|
|
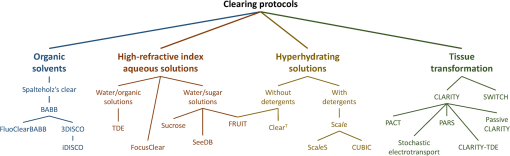
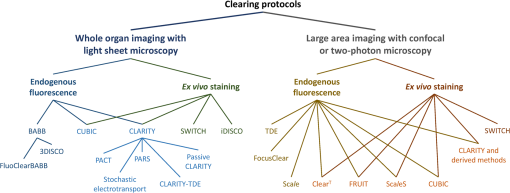
1.IntroductionCutting tissue into thin slices has been the standard practice in histology since its early days. Indeed, this approach allows solving (although roughly) two problems at once, i.e., absence of optical sectioning in conventional wide-field microscopes and tissue opacity. However, biological structures are inherently three-dimensional (3-D); for this reason, scientists have always tried to extend tissue imaging to thick specimens. On the imaging side, the invention of confocal microscopy by Minsky in 1955,1 the development of two-photon microscopy in 1990,2 and the recent revival of light-sheet microscopy,3 have provided a wide choice of microscopy methods allowing optical sectioning, i.e., 3-D resolution. On the other hand, the effectiveness and usability of these techniques in real macroscopic specimens are hampered, because biological specimens are naturally opaque. In general, the main source of this opaqueness is not absorption, which is characteristic only of a subset of tissues (most notably blood, muscles, and liver), but scattering. Indeed, every biological sample is ultimately constituted by high-refractive index molecules (lipids and proteins) immersed in a low-refractive medium (water). The “dry” refractive index of tissue, which takes into account the biomolecules contribution, has been estimated to be .4 The mismatch between this value and (1.33) results in multiple scattering events, eventually resulting in an opaque sample. The general approach pursued by almost all clearing methods is therefore to reduce as much as possible the refractive index mismatch between biomolecules and the surrounding medium. On the one hand, water can be replaced by liquids or mixtures with a higher refractive index. On the other hand, can be modified by removing some components (as lipids) or by modifying the optical properties of others (e.g., changing the hydration state of proteins). For this reason, the final refractive index of the cleared tissue () can have different values according to the protocol used, ranging from 1.385 to 1.56.6 An excellent review of tissue clearing methods, classified by the underlying mechanism, has been recently presented by Richardson and Lichtman.7 Ideally, an optical clearing method should provide high transparency of the samples, preserve endogenous fluorescence, allow exogenous molecular staining—e.g., immunohistochemistry (IHC) or fluorescence in situ hybridization—and be applicable to a wide range of samples in terms of species (rodents, primates, and humans), organs, and age. Other quite desirable properties are speed, ease of use, large reproducibility, safety, and low cost of the chemicals used. Of course, such an ideal method is unfeasible in real laboratory life, and one has to sacrifice some features to preserve those that are necessary given the microscopy technique used and the experiment to be performed. Anyway, choosing the appropriate clearing protocol might be difficult, given the plethora (more than 20) of methods presented in the literature in the last years.5,6,8–27 Here, we try to classify clearing approaches based on the microscopy technique where they can be applied, rather than on the clearing mechanism.7 We believe that this new taxonomy can be a helpful starting point for microscopists willing to work with transparent tissue. We first report the traditional taxonomy of clearing methods (Sec. 2); afterward, we describe our new classification scheme (Sec. 3). After discussing future perspectives in the field (Sec. 4), we conclude with a small checklist that might help to find the potential and the limits of newly published methods (Sec. 5). 2.Classical TaxonomyThe traditional subdivision of clearing procedures is based on the main physical mechanism underlying the clearing process (Fig. 1). In this taxonomy, four families of techniques can be identified: organic solvents, aqueous solutions with high refractive index, protein hyperhydration, and tissue transformation. 2.1.Organic SolventsThe first clearing method for fixed tissue, reported by Spalteholz a hundred years ago,20 was based on the substitution of water with a mixture of benzyl benzoate and methyl salicylate, showing a refractive index of about 1.55. Since benzyl benzoate is insoluble in water, an intermediate dehydration step based on ethanol was introduced. This general scheme (dehydration followed by incubation in high-refractive index solvents) has been reintroduced by Dodt et al.8 who exploited as index-matching solution a mixture of benzyl alcohol and benzyl benzoate (BABB, ). Since the original BABB protocol quenches the emission of fluorescent proteins after a short time period, the same group investigated alternative dehydration and clearing agents, finding that dehydration with tetrahydrofuran (THF) and clearing in dibenzyl ether (DBE, ) achieved the best results in fluorescence preservation and sample transparency (3DISCO).6,9 Notably, the authors pointed out that both THF and DBE tend to develop fluorescence-quenching peroxides, and for this reason all solutions should be previously filtered in alumina.6 Other improvements on BABB clearing, based on pH control and different dehydration agents, have been recently reported by Schwarz et al. (FluoClearBABB).26 A further extension of clearing based on organic solvents is represented by the embedding of samples in clear, organic-based, solid resins: an intriguing feature of this method is the large reduction of photobleaching.11 2.2.High-Refractive Index Aqueous SolutionsFluorescence can be more easily preserved if the sample is maintained in an aqueous, protein-friendly environment. Indeed, fluorescence quenching observed in organic solvents (at least in some protocols) stimulated the quest for water-based solutions with high refractive index. Some methods exploit high-concentration solutions of sugars, such as sucrose ()25 or fructose (SeeDB and FRUIT, ).12,13 A drawback of sugar solutions is their high viscosity; to solve this problem, alternative approaches based on organic compounds like 2,2′-thiodiethanol (TDE, )19,23,28 have been proposed. The family of water-based clearing includes also commercial solutions whose detailed composition is not known (FocusClear, ). 2.3.Hyperhydrating SolutionsIn general, the index of refraction of aqueous solutions cannot be increased above 1.48. However, this is usually not enough to correctly match the average refractive index of the dry component of the tissue.4 A workaround to this problem is to simultaneously act on tissue components (such as proteins) to lower their effective refractive index, and/or to remove other components such as lipids. Indeed, lipid removal exploiting a polyalcohol (glycerol) and a detergent (Triton X-100), and protein hyperhydration with urea are the main building blocks of the Scale method ().5 The results obtained with Scale triggered a more comprehensive screening of chemical agents (polyalcohols, detergents, and hydrophilic compounds), which resulted in a protocol called CUBIC ().22 CUBIC has been recently further optimized to afford clearing of the whole mouse body, guaranteeing also discoloration of hemoglobin.24 A further development of Scale is ScaleS, a method where glycerol is replaced by sorbitol.27 Another protocol exploiting hyperhydration (without lipid removal) is based on formamide/water mixtures (, ).21 2.4.Tissue TransformationAlthough removal of lipids from the tissue does provide increased transparency at moderate refractive indexes (, affordable by aqueous solutions), the use of strong detergents results also in a loss of protein content from the tissue. Furthermore, strongly hydrophilic molecules like urea can lead to protein denaturation, hindering epitopes potentially useful for IHC. Tissue transformation techniques try to stabilize the protein content of the sample by cross-linking proteins to a gel mesh. The pioneer methodology in the field has been CLARITY, where proteins and nucleic acids are cross-linked to a polyacrylamide gel by paraformaldehyde.14 The tissue–gel hybrid is then treated with a strong detergent [sodium dodecyl sulfate (SDS)] for lipid removal. Finally, the sample is immersed in an index-matching solution (originally FocusClear) to render it transparent. Lipid removal by SDS can be sped up by electrophoresis14 or by raising incubation temperature.15 However, strong electric field or high temperatures can damage tissue–gel hybrids on both a structural and molecular level. Stochastic implementations of electrophoresis16 and novel tissue-hybrid recipes17 improve sample stability while keeping lipid removal time quite short (few days). Continuous perfusion of the sample with SDS is another approach to improve clearing speed.18 Since the same perfusion system can be used also to deliver hydrogel monomers as well as immunohistochemical solutions (like antibodies and blocking buffers), this approach has been termed perfusion-assisted agent release in situ (PARS).18 Other improvements to the original CLARITY approach include the use of cheaper index matching solutions, such as TDE,19 histodenz,18 or diatrizoic acid.16,17 Tissue-transforming methods are usually quite complex and expensive; nonetheless, they are attractive because of the high degree of transparency and of the increased sample porosity, which helps whole-mount IHC on large portions of tissue.14,19 Diffusion of antibody inside the sample can be further facilitated by the application of stochastic electric fields.16 3.Application-Oriented TaxonomyThe classification of clearing protocols presented in the previous section (and described in more detail in Ref. 7) is definitely useful for researchers willing to develop new methods or optimize existing ones. However, we believe that this subdivision might be confusing for researchers looking for the method most suitable for their application. For this reason, we propose a different taxonomy of state of the art clearing methods (Fig. 2) based on the microscopy method used, the need for ex vivo tissue staining, and other practical considerations, such as technical complexity and expensiveness. 3.1.Whole Organ Imaging with Light-Sheet MicroscopyProviding optical sectioning in a wide-field detection scheme, light-sheet microscopy (LSM) is one of the fastest methods for volumetric imaging.3 Planar illumination can be provided either by cylindrical optics29 or by one-dimensional scanning of a line.30 The excitation beam is poorly focused, guaranteeing that the thickness of the light sheet is almost constant within the field of view of the camera. Visible light is generally used, although two-photon LSM has been demonstrated for small samples.31 The linear excitation mechanism makes this technique quite sensitive to light scattering inside the sample, which results in unwanted blur and eventually in a completely misty image. Therefore, although some optical methods to reduce scattering effects have been devised in the last years (like confocal detection32,33 and Bessel beam illumination34), LSM requires the sample to be as transparent as possible for proper imaging. Indeed, LSM of macroscopic biological specimens was coupled to tissue clearing since its first reported application in 1993.35 When choosing the appropriate clearing method for LSM, the most important feature to be taken into account is a complete, crystalline, clearing. “Average” transparency usually does not work. Here below, we list published clearing protocols suitable for LSM, according to their compatibility with endogenous fluorescence (e.g., GFP) and/or ex vivo tissue staining. A visual summary of the clearing and imaging capabilities of the various methods is reported in Fig. 3. Fig. 3Whole-brain transparencies and representative fluorescence images obtained with different methods. Images adapted with permission from Refs. 14, 1617.18.–19, 22, and 36. 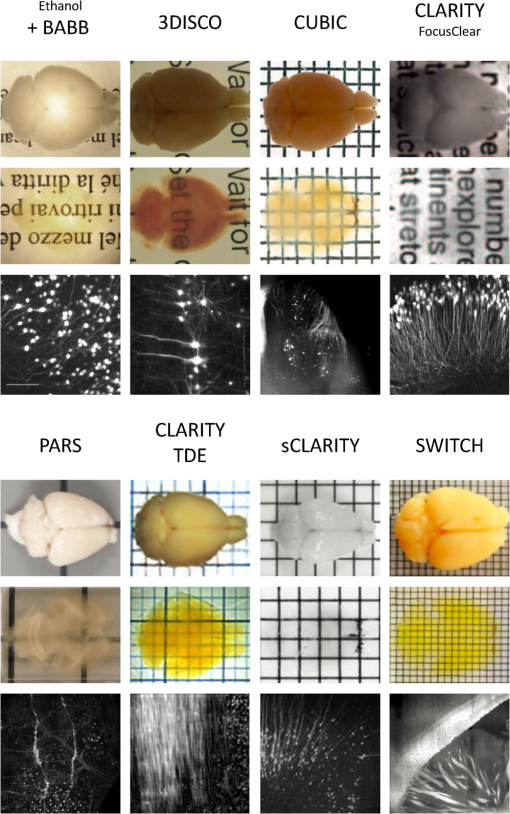 3.1.1.Endogenous fluorescenceDodt et al.8 demonstrated the possibility of applying LSM to transgenic fluorescent mice (thy1-GFP-M) with BABB clearing. However, the preservation of GFP fluorescence with this protocol was not always guaranteed. In the following years, the same group successfully tackled this issue, optimizing chemical clearing of GFP-expressing mouse brain.6,9 In addition to an accurate selection of the reagents, the importance of removing fluorescence-quenching peroxides from the solutions was underlined.11 Unfortunately, this step adds a practical complication to an otherwise very easy protocol, since it requires the use of a chromatography column setup. On the other hand, a recent article reported a modified yet simple BABB protocol providing high transparency and good preservation of fluorescence, obtained by proper pH titration of the solutions used.26 In general, organic solvents provide superior sample transparency—the best in our experience—and, if properly optimized, good preservation of endogenous fluorescence. In our opinion, the real problem with these clearing protocols is another one, often overlooked: no objective lens suitable for immersion in BABB or DBE exists on the market. (Leica produces a BABB-corrected objective, but with very short working distance, thus useless for whole-brain LSM.) Researchers commonly use air objectives, introducing very large spherical aberration that leads to a dramatic reduction in the detected signal and in the optical sectioning capabilities of LSM.37 A promising approach to recover diffraction-limited performances is the use of specialized optical elements for the correction of air objectives, as described by Dodt et al.38 If these devices would become commercially available, they could boost the use of organic solvents for clearing. On the other hand, clearing methods resulting in an overall lower refractive index can exploit objective lenses corrected up to . Organs cleared with CUBIC have a refractive index of 1.38,22 and present a quite good transparency, although not as good as with DBE. CLARITY, which uses as index-matching agent FocusClear (), can provide crystal-clear entire mouse brains,14 and indeed it has been successfully applied in combination with LSM.15,39 Tissue processing with CLARITY requires at least 1 week (while it can be performed in 1 to 2 days with DBE). Recently, a speed-up of the clearing process has been achieved using stochastic electrotransport of lipids.16 The use of electrophoresis, either “standard” or stochastic, introduces great technical complexity, unless a commercial system is used. Passive methods, relying on spontaneous diffusion of lipids, have been described.15,18 Different implementations, like passive CLARITY15 or passive clarity technique (PACT),18 differ in the concentration of SDS and in the solution used for refractive index matching. Although these approaches are much easier to reproduce than electrophoresis-based ones, some tricky experimental procedures (like sample degassing prior to polymerization) are still present. In addition, proper sample clearing can take months in this case. Faster clearing can be obtained using perfusion to deliver in situ the fat-removing solution (PARS), but at the expense of added technical complexity.18 As a final remark on CLARITY techniques, it is worth noting that many alternative cost-effective index-matching solutions, replacing the expensive FocusClear, have been reported, like Histodenz,18 TDE,19 and diatrizoic acid.16,17 3.1.2.Ex vivo tissue stainingPerforming molecular labeling on the entire mouse brain, or on large portion of it, is quite challenging because conventional protocols rely on slow spontaneous diffusion. Indeed, immunostaining of large portions of adult mouse brain (hypothalamus) reported with the CUBIC protocol required an overall staining time of about 1 week.22 Also in the highly porous tissue-gel hybrids produced by CLARITY, whole-brain IHC takes several weeks of incubation.14 However, CLARITY is, to our knowledge, the only protocol where IHC on whole adult mouse brains has been successfully demonstrated. CLARITY has shown to be compatible with fluorescence in situ hybridization, too.18 A significant speed up on whole-organ immunolabeling can be afforded exploiting the same stochastic electrotransport principle used for lipid removal. With this approach entire mouse brains can be immunostained overnight.16 Although this method is technically challenging, it can open the door to high-throughput molecular staining on large tissue blocks, driving the application of LSM on big volumes of human tissue. Large-volume immunolabeling can be also combined with organic solvent clearing (iDISCO).10 The authors reported mainly application with mouse embryos, although portions of adult mouse brain have been stained as well. This protocol is limited by the long incubation times and, from a practical point of view, by the extensive use of a toxic chemical (methanol). 3.2.Large Area Imaging with Two-Photon or Confocal MicroscopyConfocal and two-photon fluorescence microscopy (TPFM) are probably the most common imaging methods capable of optical sectioning (i.e., volumetric imaging).40–42 Typical setups are based on epi-fluorescence detection and a scanning system (usually a pair of galvo mirrors) to reconstruct the image of the sample point by point. Optical sectioning capabilities of both methods are proportional to the NA of the objective used, and in practice moderate to large numerical apertures () are used. This fact contributes to render confocal and two-photon microscopy sensible to optical aberrations.43 Thus, in general, these techniques require objective lenses corrected for the refractive index of the clearing solution employed. The sequential acquisition scheme usually employed in confocal and two-photon microscopy limits imaging speed,36 restricting the common use of these methods only portions of entire organs.19 To afford full reconstruction of whole mouse brains, significant engineering efforts are required to keep system stability over long (several weeks) imaging times.44,45 These stability requirements also apply to the sample itself; if cm-sized samples have to be imaged with point-scanning methods, the clearing method used must provide long-term sample stability and fluorescence preservation. TPFM is known to perform well deep inside scattering tissue.42 Indeed, due to the nonlinearity of excitation, diffused light does not contribute to background signal since it cannot reach enough intensity to excite fluorescence; in addition, the near-infrared wavelengths used for excitation are scattered less by biological specimens. TPFM is nowadays the standard method for in vivo imaging deep (up to 700 to ) in mouse brain. However, the penetration depth of this method in aldehyde-fixed tissue is reduced to about because of increased optical scattering.12 Nonetheless, the robustness to scattering intrinsic to TPFM allows its use for volumetric imaging also in poorly cleared specimens. To summarize, when choosing the appropriate clearing protocol for confocal or two-photon microscopy, the main requirement concerns the availability of suitable objective lenses, while for TPFM high transparency is not as crucial as for LSM. From these considerations, we can conclude that high-refractive-index organic solvents are not the best choice in this case, while direct immersion in aqueous solution can be a valid approach, although it provides only moderate transparency. Finally, if one wants to exploit serial tissue sectioning to perform reconstructions on larger areas, also tissue stability and cutting capability become important and should be taken into account. Here below, we list published clearing protocols suitable for confocal and two-photon microscopy, according to their compatibility with endogenous fluorescence (e.g., GFP) and/or ex vivo tissue staining. A visual summary of the clearing and imaging capabilities of the various methods is reported in Fig. 4. Fig. 4Brain slice transparencies and representative fluorescence images obtained with different methods. Images adapted with permission from Refs. 5, 12, 19, 21, and 27. 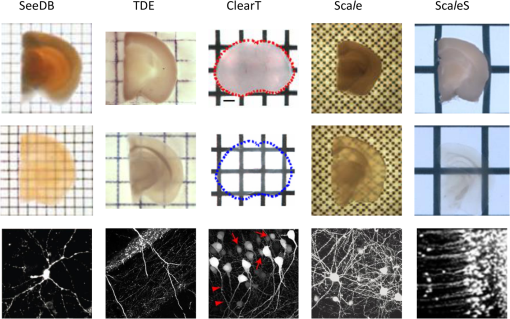 3.2.1.Endogenous fluorescenceScale5 was the first clearing method demonstrated with two-photon microscopy. Although it cannot provide high transparency across the whole mouse brain, its clearing effectiveness is well enough for TPFM. It drove the development of commercial objective lenses corrected for high refractive indexes (around 1.38), with long working distances (), and moderate to high NA (). It is based on simple immersion and operates in about 2 weeks. However, samples become highly fragile and difficult to handle, and large tissue swelling is observed. The same laboratory recently developed ScaleS, a method based on sorbitol and urea.27 Tissue processed with ScaleS presents good transparency and structural stability, potentially allowing its use also in a serial sectioning apparatus. The protocol lasts just few days, and the ultrastructure of the tissue is preserved, allowing subsequent analysis with electron microscopy. Another protocol for fast, direct clearing of small brain regions is SeeDB,12 based on saturated fructose solutions. Although the protocol is quite simple and no significant swelling has been reported, the high viscosity of the clearing solution may represent a practical limitation. Urea and fructose clearing have been combined in the FRUIT protocol.13 The authors reported that, with the addition of urea, clearing capabilities comparable to the one of SeeDB can be achieved at lower fructose concentrations, thus with a much less viscous immersion medium. can provide direct and fast clearing of embryonic tissue and brain, well preserving endogenous fluorescence.21 However, to our knowledge no application of this protocol to adult mouse brain tissue has been reported. Furthermore, formamide poses serious safety issues due to its teratogenicity. Another method demonstrated with TPFM is direct immersion in TDE.19 TDE is a nonviscous, nontoxic, and cheap medium, miscible with water in any ratio.28 The refractive index of TDE-water solutions can be finely tuned by changing their respective ratio. TDE-treated samples are rigid enough to be cut in a serial-sectioning apparatus. Furthermore, ultrastructural details are preserved, in case subsequent electron microscopy analysis would be of interest. In general, all protocols suitable for whole-brain imaging of endogenous fluorescence (see Sec. 3.1.1)—provided that the refractive index of the final immersion medium is smaller than 1.52 (like CLARITY14 and CUBIC22)—can be used as well in confocal and two-photon imaging of smaller regions. However, these approaches are usually technically complex and/or require longer incubation times, and their use in conjunction with TPFM might be an overwork. Nonetheless, the possibility to apply confocal or two-photon microscopy to CLARITY- or CUBIC-cleared samples allows imaging the same specimen with multiple techniques, in a correlative fashion.46 3.2.2.Ex vivo tissue stainingSince it relies mainly on passive diffusion, IHC is easier for smaller tissue blocks than for whole organs (see Sec. 3.1.2). Anyhow, porosity increase is a mandatory step to allow antibodies to effectively stain the entire sample. CLARITY can provide immunostaining of thick tissue slabs (1 to 2 mm) within a reasonable time.14,15,19 A recent development of this methodology, called SWITCH,17 replaced the original polyacrylamide mesh with a more stable glutaraldehyde one, allowing more than 20 IHC rounds while maintaining structural and molecular stability of the tissue. Increased porosity helps ex vivo staining also in hyperhydration-based methods, such as CUBIC,22 ScaleS,27 ,21 and FRUIT.13 A particularly intriguing feature of these methods is that brain lipid structures are preserved, allowing lipophilic labeling usually not possible with CLARITY-based methods (although it has been reported in SWITCH-processed mouse brains). 4.DiscussionThe research of methods to clear and stain large portions of biological tissue (even entire organs) has garnered large attention in the recent years.7 A prominent reason for the flourishing of these techniques is the concurrent development of high-throughput microscopy imaging systems, which allow the reconstruction of large specimens with micrometric resolution.36 Nonetheless, the landscape of clearing technologies is often depicted just in chemical terms, without taking into proper account the imaging methods to which clearing protocols should be eventually coupled. Here, we tried to describe and classify existing techniques from a microscopist’s perspective, highlighting the fact that there is no “ultimate” clearing protocol, surmounting all the others in all applications. It is up to the end user to find out the method best suited for her application, by properly evaluating the right balance between costs (technical complexity, time, monetary costs, and so on) and benefits (image quality): unfortunately, this effort is necessary and cannot be overlooked. Our hope is that the application-oriented taxonomy described in this review can help researchers to restrict the space of their survey, speeding it up. Given the strong connection between clearing and imaging, we believe that further integration between these two fields will benefit both. For instance, the development of microscope objectives corrected for immersion in high-refractive index () organic solvents would significantly expand the application of these methods in both LSM and TPFM. Also, apparently side aspects of clearing protocol such as sample stiffness and compatibility with transparent embedding have large practical implications: their optimization would be relevant for serial-sectioning applications and for specimen mounting in LSM, respectively. From an end-user’s perspective, it would be important to define common benchmarks to better compare clearing methods. Transmittance measurements of cleared samples are often reported, but some crucial parameters like the age of the animal or the kind of specimen used (whole brain, hemisphere, and slice) vary between different papers. Also, data from a significant number of samples should be reported, to take into account intersubject variability and highlight the reproducibility of the method. The transgenic animal model used is important as well: as pointed out in Ref. 27, most clearing methods are demonstrated in thy1 transgenes (usually line YFP-H or GFP-M),47 which are known for their outstanding brightness due to the large expression of fluorescent proteins. The demonstration of the method in other samples would be useful for a more realistic evaluation of fluorescence preservation. Another field where benchmarking will be beneficial is sample staining. Chemical processing of the specimen can hinder some epitopes, preventing immunostaining. Therefore, successful whole-brain or whole-body labeling methods demonstrated with a specific antibody are not guaranteed to work with another one. Large-scale screening of multiple antibodies17 can help users identifying the protocol to use. Also, in quantifying antibody penetration, defining standard sample conditions in terms of species (mouse, rat, human, and so on), age and sample size is essential for proper comparison of different methods. 5.ConclusionTissue clearing methods, together with high-throughput microscopic imaging and subsequent image analysis, are paving the way toward a systemic study of biological systems, on an organ-wide or even organism-wide scale but with cellular or subcellular resolution. We expect that further methods will be presented in the next future, providing valuable tools for biomedical researchers. Furthermore, optimization and simplification of current methods will allow their usage on more robust and reproducible basis. Anyhow, we do not expect that a single method will be dominant over the others: researchers will rather need to choose the right method based on their microscope and on their application. We would like to conclude this review of current clearing methods for fixed tissue with a checklist that might be useful to contextualize newly published methods, and to better figure out their potential:
We hope that this short checklist could help interested readers to quickly find out the pros and cons of oncoming clearing protocols, spreading the use of these technologies to advance our knowledge of biological systems such as the whole mammalian brain. AcknowledgmentsThe authors received funding support from the European Union Seventh Framework Program (FP7/2007-2013) under Grant No. 604102 (Human Brain Project) and Grant No. 284464 (LASERLAB-EUROPE). They have also been supported by the Italian Ministry for Education, University, and Research in the framework of the Flagship Project NanoMAX, and by “Ente Cassa di Risparmio di Firenze” (private foundation). ReferencesM. Minsky,
“Memoir on inventing the confocal scanning microscope,”
Scanning, 10
(4), 128
–138
(1988). http://dx.doi.org/10.1002/sca.v10:4 SCNNDF 0161-0457 Google Scholar
W. Denk, J. H. Strickler and W. W. Webb,
“Two-photon laser scanning fluorescence microscopy,”
Science, 248
(4951), 73
–76
(1990). http://dx.doi.org/10.1126/science.2321027 SCIEAS 0036-8075 Google Scholar
P. J. Keller and H. U. Dodt,
“Light sheet microscopy of living or cleared specimens,”
Curr. Opin. Neurobiol., 22
(1), 138
–143
(2012). http://dx.doi.org/10.1016/j.conb.2011.08.003 COPUEN 0959-4388 Google Scholar
S. L. Jacques,
“Optical properties of biological tissues: a review,”
Phys. Med. Biol., 58
(14), 5007
–5008
(2013). http://dx.doi.org/10.1088/0031-9155/58/14/5007 PHMBA7 0031-9155 Google Scholar
H. Hama et al.,
“Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain,”
Nat. Neurosci., 14
(11), 1481
–1488
(2011). http://dx.doi.org/10.1038/nn.2928 NANEFN 1097-6256 Google Scholar
K. Becker et al.,
“Chemical clearing and dehydration of GFP expressing mouse brains,”
PLoS One, 7
(3), e33916
(2012). http://dx.doi.org/10.1371/journal.pone.0033916 POLNCL 1932-6203 Google Scholar
D. S. Richardson and J. W. Lichtman,
“Clarifying tissue clearing,”
Cell, 162
(2), 246
–257
(2015). http://dx.doi.org/10.1016/j.cell.2015.06.067 CELLB5 0092-8674 Google Scholar
H. U. Dodt et al.,
“Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain,”
Nat. Methods, 4
(4), 331
–336
(2007). http://dx.doi.org/10.1038/nmeth1036 1548-7091 Google Scholar
A. Erturk et al.,
“Three-dimensional imaging of solvent-cleared organs using 3DISCO,”
Nat. Protoc., 7
(11), 1983
–1995
(2012). http://dx.doi.org/10.1038/nprot.2012.119 1754-2189 Google Scholar
N. Renier et al.,
“iDISCO: a simple, rapidmethod to immunolabel large tissue samples for volume imaging,”
Cell, 159
(4), 896
–910
(2014). http://dx.doi.org/10.1016/j.cell.2014.10.010 CELLB5 0092-8674 Google Scholar
K. Becker et al.,
“Reduction of photo bleaching and long term archiving of chemically cleared GFP-expressing mouse brains,”
PLoS One, 9
(12), e114149
(2014). http://dx.doi.org/10.1371/journal.pone.0114149 POLNCL 1932-6203 Google Scholar
M. T. Ke, S. Fujimoto and T. Imai,
“SeeDB: a simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction,”
Nat. Neurosci., 16
(8), 1154
–1161
(2013). http://dx.doi.org/10.1038/nn.3447 NANEFN 1097-6256 Google Scholar
B. Hou et al.,
“Scalable and Dil-compatible optical clearance of the mammalian brain,”
Front. Neuroanat., 9 19
(2015). http://dx.doi.org/10.3389/fnana.2015.00019 Google Scholar
K. Chung et al.,
“Structural and molecular interrogation of intact biological systems,”
Nature, 497
(7449), 332
(2013). http://dx.doi.org/10.1038/nature12107 Google Scholar
R. Tomer et al.,
“Advanced CLARITY for rapid and high-resolution imaging of intact tissues,”
Nat. Protoc., 9
(7), 1682
–1697
(2014). http://dx.doi.org/10.1038/nprot.2014.123 1754-2189 Google Scholar
S.-Y. Kim et al.,
“Stochastic electrotransport selectively enhances the transport of highly electromobile molecules,”
Proc. Natl. Acad. Sci. U. S. A., 112
(46), E6274
–E6283
(2015). http://dx.doi.org/10.1073/pnas.1510133112 Google Scholar
E. Murray et al.,
“Simple, scalable proteomic imaging for high-dimensional profiling of intact systems,”
Cell, 163
(6), 1500
–1514
(2015). http://dx.doi.org/10.1016/j.cell.2015.11.025 CELLB5 0092-8674 Google Scholar
B. Yang et al.,
“Single-cell phenotyping within transparent intact tissue through whole-body clearing,”
Cell, 158
(4), 945
–958
(2014). http://dx.doi.org/10.1016/j.cell.2014.07.017 CELLB5 0092-8674 Google Scholar
I. Costantini et al.,
“A versatile clearing agent for multi-modal brain imaging,”
Sci. Rep., 5 9808
(2015). http://dx.doi.org/10.1038/srep09808 Google Scholar
W. Spalteholz, Über das Durchsichtigmachen von menschlichen und tierischen Präpareten und seine theoretischen Bedingungen, S. Hirzel, Leipzig
(1914). Google Scholar
T. Kuwajima et al.,
“Clear(T): a detergent- and solvent-free clearing method for neuronal and non-neuronal tissue,”
Development, 140
(6), 1364
–1368
(2013). http://dx.doi.org/10.1242/dev.091844 Google Scholar
E. A. Susaki et al.,
“Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis,”
Cell, 157
(3), 726
–739
(2014). http://dx.doi.org/10.1016/j.cell.2014.03.042 CELLB5 0092-8674 Google Scholar
Y. Aoyagi et al.,
“A rapid optical clearing protocol using 2, 2′-thiodiethanol for microscopic observation of fixed mouse brain,”
PLoS One, 10
(1), e0116280
(2015). http://dx.doi.org/10.1371/journal.pone.0116280 POLNCL 1932-6203 Google Scholar
K. Tainaka et al.,
“Whole-body imaging with single-cell resolution by tissue decolorization,”
Cell, 159
(4), 911
–924
(2014). http://dx.doi.org/10.1016/j.cell.2014.10.034 CELLB5 0092-8674 Google Scholar
P. S. Tsai et al.,
“Correlations of neuronal and microvascular densities in murine cortex revealed by direct counting and colocalization of nuclei and vessels,”
J. Neurosci., 29
(46), 14553
–14570
(2009). http://dx.doi.org/10.1523/JNEUROSCI.3287-09.2009 JNRSDS 0270-6474 Google Scholar
M. K. Schwarz et al.,
“Fluorescent-protein stabilization and high-resolution imaging of cleared, intact mouse brains,”
PLoS One, 10
(5), 0124650
(2015). http://dx.doi.org/10.1371/journal.pone.0124650 POLNCL 1932-6203 Google Scholar
H. Hama et al.,
“Sca/eS: an optical clearing palette for biological imaging,”
Nat. Neurosci., 18
(10), 1518
(2015). http://dx.doi.org/10.1038/nn.4107 NANEFN 1097-6256 Google Scholar
T. Staudt et al.,
“2, 2′-thiodiethanol: a new water soluble mounting medium for high resolution optical microscopy,”
Microsc. Res. Tech., 70
(1), 1
–9
(2007). http://dx.doi.org/10.1002/(ISSN)1097-0029 MRTEEO 1059-910X Google Scholar
J. Huisken et al.,
“Optical sectioning deep inside live embryos by selective plane illumination microscopy,”
Science, 305
(5686), 1007
–1009
(2004). http://dx.doi.org/10.1126/science.1100035 SCIEAS 0036-8075 Google Scholar
P. J. Keller et al.,
“Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy,”
Science, 322
(5904), 1065
–1069
(2008). http://dx.doi.org/10.1126/science.1162493 SCIEAS 0036-8075 Google Scholar
T. V. Truong et al.,
“Deep and fast live imaging with two-photon scanned light-sheet microscopy,”
Nat. Methods, 8
(9), 757
–760
(2011). http://dx.doi.org/10.1038/nmeth.1652 1548-7091 Google Scholar
L. Silvestri et al.,
“Confocal light sheet microscopy: micron-scale neuroanatomy of the entire mouse brain,”
Opt. Express, 20
(18), 20582
–20598
(2012). http://dx.doi.org/10.1364/OE.20.020582 OPEXFF 1094-4087 Google Scholar
E. Baumgart and U. Kubitscheck,
“Scanned light sheet microscopy with confocal slit detection,”
Opt. Express, 20
(19), 21805
–21814
(2012). http://dx.doi.org/10.1364/OE.20.021805 OPEXFF 1094-4087 Google Scholar
F. O. Fahrbach and A. Rohrbach,
“Propagation stability of self-reconstructing Bessel beams enables contrast-enhanced imaging in thick media,”
Nat. Commun., 3 632
(2012). http://dx.doi.org/10.1038/ncomms1646 NCAOBW 2041-1723 Google Scholar
A. H. Voie, D. H. Burns and F. A. Spelman,
“Orthogonal-plane fluorescence optical sectioning—3-dimensional imaging of macroscopic biological specimens,”
J. Microsc., 170 229
–236
(1993). http://dx.doi.org/10.1111/jmi.1993.170.issue-3 Google Scholar
L. Silvestri et al.,
“Advanced optical techniques to explore brain structure and function,”
J. Innov. Opt. Health Sci., 6
(1), 123000
(2013). http://dx.doi.org/10.1142/S1793545812300029 Google Scholar
L. Silvestri, L. Sacconi and F. S. Pavone,
“Correcting spherical aberrations in confocal light sheet microscopy: a theoretical study,”
Microsc. Res. Tech., 77
(7), 483
–491
(2014). http://dx.doi.org/10.1002/jemt.v77.7 MRTEEO 1059-910X Google Scholar
H. U. Dodt et al.,
“Ultramicroscopy: development and outlook,”
Neurophoton, 2
(4), 041407
(2015). http://dx.doi.org/10.1117/1.NPh.2.4.041407 Google Scholar
W. Menegas et al.,
“Dopamine neurons projecting to the posterior striatum form an anatomically distinct subclass,”
Elife, 4 e10032
(2015). http://dx.doi.org/10.7554/eLife.10032 Google Scholar
J. A. Conchello and J. W. Lichtman,
“Optical sectioning microscopy,”
Nat. Methods, 2
(12), 920
–931
(2005). http://dx.doi.org/10.1038/nmeth815 1548-7091 Google Scholar
W. R. Zipfel, R. M. Williams and W. W. Webb,
“Nonlinear magic: multiphoton microscopy in the biosciences,”
Nat. Biotechnol., 21
(11), 1369
–1377
(2003). http://dx.doi.org/10.1038/nbt899 NABIF9 1087-0156 Google Scholar
F. Helmchen and W. Denk,
“Deep tissue two-photon microscopy,”
Nat. Methods, 2
(12), 932
–940
(2005). http://dx.doi.org/10.1038/nmeth818 1548-7091 Google Scholar
H. Jacobsen et al.,
“Refractive-index-induced aberrations in 2-photon confocal fluorescence microscopy,”
J. Microsc., 176 226
–230
(1994). http://dx.doi.org/10.1111/jmi.1994.176.issue-3 Google Scholar
T. Zheng et al.,
“Visualization of brain circuits using two-photon fluorescence micro-optical sectioning tomography,”
Opt. Express, 21
(8), 9839
–9850
(2013). http://dx.doi.org/10.1364/OE.21.009839 OPEXFF 1094-4087 Google Scholar
H. Gong et al.,
“Continuously tracing brain-wide long-distance axonal projections in mice at a one-micron voxel resolution,”
NeuroImage, 74 87
–98
(2013). http://dx.doi.org/10.1016/j.neuroimage.2013.02.005 NEIMEF 1053-8119 Google Scholar
A. L. Allegra Mascaro et al.,
“Towards a comprehensive understanding of brain machinery by correlative microscopy,”
J. Biomed. Opt., 20
(6), 061105
(2015). http://dx.doi.org/10.1117/1.JBO.20.6.061105 Google Scholar
G. Feng et al.,
“Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP,”
Neuron, 28
(1), 41
–51
(2000). http://dx.doi.org/10.1016/S0896-6273(00)00084-2 NERNET 0896-6273 Google Scholar
BiographyLudovico Silvestri received his master’s degree in physics in 2008 at the University of Pisa. He then moved to the University of Florence, where he obtained his PhD in atomic and molecular spectroscopy in 2012. From 2012 to 2014, he has been a postdoc at the European Laboratory for Non-Linear Spectroscopy. Currently, he is a researcher at the National Institute of Optics in Florence. His research activity is focused on light-sheet microscopy of cleared specimens. Irene Costantini obtained her master’s degree in medical and pharmaceutical biotechnology in 2011 at the University of Florence with Professor Cavalieri and Professor Netea. An internship was conducted in the Radboud University Nijmegen Medical Center, Netherlands. She is currently attending an international PhD in atomic and molecular photonics at LENS. Her research activity is focused on 3-D reconstruction of intact neuronal networks in whole mouse and human brains using light-sheet microscopy and two-photon fluorescence microscopy. Leonardo Sacconi received his master’s degree in physics in 2001 at the University of Florence. He then moved to Trento, where he obtained his PhD in physics. He was a visiting scientist at Cornell University, Ithaca, New York, in the group of Watt Webb. From 2005 to 2011, he started new research lines on functional imaging of neuronal networks in vivo. He is now a researcher at the National Institute of Optics in Florence. Francesco Saverio Pavone is developing new microscopy techniques for high-resolution and high-sensitivity imaging and for laser manipulation purposes. These techniques have been applied to single-molecule biophysics, single-cell imaging, and optical manipulation. Tissue imaging is another research area developed, where nonlinear optical techniques have been applied for skin and neural tissue imaging also in vivo. Currently, he is the director of the European Laboratory for Non-Linear Spectroscopy in Florence. |