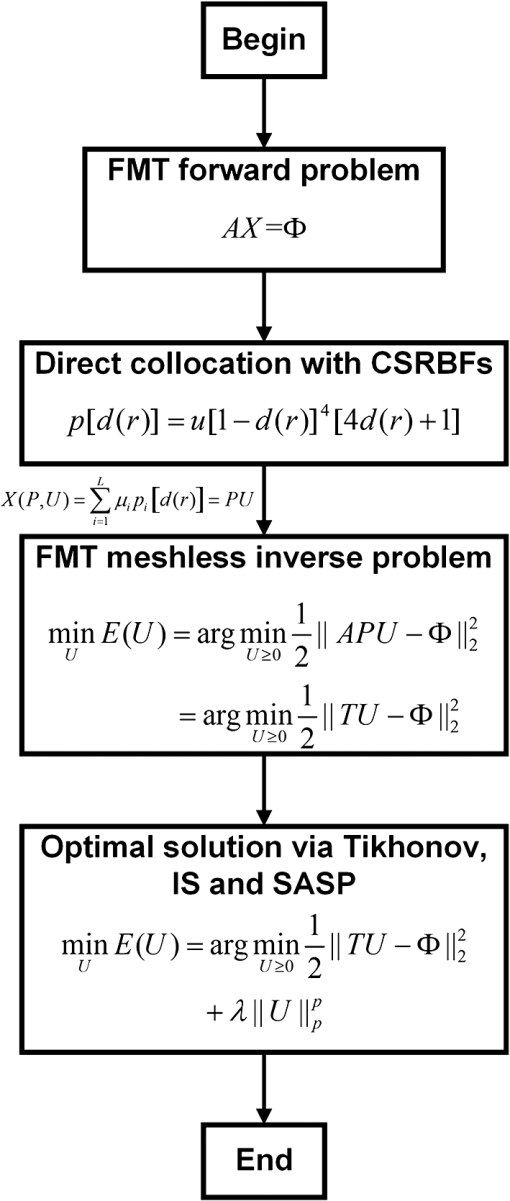
|
|
1.IntroductionIn recent years, optical molecular imaging (OMI) and related optical technique–based multimodality fusion methods have experienced notable growth and attracted considerable attention for their excellent temporal resolution, simple operation, and high cost-effectiveness. Furthermore, OMI has already been used for early stage cancer diagnosis, drug discovery, and disease treatment with the help of exogenous agents that provide detectable signals or endogenous molecules with optical signatures.1–7 In particular, the application of OMI in vivo studies has greatly advanced our understanding of biology and medicine. Fluorescence molecular tomography (FMT) is one of the various modalities of OMI which has attracted considerable attention due to its high sensitivity and low cost, aiming at noninvasive and quantitative imaging of the biodistribution of fluorophores in deep tissues by upgrading the two-dimensional (2-D) fluorescence imaging to three-dimensional (3-D) in vivo detection.8–10 Among FMT methods, it is well known that photon propagation models in biological tissues and inverse problems are the two main challenges, which have a marked effect on tomographic imaging performance.11,12 To perform quantitative FMT, the forward photon propagation model needs to be solved, which can generate a linear system linking the internal unknown fluorescent source and the boundary measurements. Then, the fluorescent source distribution is reconstructed by solving the inverse problem. Currently, different reconstruction techniques for FMT have been proposed based on numerical techniques,13,14 including the finite-difference method (FDM),15 finite-element method (FEM),16 and boundary element method (BEM).17 Among these methods, the FEM has become the most commonly used technique in FMT for its versatility, which makes it easily executable in complex heterogeneous geometrics.18 In particular, the acquired sparsity positive-definite FEM yields great numerical stability and efficiency, which has significantly contributed to the development of reconstruction algorithms.19–22 However, FEM relies on strictly designed meshes connected together by nodes in a properly predefined manner. It has limitations including time consumption in mesh generation, difficulty in the case of adaptive analysis, and significant discretization error in extremely complicated geometry (such as bones).23 In order to overcome the difficulty of 3-D meshing, the meshless method (MM) was recently proposed.24 This technique uses only a set of nodes to represent the problem domain and its boundary. Without any point connectivity or element information, the burdensome mesh discretization is avoided, resulting in improved accuracy due to the decrease in the discretization error. It has been successfully applied to solve problems in solid mechanics, heat transfer, fluid flow, and so on.25 Qin et al. have introduced MM in bioluminescence tomography (BLT)22 based on the Galerkin method. This work has shown a huge potential of MM in the field of optical imaging. Compared with the Galerkin-based methods, the compactly supported radial basis functions (CSRBFs) method possesses advantages in the following aspects: (1) it is a truly mesh-free algorithm; (2) it is space dimension independent in the sense that the order of convergence is , where is the density of the collocation points and is the spatial dimension; and (3) in the context of scattered data interpolation, CSRBFs have spectral convergence orders. In this paper, we introduce a meshless method for the reconstruction of FMT (MM-FMT) based on CSRBF, in order to improve the accuracy and efficiency of the reconstruction. The image domain is represented by a series of CSRBFs based on a direct collocation method, which guarantees the accuracy of the forward model of FMT. After formulating the inverse problem of MM-FMT based on the forward model, three different conventional algorithms, including Tikhonov regularization, L1-norm iteration shrinkage (IS),26 and sparsity adaptive matching pursuit (SAMP),27 are utilized to solve the inverse problem. Heterogeneous mouse model experiments and in vivo experiments are performed to validate the proposed method. We compare our results with those obtained by the FEM-based reconstruction methods (FEM-FMT). Results show that the proposed MM-FMT can achieve better performance. This paper is organized as follows. Section 2 describes the details of the forward model of FMT and the proposed MM-FMT. Section 3 shows the results of numerical heterogeneous mouse experiments and in vivo experiments. Finally, Section 4 discusses the results and draws the conclusion. 2.Method2.1.Photon Propagation Model and Inverse ProblemFor steady-state FMT with point excitation sources on the boundary of the imaging object, the photon propagation model in highly scattering media such as biological tissues is based on the diffusion equation (DE).28 The following coupled DEs with a Robin-type boundary condition can be utilized to describe the forward problem:29,30 where denotes the position of the point inside the image domain . The subscripts and denote the excitation and emission wavelengths, respectively. and are the photon flux densities. and are the absorption and scattering coefficients. is the diffusion coefficient, and is the anisotropy parameter. denotes the fluorophore field which is to be reconstructed, and denotes the optical reflective index mismatch. The excitation light sources are set to be isotropic point sources, which are located beneath the boundary of . denotes the amplitude of the point sources.After applying the process of numerical computation by finite-element formulation, the linear relationship between the measured outgoing photon distribution on the surface and the unknown internal photon distribution is given in a matrix vector form by where denotes the fluorescence distribution on the boundary of the image domain, denotes the system weight matrix, and denotes the intensity of the fluorescent distribution in biological tissues. Detailed descriptions can be found in Ref. 30.To solve the inverse problem of FMT, we need to infer from . The inverse problem can be directly solved by inverting the weight matrix . However, this inversion is often ill-posed in the Hadamard sense, as the dimension of the null space of is not zero. Instead, the least-square solution is used to solve Eq. (2), which can be described as follows: 2.2.Compactly Supported Radial Basis FunctionIn essence, the pioneering work concerning CSRBFs is attributed to Wu31 and Wendland32 in the mid-1990s. Generally, in the situation of 3-D image reconstruction, a CSRBF is expressed as where is a prescribed polynomial, is the distance between the center of the CSRBF and the point inside the image domain, and is the compactly supported radius, which defines the support set of CSRBF. is a constant positive-defined factor. For example,33 is a step function which is expressed asThe explicit formulae of CSRBFs which possess smooth continuous derivatives for , 1, 2, 3 are given in Table 1.34 In this paper, the smooth continuous derivatives of the formulae are set to , and the function is given by Table 1Explicit formulae of some compactly supported radial basis functions (CSRBFs).
2.3.Direct Collocation with Compactly Supported Radial Basis FunctionsFor the reconstruction of FMT, we adopt CSRBFs to represent the image domain based on a direct collocation method to guarantee the accuracy. Figure 1 shows the schematic diagram of the method. First, we distribute the CSRBFs uniformly along the boundary of the image domain. The CSRBFs should be distributed densely to ensure the integrity and accuracy of the boundary. Then we randomly distribute the CSRBFs within the image domain, and the maximal distance between the adjacent centers of CSRBFs is set to be the compactly supported radius, which can avoid the leakage space inside the image domain. Fig. 1Direct collocation based on compactly supported radial basis functions (CSRBFs) and the schematic diagram of a CSRBF. (a) The boundary of the image domain is represented by a series of CSRBFs, which are uniformly distributed on the boundary. The red solid circle denotes the boundary of the image domain, and the black dots denote the centers of CSRBFs. The green-dashed circles represent the support regions of each CSRBF. (b) After the boundary discretization, the image domain is represented by randomly distributed CSRBFs. (c) The schematic diagram of a CSRBF. 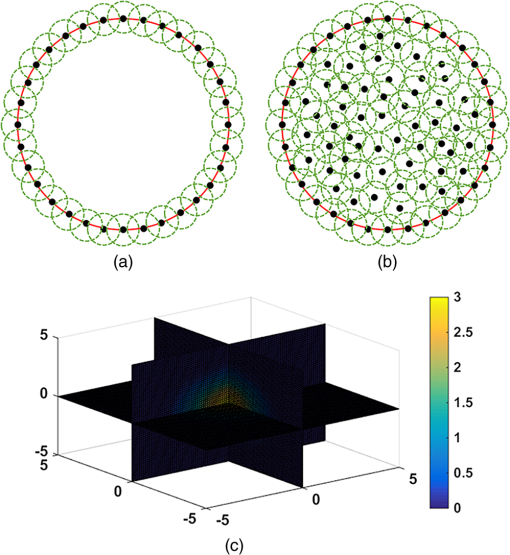 After the representation, the fluorescent distribution at node can be collocated by the CSRBFs as follows: where is the matrix of CSRBFs, is the fluorescent intensity coefficient of the CSRBFs, and is the number of CSRBFs. Substituting Eq. (8) into Eq. (3), the inverse problem of FMT leads to a meshless formulation: where is the meshless system matrix for FMT.2.4.Meshless Inverse ProblemDue to the multiple scatterings of photon propagation through heterogeneous biological tissues, the inverse problem of FMT is often highly ill-posed and ill-conditioned.20,35 Although more fluorescence information can be captured by multiple spatial patterns of illumination, the problem may still be strongly ill-posed because of the sensitivity to noise and errors caused in the data-gathering process and raw data discretization.19 To obtain a reasonable solution, penalty terms are typically introduced to the objective function, which is considered as a priori information: where is the penalty function and is a positive real number called the regularization parameter used to balance the two terms. Here, we consider the case when is with . In this case, the inverse problem with general Lp-norm regularization can be modified intoTo get the optimal solution of Eq. (11), we adopt three different conventional algorithms, namely, Tikhonov regularization (where ), IS, and SAMP (where ), as described in the previous sections. The procedure of the proposed MM-FMT is summarized in Fig. 2. 3.Experiment and ResultsIn this section, both numerical simulation studies and in vivo mouse studies have been designed to evaluate the accuracy, robustness, and efficiency of the proposed method. All of the computational processing was completed on a personal computer with a 3.20 GHz Intel Core i5 CPU and 4 GB RAM. To validate the performance of the reconstruction method, we utilize the position error (PE), the contrast-to-noise ratio (CNR), and computation time in this paper. The PE is defined to measure the accuracy of the results: where is the centroid of the reconstruction results, is the actual location of the fluorophore, and is the 3-D coordinate.The CNR is also used in this paper as a metric to indicate whether the reconstructed object could be clearly distinguished from the background.36 In this paper, we divide the image domain into two regions: volume of interest (VOI) and volume of background (VOB). VOI is defined according to the location and size of the reconstructed object, and VOB is the remaining part of the domain. CNR can be calculated by where and are the weight factors of the VOI and VOB, respectively; and are the respective mean values; and and are the respective standard deviations. In this paper, the VOI is defined based on a threshold of 30% of the maximum value.3.1.Results of the Heterogeneous Mouse ModelTo generate the heterogeneous mouse model, an adult nude mouse was used to acquire the original dataset by micro-CT. All animals used in this study were purchased from the Laboratory Animal Center, Peking University, China, and followed the experimental protocol guidelines according to the Institutional Animal Care and Use Committee (IACUC) at Peking University (Permit Number: 2011-0039). In our study, it is supposed that there were two small tumors on the surface of the animal’s liver, which were the fluorescent targets to be detected. Only the torso section of the mouse was selected as the region to be investigated, as shown in Fig. 3(a). The diameters of the tumors were both set to 2 mm. The edge-to-edge distance of the two tumors was 9 mm. The major organs and tissues (heart, lungs, liver, kidneys, and muscle) were used to construct the heterogeneous mouse model, where the corresponding optical properties were assigned according to Table 2.37 The excitation light sources were modeled as isotropic point sources located in the plane with a light intensity of 0.02 w. The measurements were taken with a 160 deg field of view, as shown in Fig. 3(b). In the experiments, a 5% Gaussian noise was added to the measurements to simulate the situation where practical fluorescence measurements were taken using a charge-coupled device (CCD) camera. Fluorescent images in a 360 deg full view were collected at every 45 deg, with eight projections adopted in total. Table 2Optical parameters of the mouse organs (units of μa and μs′: mm−1).
Fig. 3Image of the heterogeneous mouse model. (a) Three-dimensional (3-D) visualization of the model. (b) Cross-sectional image of the model in the plane , which contains the fluorophore. The white dots denote the location of the excitation light sources. T1 and T2 are two the tumors on the surface of the liver, which are the targets to be reconstructed. 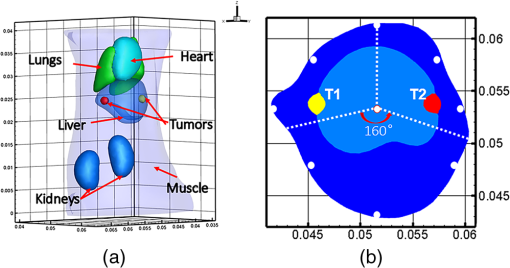 3.1.1.Evaluation of reconstruction reliabilityTo verify the reconstruction reliability of the MM-FMT, we have conducted 50 reconstructions with different numbers of CSRBFs, denoted by the number of functions (NOFs). The NOFs can have a considerable effect on the reconstruction accuracy of MM-FMT when representing the image domain. In this section, the NOF was selected with different orders of magnitude, ranging from 100 to 5500. The reconstructions were performed using IS. The regularization parameters were both set to , and the maximum iteration number was set to 200. The reconstruction results are partly delineated in Fig. 4, and the quantitative analysis is listed in Fig. 5. Fig. 43-D views of the reconstructed results by meshless method FMT (MM-FMT) with different number of functions (NOFs). NOF was selected with different orders of magnitude, ranging from 100 to 5500. The red balls inside the image domain denote the real position of the tumors. 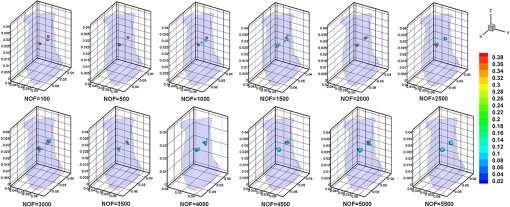 Fig. 5Influence of NOF on position error (PE), contrast-to-noise ratio (CNR), and computation time in the numerical heterogeneous mouse experiments. (a) Chart of PE for two tumors with an increase in CNR. (b) Chart of CNR and computation time with the increase of CNR. The PE, CNR, and computation time were obtained according to the table below the chart, with the range of NOF from 100 to 5500. 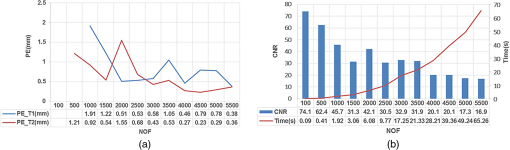 From Fig. 5, we know that when the NOF is too small (), meaning the image domain is too sparsely discretized, the two fluorescent sources cannot be reconstructed at the same time, in which case the accuracy of reconstruction is not ensured either. As NOF increases, the accuracy of the reconstruction increases. However, a certain number of artifacts were found in the results when , since the ill-posedness of the inverse problem increases as the NOF increases. This leads to a decrease in CNR and an increase in computation time. Because the CSRBFs are randomly distributed in the interior of the image domain, the reconstructions are not stable when . When , the variation of the results begins to decrease, and the reconstruction tends to be stable. As can be concluded from the above results, the reconstruction results are satisfying when . The reconstruction becomes stable when . 3.1.2.Evaluation of reconstruction efficiencyTo examine the efficiency of the MM-FMT, we tested it using the numerical heterogeneous mouse model. In this section, we used both MM-FMT and FEM-FMT methods to reconstruct the internal tumors, where Tikhonov, IS, and SAMP were performed to more sufficiently illustrate the efficiency of the MM-FMT. For the FEM-FMT, we discretized the heterogeneous mouse model into 5624 nodes and 29,306 elements. For the purpose of comparison, we set , which is equal to the number of nodes. The regularization parameters were both set to , and the maximum iteration number was set to 200. The reconstruction results for both MM-FMT and FEM-FMT via Tikhonov, IS, and SAMP are shown in Figs. 6 and 7, where Fig. 6 is the 3-D view and Fig. 7 is the cross-sectional view. The quantitative analysis of the two methods is listed in Table 3, where we include PE, CNR, and computation time. As seen in Figs. 6 and 7, both MM-FMT and FEM-FMT can achieve the reconstruction of the two tumors, except for the FEM-FMT via SAMP. The results of the MM-FMT are more condensed with smoother surfaces. The reconstruction regions of the MM-FMT are more accurate with smaller intensity variations, as can be seen from the PE and CNR values. Due to the same dimension of the reconstruction data, the computation times of the MM-FMT and FEM-FMT via the three algorithms have the same order of magnitude. In addition, the computation time of the center distribution of the MM-FMT (17.82 s) is much shorter than that of the mesh generation of the FEM-FMT (295.74 s). This can be attributed to the fact that the MM-FMT does not require the connective information of each center, so there is essentially no other additional operation for mesh generation, simplifying, smoothing, and filtering. Table 3Quantitative analysis of the meshless method fluorescence molecular tomography (MM-FMT) and finite-element method FMT (FEM-FMT) with three different methods.
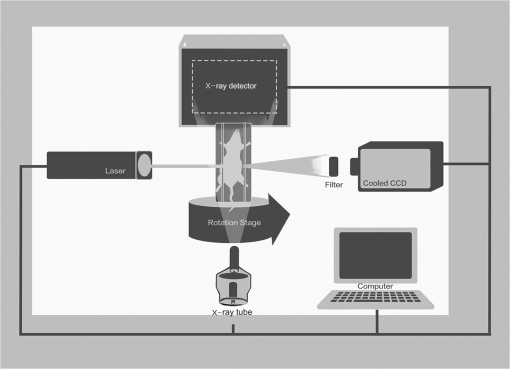
Fig. 63-D view of the reconstructed results of the heterogeneous mouse model with MM-FMT and FEM-FMT, using Tikhonov, IS, and sparsity adaptive matching pursuit (SAMP) methods. The first row lists the results of the MM-FMT, and the second row lists the results of the FEM-FMT. The columns denote the results obtained by Tikhonov, IS, and SAMP. The iso-surfaces in the figure denote the reconstruction results of the tumors. The threshold for the iso-surfaces is 30% of the maximum intensity. The red balls inside the image domain denote the real positions of the tumors. 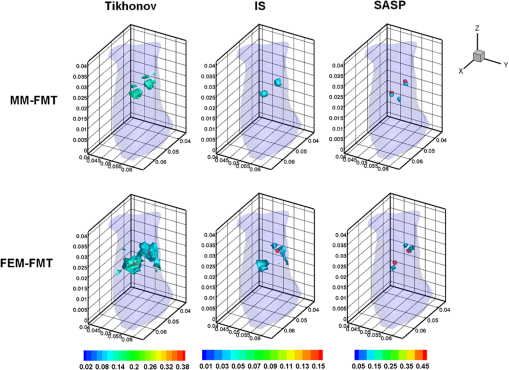 Fig. 7Cross-sectional view of the reconstructed results heterogeneous mouse model with MM-FMT and FEM-FMT. The first row lists the results of the MM-FMT, and the second row lists the results of the FEM-FMT. The columns denote the results obtained by Tikhonov, IS, and SAMP. The red circles mark the real positions of the tumors. 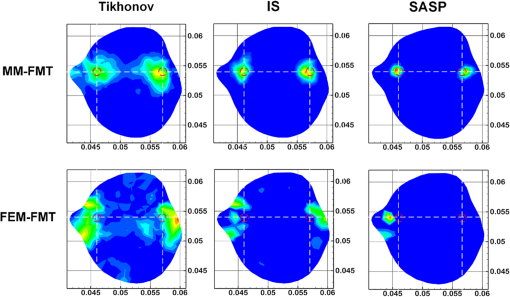 3.2.Results of the In Vivo ExperimentTo validate MM-FMT in a practical application, an in vivo mouse experiment on an adult Kunming mouse was conducted. For imaging purposes, the multimodality imaging system developed by our group38–40 was used to acquire the experimental datasets, which is an integrative platform combining fluorescence imaging with micro-CT. For micro-CT scanning, the cone-beam x-ray generator (Oxford Instruments, 90 kV UltraBright Micro-focus Source) was operated in a continuous mode with a 55 kVp tube voltage, where 360 deg projections were scanned and the signal was accepted by an x-ray detector (Hamamatsu Photonics, C7943CA-02 Flat Panel Sensor). An ultrasensitive cooled CCD camera (Princeton Instruments, PIXIS: 1024) was used to obtain the multiview fluorescent images. The excitation light was generated by a 671-nm continuous wave (CW) laser. The schematic diagram of the multimodality imaging system is shown in Fig. 8. We collected the fluorescence measurements in transillumination mode. A plastic fluorescent bead was implanted inside the hypogastrium of the mouse, which was filled with a cy5.5 solution with a concentration of 2000 nM. This fluorescent solution has a peak excitation wavelength of 671 nm with quantum efficiency of 0.23, and the emission wavelength is 710 nm.41 After imaging, the micro-CT volume and fluorescent images were acquired by the dual-modality system. In order to build the heterogeneous mouse mode, the micro-CT volume was segmented into several parts40 (muscle, heart, lungs, liver, kidneys, and bones), as shown in Fig. 9. The corresponding optical properties were assigned to different organs and tissues according to Table 4.42 Then the fluorescent images were mapped onto the 3-D micro-CT volume via a 3-D surface flux reconstruction algorithm,43 so the photon distribution of the fluorescent signals on the mouse surface was obtained, which was indispensable for reconstruction. Table 4Optical parameters of the mouse organs at 670 and 710 nm (units of μa and μs′: mm−1).
Fig. 9Anatomical structure of the mouse. (a) Transverse view. (b) Sagittal view. (c) Coronal view. The yellow square marker in (a–c) illustrates the location of the fluorescent bead. (d) 3-D visualization of the mouse. The red dot in the graph corresponds to the fluorescent source. 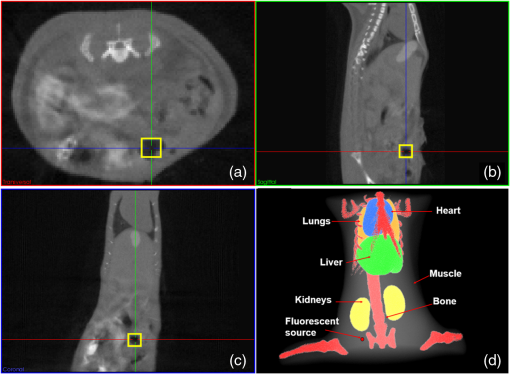 After finishing the above procedure, the MM-FMT and FEM-FMT were both performed on the mouse through three different algorithms (Tikhonov, IS, and SAMP). The image domain of the mouse was discretized into 5639 nodes and 33,627 elements for FEM-FMT and 5639 CSRBFs for MM-FMT. The 3-D reconstruction results of MM-FMT and FEM-FMT are shown in Fig. 10. The cross-sectional views are given in Fig. 11. In addition, the quantitative analysis of the reconstruction results is listed in Table 5. Table 5Quantitative analysis of the in vivo experiment with MM-and FEM-FMT.
Fig. 103-D view of the reconstruction results for the in vivo experiment with MM-FMT and FEM-FMT, via Tikhonov, IS, and SAMP. 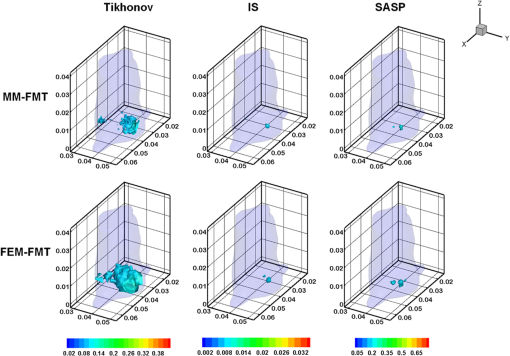 Fig. 11Cross-sectional views of the reconstruction results of in vivo experiment with MM-FMT and FEM-FMT, via Tikhonov, IS, and SAMP. The first row lists the results of MM-FMT, and the second row lists the results of FEM-FMT. The third row lists the standard CT cross-sectional slices, which involve the fluorescent source. The columns denote the results obtained by Tikhonov, IS, and SAMP. The black square markers denote the actual location of the fluorescent source. 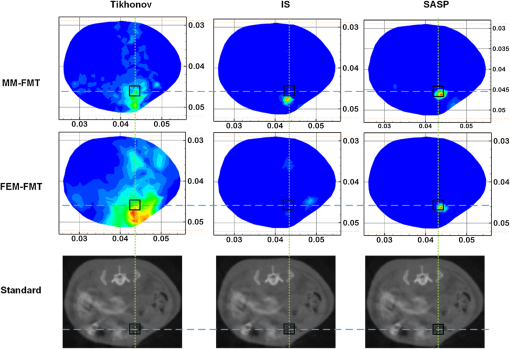 Similar to the numerical heterogeneous mouse experiments, better reconstruction results are achieved by the MM-FMT. The CT scan could determine the location of the internal fluorescent bead, since the bead was wrapped in a plastic material (Fig. 11). Compared with the real location of the fluorescent bead, the MM-FMT does not perform as well as in numerical studies, but the reconstruction accuracy is still acceptable. Quantitative analysis of the performance further demonstrates the superiority of the MM-FMT. 4.Discussion and ConclusionFMT has been rapidly developed in recent years for its advantages in the study of physiological and pathological processes in vivo at cellular and molecular levels. Research of the inverse problem of FMT is critical as the solution to the inverse problem, which is central to the accuracy and efficiency of the reconstruction. However, the application scope of the existing analytical and statistical techniques is currently limited by the different weaknesses inherent in each method. Developing more powerful numerical techniques is needed for better applicability and higher efficiency of FMT, particularly in the case of more complicated geometrics. In this paper, we presented a CSRBF-based meshless method to solve the inverse problem in heterogeneous biological tissues. Compared with the conventional FEM analysis, the MM-FMT can obtain better reconstruction performance. It has been shown in Section 3.1.1 that the NOF is serious for the reconstruction. When the NOF is small, the computation cost is reduced, but the accuracy is undesirable. When the NOF increases, the computation cost and the accuracy both increase. When , the reconstruction results obtained by the MM-FMT demonstrate good performance (Figs. 4 and 5). In this paper, we chose the larger NOF to ensure accuracy. In future work, we will do some research to find the optimal NOF for the application of FMT. Compared with FEM-FMT, better accuracy can be obtained with MM-FMT (Figs. 6 and 7 and Table 3). This is due to the fact that the meshless method can express the image domain continuously, avoiding the discretization error as in FEM to a certain degree. Besides, the reconstruction results of the in vivo experiment validate the feasibility of MM-FMT in a practical application. In this paper, the MM-FMT was performed via three conventional methods including Tikhonov, IS, and SAMP. More effective reconstruction methods for FMT can be combined with MM-FMT to achieve better performance. In this paper, we distributed the center of CSRBFs with direct collocation. The adaptive collocation method can be further evaluated to more accurately and effectively express the image domain. In conclusion, we have presented a meshless reconstruction based on CSRBFs to increase the accuracy of FMT. By choosing an appropriate number of CSRBFs to discretize the image domain, the image to be constructed can be more accurately expressed, thus avoiding the limitations of conventional FEM. The results of the numerical heterogeneous mouse experiments and in vivo experiment demonstrate the superiority of the MM-FMT. AcknowledgmentsThis paper is supported by the National Natural Science Foundation of China under Grant Nos. 81571836, 81227901, 61231004, and 61501462, the National Basic Research Program of China (973 Program) under Grant No. 2011CB707700, the Instrument Developing Project of the Chinese Academy of Sciences under Grant No. YZ201359, the Fundamental Research Funds for the Central Universities under Grant Nos. 2014JBM026 and 2015JBM026, and the China Postdoctoral Science Foundation under Grant No. 2015M570032 and 2015T80155. ReferencesV. Ntziachristos,
“Going deeper than microscopy: the optical imaging frontier in biology,”
Nat. Methods, 7
(8), 603
–614
(2010). http://dx.doi.org/10.1038/nmeth.1483 Google Scholar
F. G. Blankenberg and H. W. Strauss,
“Recent advances in the molecular imaging of programmed cell death: Part II--non-probe-based MRI, ultrasound, and optical clinical imaging techniques,”
J. Nucl. Med., 54
(1), 1
–4
(2013). http://dx.doi.org/10.2967/jnumed.112.111740 Google Scholar
S. Keereweer et al.,
“Optical image-guided surgery—where do we stand?,”
Mol. Imaging Biol., 13
(2), 199
–207
(2011). http://dx.doi.org/10.1007/s11307-010-0373-2 Google Scholar
C. Chi et al.,
“Intraoperative imaging-guided cancer surgery: from current fluorescence molecular imaging methods to future multi-modality imaging technology,”
Theranostics, 4
(11), 1072
–1084
(2014). http://dx.doi.org/10.7150/thno.9899 Google Scholar
C. W. Chi et al.,
“Use of indocyanine green for detecting the sentinel lymph node in breast cancer patients: from preclinical evaluation to clinical validation,”
PLoS One, 8
(12),
(2013). http://dx.doi.org/10.1371/journal.pone.0083927 POLNCL 1932-6203 Google Scholar
J. K. Willmann et al.,
“Molecular imaging in drug development,”
Nat. Rev. Drug Discov., 7
(7), 591
–607
(2008). http://dx.doi.org/10.1038/nrd2290 Google Scholar
Z. Hu et al.,
“In vivo nanoparticle-mediated radiopharmaceutical-excited fluorescence molecular imaging,”
Nat. Commun., 6 7560
(2015). https://doi.org/10.1038/ncomms8560 Google Scholar
A. Ale et al.,
“FMT-XCT: in vivo animal studies with hybrid fluorescence molecular tomography-X-ray computed tomography,”
Nat. Methods, 9
(6), 615
–620
(2012). http://dx.doi.org/10.1038/nmeth.2014 Google Scholar
F. Leblond et al.,
“Toward whole-body optical imaging of rats using single-photon counting fluorescence tomography,”
Opt. Lett., 36
(19), 3723
–3725
(2011). http://dx.doi.org/10.1364/OL.36.003723 Google Scholar
V. Ntziachristos et al.,
“Fluorescence molecular tomography resolves protease activity in vivo,”
Nat. Med., 8
(7), 757
–760
(2002). http://dx.doi.org/10.1038/nm729 Google Scholar
V. Ntziachristos,
“Fluorescence molecular imaging,”
Annu. Rev. Biomed. Eng., 8 1
–33
(2006). http://dx.doi.org/10.1146/annurev.bioeng.8.061505.095831 Google Scholar
C. Darne, Y. Lu and E. M. Sevick-Muraca,
“Small animal fluorescence and bioluminescence tomography: a review of approaches, algorithms and technology update,”
Phys. Med. Biol., 59
(1), R1
–64
(2014). http://dx.doi.org/10.1088/0031-9155/59/1/R1 Google Scholar
A. P. Gibson, J. C. Hebden and S. R. Arridge,
“Recent advances in diffuse optical imaging,”
Phys. Med. Biol., 50
(4), R1
–43
(2005). http://dx.doi.org/10.1088/0031-9155/50/4/R01 Google Scholar
C. C. Leng and J. Tian,
“Mathematical method in optical molecular imaging,”
Sci. China Inform. Sci., 58
(3),
(2015). http://dx.doi.org/10.1007/s11432-014-5222-5 Google Scholar
A. D. Klose et al.,
“In vivo bioluminescence tomography with a blocking-off finite-difference SP3 method and MRI/CT coregistration,”
Med. Phys., 37
(1), 329
–338
(2010). http://dx.doi.org/10.1118/1.3273034 Google Scholar
W. Cong et al.,
“Practical reconstruction method for bioluminescence tomography,”
Opt. Express, 13
(18), 6756
–6771
(2005). http://dx.doi.org/10.1364/OPEX.13.006756 Google Scholar
S. Tang et al.,
“Combined multiphoton microscopy and optical coherence tomography using a 12-fs broadband source,”
J. Biomed. Opt., 11
(2), 020502
(2006). http://dx.doi.org/10.1117/1.2193428 Google Scholar
C. H. Qin et al.,
“Recent advances in bioluminescence tomography: methodology and system as well as application,”
Laser Photonics Rev., 8
(1), 94
–114
(2014). http://dx.doi.org/10.1002/lpor.201280011 Google Scholar
Y. An et al.,
“A novel region reconstruction method for fluorescence molecular tomography,”
IEEE Trans. Biomed. Eng., 62
(7), 1818
–1826
(2015). http://dx.doi.org/10.1109/TBME.2015.2404915 Google Scholar
J. Shi et al.,
“An adaptive support driven reweighted L1-regularization algorithm for fluorescence molecular tomography,”
Biomed. Opt. Express, 5
(11), 4039
–4052
(2014). http://dx.doi.org/10.1364/BOE.5.004039 Google Scholar
D. Han et al.,
“Efficient reconstruction method for L1 regularization in fluorescence molecular tomography,”
Appl. Opt., 49
(36), 6930
–6937
(2010). http://dx.doi.org/10.1364/AO.49.006930 Google Scholar
J. Ye et al.,
“Reconstruction of fluorescence molecular tomography via a nonmonotone spectral projected gradient pursuit method,”
J. Biomed. Opt., 19
(12), 126013
(2014). http://dx.doi.org/10.1117/1.JBO.19.12.126013 Google Scholar
C. Qin et al.,
“Galerkin-based meshless methods for photon transport in the biological tissue,”
Opt. Express, 16
(25), 20317
–20333
(2008). http://dx.doi.org/10.1364/OE.16.020317 Google Scholar
G. R. Liu and Y. T. Gu, An Introduction to Meshfree Methods and Their Programming, Springer, Dordrecht; New York
(2005). Google Scholar
A. Gelas et al.,
“Compactly supported radial basis functions based collocation method for level-set evolution in image segmentation,”
IEEE Trans. Image Process., 16
(7), 1873
–1887
(2007). http://dx.doi.org/10.1109/TIP.2007.898969 Google Scholar
D. Han et al.,
“A fast reconstruction algorithm for fluorescence molecular tomography with sparsity regularization,”
Opt. Express, 18
(8), 8630
–8646
(2010). http://dx.doi.org/10.1364/OE.18.008630 Google Scholar
J. Ye et al.,
“Fast and robust reconstruction for fluorescence molecular tomography via a sparsity adaptive subspace pursuit method,”
Biomed. Opt. Express, 5
(2), 387
–406
(2014). http://dx.doi.org/10.1364/BOE.5.000387 Google Scholar
Y. Tan and H. Jiang,
“DOT guided fluorescence molecular tomography of arbitrarily shaped objects,”
Med. Phys., 35
(12), 5703
–5707
(2008). http://dx.doi.org/10.1118/1.3020594 Google Scholar
A. Joshi, W. Bangerth and E. Sevick-Muraca,
“Adaptive finite element based tomography for fluorescence optical imaging in tissue,”
Opt. Express, 12
(22), 5402
–5417
(2004). http://dx.doi.org/10.1364/OPEX.12.005402 Google Scholar
J. H. Lee et al.,
“Fully adaptive finite element based tomography using tetrahedral dual-meshing for fluorescence enhanced optical imaging in tissue,”
Opt. Express, 15
(11), 6955
–6975
(2007). http://dx.doi.org/10.1364/OE.15.006955 Google Scholar
Z. Wu,
“Compactly supported positive definite radial functions,”
Adv. Comput. Math., 4
(1), 283
–292
(1995). http://dx.doi.org/10.1007/BF03177517 Google Scholar
H. Wendland,
“Piecewise polynomial, positive definite and compactly supported radial functions of minimal degree,”
Adv. Comput. Math., 4
(1), 389
–396
(1995). http://dx.doi.org/10.1007/BF02123482 Google Scholar
A. Iske, Scattered Data Modelling Using Radial Basis Functions, Springer, Berlin Heidelberg
(2002). Google Scholar
S. L. Ho et al.,
“A fast global optimizer based on improved CS-RBF and stochastic optimal algorithm,”
IEEE Trans. Magn., 42
(4), 1175
–1178
(2006). http://dx.doi.org/10.1109/TMAG.2006.871429 IEMGAQ 0018-9464 Google Scholar
J. Shi et al.,
“Enhanced spatial resolution in fluorescence molecular tomography using restarted L1-regularized nonlinear conjugate gradient algorithm,”
J. Biomed. Opt., 19
(4), 046018
(2014). http://dx.doi.org/10.1117/1.JBO.19.4.046018 Google Scholar
C. Chen et al.,
“Diffuse optical tomography enhanced by clustered sparsity for functional brain imaging,”
IEEE Trans. Med. Imaging, 33
(12), 2323
–2331
(2014). http://dx.doi.org/10.1109/TMI.2014.2338214 Google Scholar
H. Yi et al.,
“Reconstruction algorithms based on l1-norm and l2-norm for two imaging models of fluorescence molecular tomography: a comparative study,”
J. Biomed. Opt., 18
(5), 056013
(2013). http://dx.doi.org/10.1117/1.JBO.18.5.056013 Google Scholar
C. Qin, S. Zhu and J. Tian,
“New optical molecular imaging systems,”
Curr. Pharm. Biotechnol., 11
(6), 620
–627
(2010). http://dx.doi.org/10.2174/138920110792246519 Google Scholar
S. Zhu et al.,
“Cone beam micro-CT system for small animal imaging and performance evaluation,”
Int. J. Biomed. Imaging, 2009 960573
(2009). http://dx.doi.org/10.1155/2009/960573 Google Scholar
W. Ping et al.,
“Bioluminescence tomography by an iterative reweighted (l)2 norm optimization,”
IEEE Trans. Biomed. Eng., 61
(1), 189
–196
(2014). http://dx.doi.org/10.1109/TBME.2013.2279190 Google Scholar
F. Gao et al.,
“A linear, featured-data scheme for image reconstruction in time-domain fluorescence molecular tomography,”
Opt. Express, 14
(16), 7109
–7124
(2006). http://dx.doi.org/10.1364/OE.14.007109 Google Scholar
G. Alexandrakis, F. R. Rannou and A. F. Chatziioannou,
“Tomographic bioluminescence imaging by use of a combined optical-PET (OPET) system: a computer simulation feasibility study,”
Phys. Med. Biol., 50
(17), 4225
–4241
(2005). http://dx.doi.org/10.1088/0031-9155/50/17/021 Google Scholar
X. Chen et al.,
“3D reconstruction of light flux distribution on arbitrary surfaces from 2D multi-photographic images,”
Opt. Express, 18
(19), 19876
–19893
(2010). http://dx.doi.org/10.1364/OE.18.019876 Google Scholar
BiographyYu An is a PhD candidate in the department of Biomedical Engineering, School of Computer and Information, Beijing Jiaotong University. His research interests are mainly focused on the algorithms of fluorescence molecular tomography and hardware development in multimodality system. Jie Liu is a professor in the department of Biomedical Engineering, School of Computer and Information, Beijing Jiaotong University. His research interests are mainly focused on the medical image processing and analysis. Chongwei Chi is an assistant professor in the Key Laboratory of Molecular Imaging, Institute of Automation, Chinese Academy of Science. His work includes image guided surgery and the application of fluorescence molecular tomography. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||