Scholar
|