|
|
1.IntroductionVisualization of neural connectivity is essential for understanding the structure-function of the brain. The combination of optical imaging and fluorescent labeling techniques has been a promising tool for neuroimaging with high resolution.1,2 However, the high scattering of biological tissues limits light penetration, which makes it difficult to image large tissues.3–5 Recently, various tissue optical clearing methods have been developed to reduce the scattering of tissues and greatly enhance the imaging depth,5–21 such as Scale,6 3DISCO,7–10 CLARITY,11–13 ,14 SeeDB,15–18 CUBIC,19 PACT-PARS,20 FRUIT,21 TDE,22,23 ScaleS,24 and so on. They have been combined with various optical imaging techniques to obtain images throughout large-volume samples.6–21 The researchers applied these methods to study the structure of different neuronal tissue blocks, such as mouse embryo,14–18 spinal cord,8–10 or brain.6–21 These optical clearing methods have brought about a vast breakout for neuroscience, but still face some challenges, i.e., complex implementation and a long clearing process. In addition, imaging larger tissue blocks also suffers from limitations of optical imaging techniques. Up until now, the combination of mechanical sectioning and optical imaging has still been an effective method to map the brain atlas.25–29 Conventional manual sectioning25,26 is not only extremely laborious, but also inevitably leads to information loss, and is prone to error because the sample needs to be sliced thin enough. The developed micro-optical sectioning tomography based on automated ultrathin sectioning provides an innovative technique for mapping the neural networks of whole-mount mouse brain at the signal cell level, but it is also time consuming because it takes 10 days to obtain the whole data of one mouse brain.27,28 Automated block sectioning with serial two-photon imaging (STP)29 imaged the whole brain with intermittent sampling of evenly spaced sections and might save time for mechanical sectioning. Costantini et al.22,23 employed 2,2′-thiodiethanol (TDE) to clear the brain samples to improve the imaging depth before performing STP. If the clearing process is rapid enough, the thickness of slices could be increased, and the loss of information would be reduced. Moreover, it is promising to perform imaging, sectioning, and clearing simultaneously, which would allow for the reconstruction of a large brain area with reduced tissue slicing and decreased acquisition time. However, rapid optical clearing methods have received little attention. We have conducted extensive research on skin optical clearing, including mechanism studies, development of new methods, and multiapplications.5,30–37 In previous work, sugar and sugar-alcohol have been found to have good clearing potential on skin tissues with good solubility of collagen, and could make skin transparent quickly.32–36 It is well known that collagen also exists in brain tissues and that sugar and sugar-alcohol have also been used as clearing agents of brain tissues.6,11–13,15–21,24,38 In this study, we set out to develop a rapid optical clearing method based on pure sugar/sugar-alcohol solution. By means of molecular dynamical (MD) simulation, the optical clearing potential of the agents was predicted. Then, the performance on transparency of mouse brain samples, imaging depth, fluorescence preservation, and linear deformation was further evaluated. This developed optical clearing method is expected to be used to improve image quality. 2.Materials and Methods2.1.Preparation of Brain Sections and Clearing AgentsAdult wild-type mice (C57BL/6J) and Thy1-green fluorescent protein (GFP) mice (line M, Jackson Laboratory) were used in this study. Mice were anesthetized with a mixture of 2% -chloralose and 10% urethane () through intraperitoneal injection, and perfused intracardially with 0.01 M PBS (Sigma) followed by 4% PFA (Sigma). The brains were postfixed overnight at 4°C in 4% PFA. The mouse brains were rinsed with PBS several times and were sliced into coronal sections with a vibratome (Leica VT 1000s). For prodium iodide (PI) staining, which is widely used in fluorescence microscopy as a popular red-fluorescent nuclear labeling dye, the sections were incubated in PI dilution () for 1 h before clearing. Animal care and experimental protocols were approved by the Experimental Animal Management Ordinance of Hubei Province, P. R. China. MD simulation was used to predict the optical clearing potential of the agents. The detailed protocols were described in our previous work.36 After the simulation, the number of hydrogen bonds between each agent and the peptide was counted every 1 ps during the entire simulation, and their sum was calculated as the total number. The average number of hydrogen bonds was obtained by dividing the total number by the production run time as well as by the number of molecules.36 The three saturated solutions at 20°C, sorbitol, sucrose, and fructose, were used to clear brain sections with simple immersion or topical treatment. Eighty percent glycerol, which is a common mounting medium, was also used here. was a relatively rapid method for neuronal and non-neuronal tissue, which needs three steps to make the tissue transparent.14 Here, we used the higher concentration mixture (50% formamide and 20% PEG8000) in to compare the fluorescence preservation and imaging depth with the above agents. The viscosities of agents were measured respectively with the viscometer (NDJ-5S, JNT, Shanghai, China) at room temperature. The concentration and viscosity of all the agents are listed in Table 1. Table 1The concentration and viscosity of clearing agents used in this study. 2.2.Microscopy and ImagingBefore and after treatment with different agents, the samples were put on a 1951 United States Air Force (USAF) resolution test target. Due to the inhomogeneity of brain tissue, the middle region (striatum) of the brain section was put on the central area of 1951 USAF target. White-light images were taken by a CCD camera (PixelFly USB, PCO computer, Germany) attached to a stereo microscope (SZ61TR, Olympus, Japan). The brain sections were mounted with two cover glasses for fluorescence imaging. The fluorescence images were acquired with confocal fluorescence microscopy (LSM710, Zeiss, Germany) equipped with the Plan-Apochromat dry objective (W.D. 0.55 mm). 2.3.Quantification AnalysisThe obtained images were analyzed using ImageJ software and calculated with MATLAB. For fluorescence quantification, the elliptical-selection tool was used to select an elliptical area in the soma of a neuron, and the histogram tool was used to measure the mean fluorescence intensity of the ellipse area, which served as the fluorescence intensity of the neuron. The fluorescence intensity of a neuron is supposed to be “A” before clearing and “B” after clearing. We calculated the fluorescence change of a neuron during clearing as “B/A.” For each group, a mean value of fluorescence change of 10 neurons was calculated. For imaging depth, the decay of image contrast value with depth in brain slice before and after clearing was used for quantification. The imaging contrast was calculated with Eq. (1), where is the grayscale value for each pixel, is the average intensity of the image, and is the number of total pixels. The imaging depth was determined at the point where the contrast value had fallen to of the maximum contrast at the tissue surface. The imaging depth after clearing was divided by the depth before clearing, which served as the increase of imaging depth for different agents. For size change measurement, the length of straight lines connecting the feature points of the images from dorsal to ventral and from lateral to middle were measured, and served as deformation in the longitudinal and horizontal directions, respectively. Statistical analysis was performed using one-way ANOVA followed by Tukey’s post hoc test. 3.ResultsTo develop a simple and rapid method to clear the brain section and improve optical imaging depth, we screened out several compounds, such as glycerol, sorbitol, sucrose, and fructose, which are safe and commonly used in skin optical clearing. The optical clearing potential was assessed with MD simulation, and the rapid clearing process was also recorded with the CCD camera. The fluorescence preservation for GFP and PI were observed and analyzed quantitatively, and the imaging depth was calculated to assess the improvement. Then the tissue size and cell morphology were investigated to find the structure preservation of the clearing methods. 3.1.Transparency of Samples with Different AgentsIn this study, the optical clearing potential of some commonly used chemical agents with rich hydroxyl groups was investigated with MD simulation, including sorbitol, sucrose, fructose, and glycerol. The propensity to form hydrogen bonds was employed to predict the clearing potential,36,39–40 as shown in Table 2. Table 2Numbers of hydrogen bonds of several agents with MD simulation.
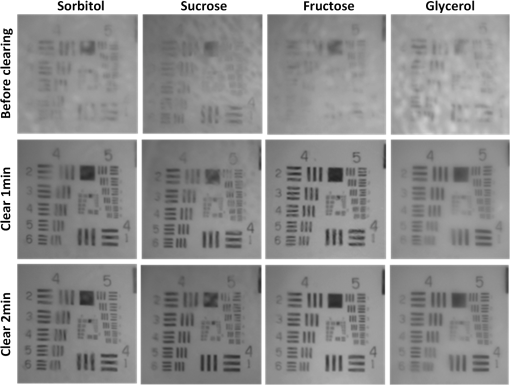
The simulation results show that sorbitol, sucrose, and fructose form more bonds than glycerol. Therefore, it can be deduced that the former three agents have stronger ability to disrupt the hydration shell around the collagen and better optical clearing potentials. Figure 1 shows the white-light images of -thickness brain sections to demonstrate the rapid clearing process within 2 min. The results show that the striatum regions of the brain, which have abundant nerve fiber, become transparent rapidly by simple immersion or topical treatment within 2 min with sorbitol, sucrose, fructose, and glycerol. After the 2-min treatment, the resolution of the 1951 USAF target measured through brain slice is , , , and for sorbitol, sucrose, fructose, and glycerol, respectively. This is to say that the former three agents can achieve better transparency than the latter one, and fructose presents the best clearing effect. 3.2.Increase of Imaging Depth with Different AgentsTo investigate the improvement of imaging depth for different agents, the GFP fluorescence images of brain samples before and after clearing were acquired with same parameter settings with confocal microscopy. Figure 2(a) shows the maximum projections of the transverse () and orthogonal () image stacks by taking sorbitol as an example. The projection images indicate the increased imaging depth after clearing. Fig. 2(a) Maximum projections of transverse () and orthogonal () image stacks. (b) Increase of imaging depth with different agents. Scale bar, . *, significant (). 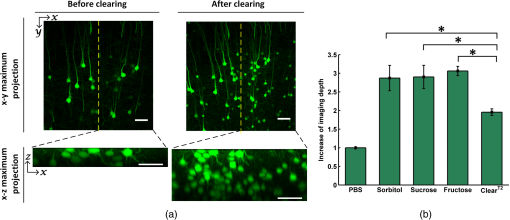 To quantify the improvement of imaging depth induced by the agents, the contrast of the images was calculated according to Eq. (1), and the imaging depth was obtained based on the contrast decay, as shown in Fig. 2(b). The results show that the imaging depth has been increased almost threefold for sorbitol, sucrose, and fructose, which is higher than with significant differences. In other words, treatment with the three clearing agents can significantly improve the optical imaging depth. 3.3.Fluorescence Preservation of GFP and Prodium Iodide with Different AgentsThe genetically encoded fluorescent proteins, such as GFP, are important biomarkers for visualizing both structural and functional details in tissue. Thus, it is particularly important to keep an eye on the fluorescence preservation of the endogenous proteins. To study the influence of the three agents on GFP fluorescence, the images of brain sections were acquired with same parameter settings with confocal microscopy before and after clearing. As shown in Fig. 3(a), the GFP fluorescence is preserved well for all the agents, including sorbitol, sucrose, fructose, and . From the images of the maximum projection of the same thickness, it can be seen that the fluorescent signals in deep layers of the tissue became stronger, and some new signals in deeper tissues could be obtained after clearing. The quantitative results shown in Fig. 3(b) shows that sorbitol and sucrose evidently enhance the mean fluorescence of GFP as well as , while fructose keeps the mean value almost as before clearing. Hence, sorbitol and sucrose could preserve GFP fluorescence better than fructose. Fig. 3Fluorescence preservation of GFP and PI. (a) Fluorescence images of GFP labeled neurons. (b) Mean fluorescence change of GFP treated with different agents. (c) Fluorescence images of PI stained cells. (d) Mean fluorescence change of PI treated with different agents. Scale bar, . Each image is a maximum projection of the -stacks (thickness: ). 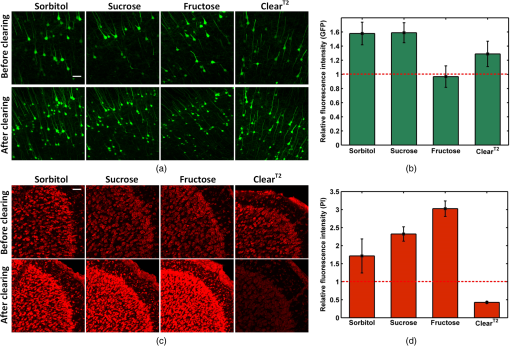 It is also examined whether the clearing agents are compatible with PI, which is a common nuclear staining dye in biological research. Figure 3(c) shows the fluorescence images of PI stained brain slices cleared with different agents. It can be seen that the fluorescence intensity for sorbitol, sucrose, and fructose preserve well, while for , fluorescence quenching is obvious. For further analysis, we quantified the fluorescence preservation of the PI signal with ImageJ software as described earlier. As shown in Fig. 3(d), the mean fluorescence intensity of PI stained cells for sorbitol, sucrose, and fructose increase by 1.5, 2.3, and 3 times, respectively, while results in a 58% loss of PI fluorescence intensity. In general, sorbitol and sucrose evidently increase both GFP and PI fluorescence intensity. Fructose can greatly increase the fluorescence intensity of PI, while slightly decreasing the fluorescence intensity of GFP. In addition, it can be seen from the images in Figs. 3(a) and 3(c) that the visual fields after clearing for sorbitol, sucrose, and fructose are larger than before, which shows sample shrinkage. This might be induced by dehydration due to the hyperosmosis and high concentration of these agents. 3.4.Tissue Size and Cell Morphology with Different AgentsDuring the clearing process, the changes of sample size are inevitable due to the dehydration or hydration of different agents. Cell morphology preservation is of particular importance to analyze fine structures of biological tissues. The consistency of the contraction or expansion rate in different directions of the sample determines the basic structure preservation. The contraction rates after clearing in the horizontal and longitudinal directions were calculated, respectively. As shown in Fig. 4(a), for the three clearing agents, the contraction rates in the horizontal and longitudinal directions are very close, and there is no significant difference between the two directions. In the case of , the horizontal contraction is significantly smaller than longitudinal, and this inconsistent contraction in the two directions will lead to morphologic deformation. Tissue contraction is more beneficial for morphology preservation since swelling might induce structure rupture. Fig. 4(a) Contraction rates of samples in two directions of different agents. Horizontal indicates the direction from left to right, and longitudinal indicates the direction from dorsal to ventral. N.S., non-significant (). *, significant (). (b) Morphology preservation of pyramidal neurons (cortex) before and after optical clearing with sorbitol solution. Scale bar, . Each image is a maximum projection of stacks (thickness: ). 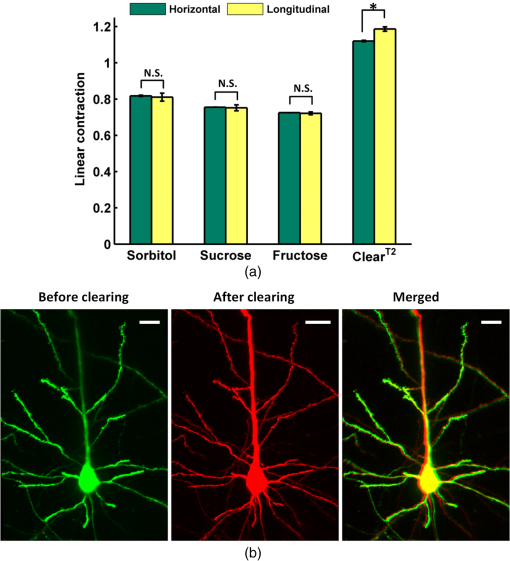 To evaluate the cell morphology preservation, we imaged the pyramidal neurons in the cortex in tissue sections before and after clearing with the agents. After clearing with sorbitol, the neuronal morphology remained intact, as shown in Fig. 4(b). Sucrose and fructose show fine structure preservation as well as sorbitol (data not shown). 4.DiscussionIn this study, a rapid and PI-compatible optical clearing method for brain tissue was developed based on sugar and sugar-alcohol, and then the effectiveness was assessed from several aspects. According to predictions with MD simulation, the optical clearing potential for the three agents was in order of sucrose, fructose, and sorbitol. While the experiments on brain slices indicated that fructose had the best clearing ability, this does not fully correspond to the simulated results. This is due to the MD simulation only predicting the interaction of optical clearing agents and collagen, while there are rich lipids besides collagen in brain tissue. It is suggested that collagen loosening is not the only mechanism for tissue optical clearing with sugar or sugar-alcohol, but the other mechanisms, including lipid dissolving, refractive index matching, and dehydration, could make an nonnegligible contribution to the transparency of mouse brain samples. The previous investigation showed that was a relatively rapid clearing method for neuronal tissue, and needed three steps to clear the larger tissue.14 Here, the higher concentration solution (50% formamide and 20% PEG8000) for the latter two steps of , which had been proved to be effective to maintain the fluorescent signal of genetically encoded proteins, immunohistochemistry labeling, and lipophilic dyes, was used to compare.14 However, the experimental results in this work demonstrated that it was unsuitable for PI-labeling samples. It is supposed that the PI and formamide in both had amino groups. They might generate chemical reactions and induce fluorescence quenching of the PI signal. The experimental results indicated that sorbitol and sucrose could enhance the mean fluorescence intensity of both GFP and PI successfully, while the fructose only did well on PI. In consideration of the enhancement of fluorescence intensity and imaging depth caused by different solutions, it is suggested that sorbitol and sucrose are good choices for GFP samples, fructose is the best for PI labeling samples, and sucrose is optimal for samples with both markers. It should be noted that though the sugar and sugar-alcohol induced certain contraction of samples, the deformation was consistent in different directions, which did not affect obtaining the cell morphology and fine structures. Therefore, sugar- and sugar-alcohol–induced rapid optical clearing are effective for improving imaging performance for thin brain samples. Combining this method with conventional sectioning and imaging, the thickness of slices could be increased to reduce the loss of information. This method will be faced with challenges if it is combined with auto-sectioning and imaging for the reconstruction of a large brain area. The agents are very viscous, with viscosities of 217, 659, and for sorbitol, sucrose, and fructose, respectively. The high viscosity of the agents and the deformation during the procedure might prevent precise cutting. One possible solution is to ensure intact reconstruction along the -axis by overlapping optical and mechanical sectioning. Another is to optimize the optical clearing method by decreasing the viscosity of the agent. For instance, Pavone’s group used TDE, a low-viscosity agent, to clear the samples and combined with STP to image large-volume tissues,22,23 Myers and Chandrashekar’s group introduced DMSO and an aqueous buffer into sorbitol solution and achieved low viscosity,41 which both provide us with good references. Moreover, the construction of a certain slicing and imaging system is to be developed. As the urgent demand for large-volume tissue reconstruction continues, we believe that a convenient and rapid clearing method combined with sectioning and imaging techniques will provide a new perspective. 5.ConclusionIn this work, a convenient and rapid clearing method was proposed based on sugar and sugar-alcohol with MD simulation and experimental demonstration. The performance on tissue transparency, imaging depth, fluorescence preservation, and linear deformation was evaluated. The results show that sorbitol, sucrose, and fructose could make mouse brain sections transparent within 1 to 2 min and induce about threefold enhancement in imaging depth. The former two agents are good choices for GFP samples, fructose is the best for PI labeling samples, and sucrose is optimal for samples with both markers. Even though the three agents caused some shrinkage in samples, the contraction in the horizontal and longitudinal directions were almost the same, and the morphology of fine structures is preserved well. The results indicate that the rapid and PI-compatible clearing method is efficient for brain tissue slices, which will be helpful to broaden the applications of optical imaging techniques. The rapid optical clearing method is expected to perform optical imaging with sectioning and clearing, and provides a new perspective for large-volume reconstruction. AcknowledgmentsThis study was supported by the National Natural Science Foundation of China (Grant No. 31571002), the Science Fund for Creative Research Group of China (Grant No. 61421064), and the seed project of Wuhan National Laboratory for Optoelectronics. The authors are thankful to Tonghui Xu at Britton Chance Center for Biomedical Photonics for providing the Thy1-GFP-M line mice. We also thank the Optical Bioimaging Core Facility of WNLO-HUST for the support in data acquisition. ReferencesS. G. Parra et al.,
“Multiphoton microscopy of cleared mouse organs,”
J. Biomed. Opt., 15 036017
(2010). http://dx.doi.org/10.1117/1.3454391 JBOPFO 1083-3668 Google Scholar
W. R. Zipfel, R. M. Williams and W. W. Webb,
“Nonlinear magic: multiphoton microscopy in the biosciences,”
Nat. Biotechnol., 21
(11), 1369
–1377
(2003). http://dx.doi.org/10.1038/nbt899 NABIF9 1087-0156 Google Scholar
V. V. Tuchin, I. L. Maksimova and D. A. Zimnyakov,
“Light propagation in tissues with controlled optical properties,”
J. Biomed. Opt., 2
(4), 401
–417
(1997). http://dx.doi.org/10.1117/12.281502 JBOPFO 1083-3668 Google Scholar
V. V. Tuchin, Optical Clearing of Tissues and Blood, PM154 SPIE Press, Bellingham, WA
(2006). Google Scholar
D. Zhu et al.,
“Recent progress in tissue optical clearing,”
Laser Photon. Rev., 7
(5), 732
–757
(2013). http://dx.doi.org/10.1002/lpor.201200056 Google Scholar
H. Hama et al.,
“Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain,”
Nat. Neurosci., 14
(11), 1481
–1488
(2011). http://dx.doi.org/10.1038/nn.2928 NANEFN 1097-6256 Google Scholar
H. U. Dodt et al.,
“Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain,”
Nat. Methods, 4
(4), 331
–336
(2007). http://dx.doi.org/10.1038/nmeth1036 1548-7091 Google Scholar
A. Erturk et al.,
“Three-dimensional imaging of the unsectioned adult spinal cord to assess axon regeneration and glial responses after injury,”
Nat. Med., 18
(1), 166
–171
(2012). http://dx.doi.org/10.1038/nm.2600 1078-8956 Google Scholar
A. Erturk et al.,
“Three-dimensional imaging of solvent-cleared organs using 3DISCO,”
Nat. Protoc., 7
(11), 1983
–1995
(2012). http://dx.doi.org/10.1038/nprot.2012.119 1754-2189 Google Scholar
A. Ertürk and F. Bradke,
“High-resolution imaging of entire organs by 3-dimensional imaging of solvent cleared organs (3DISCO),”
Exp. Neurol., 242 57
–64
(2013). http://dx.doi.org/10.1016/j.expneurol.2012.10.018 EXNEAC 0014-4886 Google Scholar
K. Chung et al.,
“Structural and molecular interrogation of intact biological systems,”
Nature, 497
(7449), 332
–337
(2013). http://dx.doi.org/10.1038/nature12107 Google Scholar
K. Chung and K. Deisseroth,
“CLARITY for mapping the nervous system,”
Nat. Methods, 10
(6), 508
–513
(2013). http://dx.doi.org/10.1038/nmeth.2481 1548-7091 Google Scholar
R. Tomer et al.,
“Advanced CLARITY for rapid and high-resolution imaging of intact tissues,”
Nat. Protoc., 9
(7), 1682
–1697
(2014). http://dx.doi.org/10.1038/nprot.2014.123 1754-2189 Google Scholar
T. Kuwajima et al.,
“ClearT: a detergent- and solvent-free clearing method for neuronal and non-neuronal tissue,”
Development, 140
(6), 1364
–1368
(2013). http://dx.doi.org/10.1242/dev.091844 Google Scholar
M. T. Ke, S. Fujimoto and T. Imai,
“SeeDB: a simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction,”
Nat. Neurosci., 16
(8), 1154
–1161
(2013). http://dx.doi.org/10.1038/nn.3447 NANEFN 1097-6256 Google Scholar
M. T. Ke and T. Imai,
“Optical clearing of fixed brain samples using SeeDB,”
Curr. Protoc. Neurosci., 66 2.22.1
–2.22.19
(2014). Google Scholar
M. T. Ke and T. Imai,
“3D fluorescence imaging of the brain using an optical clearing agent, SeeDB,”
Jikken-Igaku, 32 449
–455
(2014). Google Scholar
M. T. Ke, S. Fujimoto and T. Imai,
“Optical clearing using SeeDB,”
Bio-protocol, 4
(3),
–e1042
(2014). Google Scholar
E. A. Susaki et al.,
“Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis,”
Cell, 157
(3), 726
–739
(2014). http://dx.doi.org/10.1016/j.cell.2014.03.042 CELLB5 0092-8674 Google Scholar
B. Yang et al.,
“Single-cell phenotyping within transparent intact tissue through whole-body clearing,”
Cell, 158
(4), 945
–958
(2014). http://dx.doi.org/10.1016/j.cell.2014.07.017 CELLB5 0092-8674 Google Scholar
B. Hou et al.,
“Scalable and DiI-compatible optical clearance of the mammalian brain,”
Front. Neuroanat., 9 19
(2015). http://dx.doi.org/10.3389/fnana.2015.00019 Google Scholar
I. Costantini et al.,
“A new versatile clearing method for brain imaging,”
Proc. SPIE, 9305 930514
(2015). http://dx.doi.org/10.1117/12.2076908 PSISDG 0277-786X Google Scholar
I. Costantini et al.,
“A versatile clearing agent for multi-modal brain imaging,”
Sci. Rep., 5 9808
(2015). http://dx.doi.org/10.1038/srep09808 SRCEC3 2045-2322 Google Scholar
H. Hama et al.,
“ScaleS: an optical clearing palette for biological imaging,”
Nat. Neurosci., 18
(10), 1518
–1529
(2015). http://dx.doi.org/10.1038/nn.4107 NANEFN 1097-6256 Google Scholar
B. Zingg et al.,
“Neural networks of the mouse neocortex,”
Cell, 156
(5), 1096
–1111
(2014). http://dx.doi.org/10.1016/j.cell.2014.02.023 CELLB5 0092-8674 Google Scholar
K. Miyamichi et al.,
“Cortical representations of olfactory input by trans-synaptic tracing,”
Nature, 472 191
–196
(2011). http://dx.doi.org/10.1038/nature09714 Google Scholar
A. Li et al.,
“Micro-optical sectioning tomography to obtain a high-resolution atlas of the mouse brain,”
Science, 330
(6009), 1404
–1408
(2010). http://dx.doi.org/10.1126/science.1191776 SCIEAS 0036-8075 Google Scholar
H. Gong et al.,
“Continuously tracing brain-wide long-distance axonal projections in mice at a one-micron voxel resolution,”
NeuroImage, 74 87
–98
(2013). http://dx.doi.org/10.1016/j.neuroimage.2013.02.005 NEIMEF 1053-8119 Google Scholar
T. Ragan et al.,
“Serial two-photon tomography for automated ex vivo mouse brain imaging,”
Nat. Methods, 9 255
–258
(2012). http://dx.doi.org/10.1038/nmeth.1854 1548-7091 Google Scholar
J. Wang et al.,
“An innovative transparent cranial window based on skull optical clearing,”
Laser Phys. Lett., 9
(6), 469
–473
(2012). http://dx.doi.org/10.7452/lapl.201210017 1612-2011 Google Scholar
J. Wang et al.,
“Review: tissue optical clearing window for blood flow monitoring,”
IEEE J. Sel. Top. Quant., 20
(2), 6801112
(2014). http://dx.doi.org/10.1109/JSTQE.2013.2286072 IJSQEN 1077-260X Google Scholar
Z. Mao et al.,
“Influence of alcohols on the optical clearing effect of skin in vitro,”
J. Biomed. Opt., 13
(2), 021104
(2008). http://dx.doi.org/10.1117/1.2892684 JBOPFO 1083-3668 Google Scholar
X. Wen et al.,
“Controling the scattering of intralipid by using optical clearing agents,”
Phys. Med. Biol., 54
(22), 6917
–6930
(2009). http://dx.doi.org/10.1088/0031-9155/54/22/011 PHMBA7 0031-9155 Google Scholar
X. Wen et al.,
“In vivo skin optical clearing by glycerol solutions: mechanism,”
J. Biophotonics, 3
(1–2), 44
–52
(2010). http://dx.doi.org/10.1002/jbio.200910080 Google Scholar
T. Yu et al.,
“Quantitative analysis of dehydration in porcine skin for assessing mechanism of optical clearing,”
J. Biomed. Opt., 16
(9), 095002
(2011). http://dx.doi.org/10.1117/1.3621515 JBOPFO 1083-3668 Google Scholar
J. Wang et al.,
“Sugar-induced skin optical clearing: from molecular dynamics simulation to experimental demonstration,”
IEEE J. Sel. Top. Quantum Electron., 20
(2), 7101007
(2014). http://dx.doi.org/10.1109/JSTQE.2013.2289966 Google Scholar
R. Shi et al.,
“Accessing to arteriovenous blood flow dynamics response using combined laser speckle contrast imaging and skin optical clearing,”
Biomed. Opt. Express, 6
(6), 1977
–1989
(2015). http://dx.doi.org/10.1364/BOE.6.001977 BOEICL 2156-7085 Google Scholar
P. S. Tsai et al.,
“Correlations of neuronal and microvascular densities in murine cortex revealed by direct counting and colocalization of nuclei and vessels,”
J. Neurosci., 29
(46), 14553
–14570
(2009). http://dx.doi.org/10.1523/JNEUROSCI.3287-09.2009 JNRSDS 0270-6474 Google Scholar
J. Hirshburg et al.,
“Correlation between collagen solubility and skin optical clearing using sugars,”
Lasers Surg. Med., 39
(2), 140
–144
(2007). http://dx.doi.org/10.1002/lsm.20417 LSMEDI 0196-8092 Google Scholar
J. M. Hirshburg et al.,
“Molecular basis for optical clearing of collagenous tissues,”
J. Biomed. Opt., 15
(5), 055002
(2010). http://dx.doi.org/10.1117/1.3484748 JBOPFO 1083-3668 Google Scholar
M. N. Economo et al.,
“A platform for brain-wide imaging and reconstruction of individual neurons,”
Elife, 5 e10566
(2016). http://dx.doi.org/10.7554/eLife.10566 Google Scholar
BiographyTingting Yu received her PhD degree in biomedical photonics from Huazhong University of Science and Technology (HUST) in 2015. She is currently a postdoc at Wuhan National Laboratory for Optoelectronics, HUST. Her research activity is focused on in vitro tissue optical clearing of neuronal tissues and neuroimaging with various optical imaging systems, including light-sheet microscopy and single/two-photon microscopy. Dan Zhu received the PhD degree in physical electronics from Huazhong University of Science and Technology (HUST) in 2001. After having completed a postdoctoral fellowship in biomedical engineering at HUST, she got the position of an associate professor in biomedical engineering in 2003, and a professor in 2007. She is currently a full professor of Wuhan National Optoelectronics Laboratory and distinguished professor of Huazhong Scholars. Recently, she has been focusing on optical clearing for skin, skull, and neuronal tissues. She is also the secretary general of the Biomedical Photonics Committee of the Chinese Optical Society. |