|
|
1.IntroductionCancer has been extensively studied to unveil its causes and to develop therapeutic tools for cure and prevention.1,2 There are multitudes of factors of its occurrence but the major, known, factors lie in the genetic mutations, which lead to aberrant signaling pathways, causing these cells to become malignant.2 Most studies have focused on cancer genomics to unravel malignant transformation and cancer invasion; nonetheless, it has emerged that genetics and molecular biology alone have not been able to address the problem completely.3–5 Therefore, efforts have been made to approach cancer from a mechanical point of view, which correlates biological processes with cell mechanics.6–12 Cells constantly sense mechanical forces from their microenvironment [extracellular matrix (ECM) and neighboring cells] that play a pivotal role in cell functions.7,13 Mechanical properties of cancer microenvironment are crucial, as they may have a role in cancer initiation, promotion, or they may cause the cancer to revert back.14,15 At the very early stage, when a cell turns into cancerous, a very strong interplay between microenvironment and the cell gets initiated, leading to cell cytoskeleton alterations, ECM stiffening, tumor progression, and metastasis.16,17 In the past decade, the mechanical interaction of a cell with its microenvironment has garnered much attention and witnessed considerable progress.18 The microenvironment is the driving element in cell proliferation,19 viscoelasticity,4,7,20 functionality,21 and cell signaling.22,23 The mechanical interaction of the cell with the surrounding microenvironment is a blend of cell–matrix and cell–cell interactions. Both of these processes have prime importance for the fundamental understanding of metastasis and cell behavior in malignant conditions. This article is focused on cell–cell interaction dynamics for breast cancer cells. Cell–cell interactions are complex and include cell–cell interactions in normal tissues, primary tumor, interactions during transit at the metastasis stage, and secondary tumor site. The cell–cell mechanical interaction has variant spatial and temporal tendencies and the precise knowledge of these interactions is still lacking. Initiation, detachment, and organ-specific affinity of cancer cells to host cells in terms of mechanical interaction can reveal deeper understanding of cancer progression and metastasis. Many studies have highlighted cell–cell interactions in terms of adhesion and other chemophysical properties, but few studies have been devoted to their mechanistic nature. The interaction between MDA-MB-231 cells and noncancerous Madin-Darby canine kidney (MDCK) epithelial cells has been studied in time-lapse three-dimensional (3-D) configuration by Ivers et al.,24 demonstrating that microenvironment has a strong impact on the growth and dynamics of cancer cells by stressing the organ-specific affinity of cancer cells to host cells. Using the optical tweezers (OT) pulling method, Hu et al.25 categorized stroma–cell interactions as tightly adherent, loosely adherent, and freely suspended. They also observed dynamic signaling pathways during cell–cell interactions coupling OT to fluorescence microscopy.26 The effect of neighboring cells on cell stiffness was recently investigated for human mammary epithelial cells, representing four different phases of cancer progression, by means of atomic force microscopy (AFM).27 This work reports that cell–cell contact strongly influences the cell stiffness of normal cells, while it has little effect on the cancer cells. In another recent study, Kamińska et al.28 have reported a body of evidence indicating that the interaction between normal and neoplastic cell contributes to tumor growth. In this study, we employed OT to measure cell stiffness by using a similar approach to that of AFM cell indentation. AFM and OT indentation regimes (force and loading rate) are very different, making these techniques complementary. As shown in some recent papers,29,30 the use of both OT and AFM enables the separation of the viscous and elastic components and provides a more robust method to investigate cell stiffness. We investigated the cell stiffness of three types of human breast cells (non-neoplastic, tumorigenic, and metastatic) in isolated and cell–cell contact conditions. The key results of our work are as follows:
2.Materials and Methods2.1.Cells Sample PreparationMDA-MB-231, MCF-7, and HBL-100 cell lines (ATCC numbers HTB-26, HTB-22, and HTB-124, respectively) were cultured in adhesion using low glucose Dulbecco’s modified Eagle medium with l-glutamine (MDA-MB-231 and MCF-7) or Roswell Park Memorial Institute 1640 medium with l-glutamine (HBL-100), supplemented with 10% (v/v) fetal calf serum, of penicillin–streptomycin, and 1 mM gentamycin. Cell cultures were maintained in flasks at 37°C in 5% . Cell splitting was performed every 2 to 3 days, as soon as the cultures reached confluence, using diluted 0.05% trypsin–EDTA. The day before the experiments, the cells were seeded overnight on 18-mm glass coverslips at a density of and in 2 ml of medium. Before starting the experiment, cells were washed three times in phosphate-buffered saline (PBS) and rinsed with medium before every measurement session. All reagents for cell culture were purchased from Gibco Lifetechnology; cell culture flasks and petri dishes were purchased from Sigma-Aldrich.32 2.2.Optical Tweezers IndentationWe employed a modified Thorlabs OKBT modular setup for force measurement by the vertical cell indentation technique, previously described by Yousafzai et al.33 Briefly, an infrared (IR) laser (single-mode Yb fiber laser YLM-5, 1064 nm, max 5 W, IPG Photonics GmbH) is focused into the sample chamber through Nikon lens [NA 1.25 oil immersion, working distance (WD) 0.3] to produce a stable trap (Fig. 1). During force measurements, the cells cultured on a coverslip are kept at physiological temperature (37°C) using a temperature controller unit (RS components). The sample is imaged through CMOS camera (Thorlabs-DCC1240C). The trapped bead is positioned above the cell using a nanopiezo cube, PS, (Thorlabs, NanoMax 3-axis flexure stage), allowing to control the sample displacement with resolution. The position of the trapped bead is monitored using back focal plane interferometry34 of the IR beam scattered by the trapped bead and collected by a second microscope lens (Nikon , NA 0.25, WD 7). The interference pattern is imaged by an additional convergent lens onto the quadrant photo detector (Thorlabs, PDQ80A, detector size 7.8 mm), which senses the lateral and axial displacement of the trapped bead. A laser power of 500 mW was used, which corresponds to 25 mW at the sample plane and was shown to be not damaging the cells and provide stable trap with a stiffness of . The cells were indented vertically by moving the cell holder, coupled with PS, against the trapped bead by a sinusoid signal (amplitude and one period with frequency of 0.2 Hz) as shown in Fig. 1, and the displacement of the bead in the trap was acquired at a 10-KHz sampling frequency. Fig. 1(a) OT indentation and force measurement setup. IR trapping laser path (solid line) and bright-field imaging path (dashed line). QPD, quadrant photodiode; PS, 3-axis nanopiezo stage; TL, tube lens; L1 to L3, convergent lenses; M1 to M3, mirrors; and DCM, 1-2 dichroic mirrors. Inset: The cell is positioned below the trapped bead and the stage is moved up by stage displacement (SD). The cell interacts with the bead displacing it by bead displacement (BD) and causing the cell indentation. (b) Linearized force–indentation plot for elastic modulus calculation. Inset: By subtracting BD from SD, indentation is obtained. 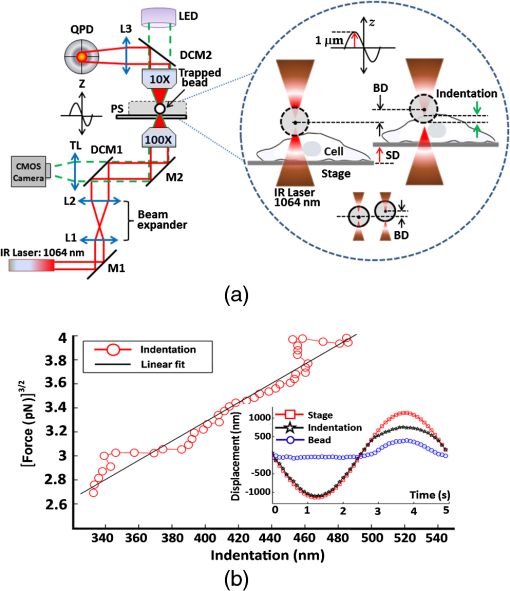 2.3.Cell Site Selection and Cell NeighboringCells are indented at three locations, at the center above the nucleus (L1), out of the cell nucleus (L2), and near the leading edge (L3), as shown in Fig. 2. Each location was indented at least two times and the results were averaged for each location. Fig. 2Differential interference contrast (DIC) image of MDA-MB-231 cells cultured on glass substrate. Cell is categorized as (a) isolated cell: when there is no interaction with any other cell and (b) connected cell: when the cell interacts with two or more cells. Each cell is indented at three locations: L1—above nucleus, L2—intermediate position (cytoplasm), and L3—near the leading edge (a). Scale bars are . 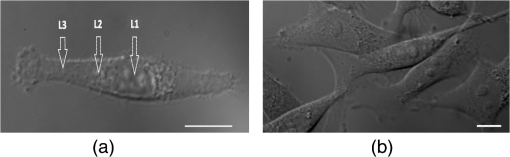 All three cell lines were cultured on the same type of glass coverslips, which act as a hard substrate to the cells (approximately 70 GPa of stiffness). Since the cell senses its surrounding, to understand the effects of neighboring cells on the mechanical properties of the cell under study, the cells were cultured with varying densities (Sec. 2.1) to get cell–cell contact [Fig. 2(b)]. An isolated cell is not touching any other cell, while a connected cell has at least two connecting sites with the neighboring cells. The surrounding cells act as mechanical cage to the cell under study. 2.4.Cell Stiffness CalculationWe investigated the elastic modulus of the three cell lines at three different locations of the cell (Sec. 2.3) using OT vertical indentation having a trap stiffness of . We used silica beads as the handle to indent cells by moving the stage (cell) against the bead in a sinusoidal ( and ) form and the corresponding loading rate was . The elastic modulus was calculated using the Hertz model.29 The apparent elastic modulus, , is where is the bead radius, is the force, () is the Poisson ratio, and Id is the indentation, calculated as (see inset Fig. 1(b)) The elastic modulus formula [Eq. (1)] can be expressed as a function of indentation, Id, as Linearizing the preceding equation in terms of Id leads to where is a constant including all the other constants in Eq. (3). By fitting linearly the curve, one can easily determine the value of the elastic modulus .2.5.Immunostaining and Confocal Microscopy ImagingCells were fixed in 4% paraformaldehyde containing 0.15% picric acid in PBS, saturated with 0.1 M glycine, permeabilized with 0.1% Triton X-100, saturated with 0.5% BSA in PBS (all from Sigma-Aldrich, St. Louis, Missouri) and then incubated for 40 min with Alexa Fluor 488 phalloidin (Invitrogen, Life Technologies, Gaithersburg, Maryland). Cells were further washed, before mounting in Vectashield (VECTOR Laboratories, Burlingame, California) incubated with 4′,6-diamidino-2-phenylindole () in PBS for 5 min. All the incubations were performed at room temperature (20°C to 22°C). Cells were examined using a Leica DMIRE2 confocal microscope (Leica Microsystems GmbH, Germany) equipped with DIC and fluorescence optics, diode laser 405 nm, Ar/ArKr 488 nm, and He/Ne lasers. The fluorescence images () were collected with a and 1.3 NA oil immersion objective. 3.Results3.1.Stiffness at Different Subcellular RegionsA wide range of approaches like AFM, optical starching, and micropipette aspiration have been used to study viscoelastic properties of cellular subregions.27,35–37 We used OT for the first time to study cell stiffness at three different cell locations. All three cell lines in isolated condition are indented at the three subregions as indicated in Fig. 2(a). The results [Fig. 3(a) and Table 1] show a descending trend in elastic modulus from nuclear region toward the leading edge. All three cell lines exhibit higher stiffness at the center (L1) than the intermediate position (L2) yet having higher value than position L3 near the leading edge. The elastic modulus values at the three locations are statistically significant and in agreement with studies carried out by other groups using different biophysical methods.27,36,37 Fig. 3(a) Isolated cells: all the three cell lines have descending character for stiffness, higher at the center and decreasing toward the leading edge. (b) Connected cells: stiffness decreases from nuclear region to the leading edge. Cell lines exhibit the same stiffness in the cytoplasm and leading edge indicating that cell cytoskeleton organization during interactions becomes more dynamic. MDA-MB-231 shows the same stiffness as HBL-100. (c) Elastic moduli variation in isolated (I) and connected (C) conditions in nuclear (L1), cytoplasm (L2), and leading edge (L3) regions. MDA-MB-231 exhibits a more prominent variation in connected conditions as compared to HBL-100 and MFC-7. The symbols represent H: HBL-100 and M: MCF-7. (-test: *, **, and ***). 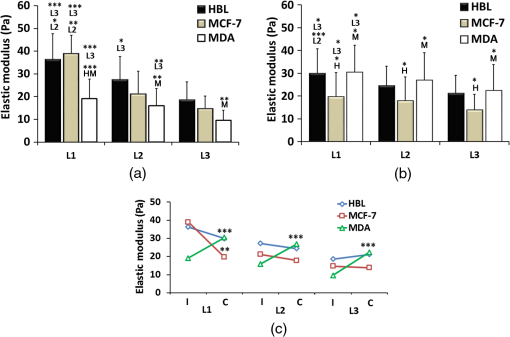 Table 1Elastic modulus measurements (±standard error of the mean) for all three cell lines measured in isolated and connected conditions in three different cellular subregions (L1, L2, and L3).
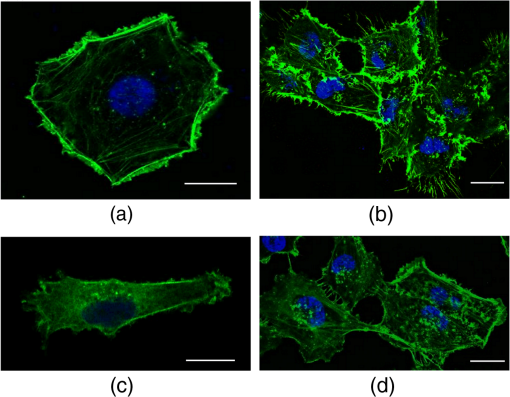
Stiffness values for single cells indented at the center (L1) show that the most aggressive cells (MDA-MB-231) are less stiff as compared to HBL-100 and MCF-7. These values agree with other results reported in the literature, showing that metastatic cells are softer as compared to their noninvasive counter parts.4,18,27,35,38 The HBL-100 and MCF-7 cells are twofold stiffer than MDA-MB-231 at the nuclear region (L1) and can be used to distinguish between cells with different aggressiveness levels. These results also act as first test for the feasibility of our successive experiments on cell stiffness as a function of cell subregions and of cell–cell interaction. When comparing different cellular subregions, for HBL-100 cells, the stiffness at the nuclear region is 25% higher than that corresponding to the cytoplasm region and 47% higher than that corresponding to the leading edge. For MCF-7 cells, the stiffness decreases as 46% and 59%, whereas for MDA-MB-231 it diminishes by 16% and 47%, respectively, for cytoplasm and the leading edge. All three cell lines have significantly different stiffness values at the nuclear region and the leading edge, showing a larger contribution of the nucleus toward mechanical architecture.37 Our results stress that for single-cell force measurement, the region of interest affects mechanical observation and hence should be considered for mechanical studies of the cells. 3.2.Cell–Cell Contact Alters Cell StiffnessTo investigate the stiffness variation during interactions with neighboring cells, we analyzed cells from three different cell lines in contact conditions [Fig. 2(b)]. Prominent variation in stiffness was observed as summarized in Table 1 and shown in Fig. 3(b). Analyzing the cellular regional variation, one can note that the cell stiffness of HBL-100 and MCF-7 cells decreases by (17% and 10%) and (30% and 30%), while for MDA-MB-231 it decreases by 13% and 26% at the cytoplasm and leading edge regions as compared with the nuclear region. Also, in contact conditions, cell stiffness at the leading edge has a significantly lower value than at the nuclear region. However, when compared to the isolated condition, one observes that the stiffness of the MD-MB-231 cells in connected condition increases and becomes comparable with that of the HBL-100 cells for all the three locations [Fig. 3(b)]. This behavior is opposite to the behavior of the other two cell lines. Comparing the stiffness values corresponding to regional locations in isolated and connected conditions, we find a decreasing trend for HBL-100 and MCF-7, whereas MDA-MB-231 stiffness is increased, as shown in Fig. 3. For HBL-100 and MCF-7, stiffness at nuclear region decreases by 17% and 49%, respectively. MDA-MB-231 responds differently to cell neighboring and becomes stiffer in contact condition. Stiffness increases by 39%, 41%, and 57% in L1, L2, and L3 locations, respectively. The higher increment at the leading edge in contact condition suggests that MD-MB-231 orchestrates more cytoskeletal components at the leading edge and applies greater force on the membrane. HBL-100 and MDA-MB-321 cells do not express E-cadherins, which are involved in cell–cell tight junctions, whereas MCF-7 does express them; hence, they experience different interaction mechanisms.27,35 Furthermore, HBL-100 (nontumorigenic) and MCF-7 (tumorigenic) are noninvasive, but MDA-MB-321 is invasive; therefore, their response to the interacting cells might be different. Lee et al.35 have shown that MDA-MB-231 cells are more mobile in the active interaction environment of MCF-10A (nontumorigenic epithelial cell line and normal counterpart of MCF-7) monolayer as compared to noninvasive MCF-7 cells. They showed that invasive cells (MDA-MB-321) are softer and more sensitive to physical forces, if applied on the lateral surfaces of the cells. Our results show that MDA-MB-231 cells, though softer in isolated conditions, feel stressed in interacting environment whereas the MCF-7 and HBL-100 turns softer during cell–cell interactions, in agreement with the results reported by Kristal-Muscal et al.38 They demonstrated that the cells with metastatic potential (MDA-MD-231) generate greater forces despite their soft character as compared to low metastatic potential cell (MDA-MB-468) and benign cells (MCF-10A), which do not indent the substrate at all.33 4.DiscussionCancer cell metastasis formation is a complicated mechanism. The evidence indicates that in the cancer microenvironment, ECM becomes stiffer as compared to its normal state, but cells behave softer39 and exhibit increased actomyosin cortex contractility.33,34 Furthermore, some studies show that by altering the mechanics of cell microenvironment, cancerous cells may act as normal ones.14,15 Interesting results on the effect of cell–cell contact on cell motility and cell adhesion have been reported for MDA-MB-231 cells.35 These results indicate that the motility of MDA-MB-231 cells, which are mechanically soft, is dramatically enhanced when they are surrounded by breast epithelial MCF-10A cells, which are mechanically stiff, compared with single-cell motility within a cluster of MDA-MB-231 cells (high density) or single MDA-MB-231 cells (low density) in the absence of non-neoplastic MCF-10A cells. Another interesting result is the cell-substrate contact/interaction effect on the cell’s ability to apply forces,38 showing that cells of higher metastatic potential develop stronger forces on gels stiff enough to provide grip handles yet soft enough to indent, whereas benign cells did not indent substrates at all. Our work was focused on cell stiffness as a marker to distinguish between cells at different stages of cancer progression. It is known that the values obtained for the cell stiffness might vary depending on the measurement method; therefore, employing complementary techniques (e.g., OT and AFM) increases the reliability.30 Using OT, we show that for the isolated cell condition, the metastatic cells are softer than tumorigenic and non-neoplastic cells and cell stiffness decreases from the nuclear region toward the cell edge for all the three cell models. The results are confirmed by measurements using peak force AFM in our recent work30 and in that of another group.31 These results are in line with the general observation that malignant cells are softer than normal cells across a range of different cancers.13 However, our results appear partially in contradiction with those obtained by Guo et al.27 Using AFM to measure cell stiffness, they report that metastatic cells are slightly stiffer than tumorigenic cells and only minor or negligible differences in stiffness differentiate the cellular regions. In our opinion, the difference might be attributed to the differences between the cell models (only the metastatic cells MDA-MB-231 are identic) and the difference between the indentation regimes (indentation force, indentation range, and loading rate). Moreover, the experiments by Guo et al. were performed at room temperature, while ours were conducted at the physiological temperature (37°C). It is well known that temperature strongly influences cell health, behavior, and properties (mechanical properties included). We found that for the metastatic cells MDA-MB-231 cell neighboring induces a significant increase in stiffness. This trend is confirmed for all three locations of measurements, above the nucleus, the cytoplasm, or near the cell edge. The cell stiffness for the other two cell lines (HBL-100 and MCF-7) decreases significantly for the nuclear region and shows minor changes for the other two regions. To our knowledge, this result points to the importance of cell environment when defining cell stiffness. The result is partially in contradiction with that showing the microenvironment condition has little or no effect on immortal and metastatic cells reported by Guo et al., probably due to the same aforementioned factors for the cell stiffness variation with the cell regions. Trying to associate the changes of cell stiffness with the cytoskeleton rearrangements, we analyzed F-actin organization in isolated and connected cells. Some sample images using immunofluorescence confocal microscopy are shown in Fig. 4 for HBL-100 and MDA-MB-231 cells. While for the HBL-100 cells, we have not observed a significant difference between the actin fibers organization in isolated and connected cells, the actin stress fibers seem to be enforced for the connected MDA-MB-231 cells with respect to the isolated cells. This result has to be further confirmed by a more thorough analysis of the indentation regions using higher spatial resolution or combined techniques. In fact, combining immunofluorescence confocal microscopy and peak-force modulated AFM, Calzado-Martin et al. recently showed that actin stress fibers are present at apical regions of healthy cells, while in tumorigenic cells they appear only at basal regions, where they cannot contribute to stiffness as probed by AFM. These results substantiate that actin stress fibers provide a dominant contribution to stiffness in healthy cells, while the elasticity of tumorigenic cells appears not predominantly determined by these structures. The combination of these two techniques could be applied in the future to investigate also the differences in actin fibers organization for isolated and connected cells. 5.ConclusionsWe report an OT approach to investigate on the change of the cell stiffness determined by cell neighboring, and we show that the metastatic cells are those which are influenced the most. By indenting cells axially on the top, we demonstrate that stiffness measurements are sensitive to the cellular subregions as well as the interacting microenvironment. Altogether, our results point to the importance of cell microenvironment when defining cell stiffness. In future studies, we will apply this technique also to investigate effect of ECM on cell–cell interaction and to study the related mechanotransduction pathways. More rigorous mechanical studies on cancer cells corresponding to different range of forces and different microenvironmental conditions may further unravel aspects of cancer progression and metastasis formation. Models of cell cytoplasm behavior and reorganization, like the poroelastic material model,40 will help a better understanding of the relation between the cytoskeleton structure and cell stiffness. Cells in 3-D rather than 2-D cell neighboring would be very interesting to be investigated and probably the results will show more variations in terms of cell stiffness. Engineering 3-D cell structures is already a reality,41 but measuring cell stiffness is unfortunately still impossible with OT or AFM in these conditions. Different probing approaches should be developed, which represents the challenge for the future. AcknowledgmentsWe would like to gratefully acknowledge the Applied Physics Section of the International Centre for Theoretical Physics, Trieste, for partial financial support to build the OT setup, and partial financial support by the European Research Council (ERC) Ideas Program through a senior grant to Giacinto Scoles, titled Molecular Nanotechnology for Life Science Applications: Quantitative Interactomics for Diagnostics, Proteomics and Quantitative Oncology (MONALISA QUIDPROQUO) Grant No. 269051. ReferencesD. Hanahan and R. A. Weinberg,
“The hallmarks of cancer,”
Cell, 100
(1), 57
–70
(2000). http://dx.doi.org/10.1016/S0092-8674(00)81683-9 CELLB5 0092-8674 Google Scholar
D. Hanahan and R. A. Weinberg,
“Hallmarks of cancer: the next generation,”
Cell, 144
(5), 646
–674
(2011). http://dx.doi.org/10.1016/j.cell.2011.02.013 CELLB5 0092-8674 Google Scholar
R. Siegel et al.,
“Cancer statistics, 2014,”
CA: Cancer J. Clin., 64
(1), 9
–29
(2014). http://dx.doi.org/10.1017/S0009840X13002084 Google Scholar
S. Suresh,
“Biomechanics and biophysics of cancer cells,”
Acta Mater., 55
(12), 3989
–4014
(2007). http://dx.doi.org/10.1016/j.actamat.2007.04.022 ACMAFD 1359-6454 Google Scholar
G. P. Gupta and J. Massagué,
“Cancer metastasis: building a framework,”
Cell, 127
(4), 679
–695
(2006). http://dx.doi.org/10.1016/j.cell.2006.11.001 CELLB5 0092-8674 Google Scholar
H. Huang et al.,
“Cell stiffness and receptors: evidence for cytoskeletal subnetworks,”
Am. J. Physiol. Cell Physiol., 288
(1), C72
–C80
(2005). http://dx.doi.org/10.1152/ajpcell.00056.2004 Google Scholar
I. Schoen, B. L. Pruitt and V. Vogel,
“The Yin-Yang of rigidity sensing: how forces and mechanical properties regulate the cellular response to materials,”
Annu. Rev. Mater. Res., 43 589
–618
(2013). http://dx.doi.org/10.1146/annurev-matsci-062910-100407 ARMRCU 1531-7331 Google Scholar
Q. Wen and P. A. Janmey,
“Effects of non-linearity on cell–ECM interactions,”
Exp. Cell. Res., 319
(16), 2481
–2489
(2013). http://dx.doi.org/10.1016/j.yexcr.2013.05.017 ECREAL 0014-4827 Google Scholar
T. R. Geiger and D. S. Peeper,
“Metastasis mechanisms,”
BBA, 1796
(2), 293
–308
(2009). http://dx.doi.org/10.1016/j.bbcan.2009.07.006 Google Scholar
M. Makale,
“Cellular mechanobiology and cancer metastasis,”
Birth Defects Res. Part C, 81
(4), 329
–343
(2007). http://dx.doi.org/10.1002/(ISSN)1542-9768 Google Scholar
N. Guz et al.,
“If cell mechanics can be described by elastic modulus: study of different models and probes used in indentation experiments,”
Biophys. J., 107
(3), 564
–575
(2014). http://dx.doi.org/10.1016/j.bpj.2014.06.033 BIOJAU 0006-3495 Google Scholar
P. Schedin and A. Elias,
“Multistep tumorigenesis and the microenvironment,”
Breast Cancer Res., 6
(2), 93
–101
(2004). http://dx.doi.org/10.1186/bcr772 BCTRD6 Google Scholar
C. T. Mierke,
“The fundamental role of mechanical properties in the progression of cancer disease and inflammation,”
Rep. Prog. Phys., 77
(7), 076602
(2014). http://dx.doi.org/10.1088/0034-4885/77/7/076602 RPPHAG 0034-4885 Google Scholar
M. J. Bissell and W. C. Hines,
“Why don’t we get more cancer? A proposed role of the microenvironment in restraining cancer progression,”
Nat. Med., 17
(3), 320
–329
(2011). http://dx.doi.org/10.1038/nm.2328 1078-8956 Google Scholar
D. E. Ingber,
“Can cancer be reversed by engineering the tumor microenvironment?,”
Semin. Cancer Biol., 18
(5), 356
–364
(2008). http://dx.doi.org/10.1016/j.semcancer.2008.03.016 SECBE7 1044-579X Google Scholar
D. Wirtz, K. Konstantopoulos and P. C. Searson,
“The physics of cancer: the role of physical interactions and mechanical forces in metastasis,”
Nat. Rev. Cancer, 11
(7), 512
–522
(2011). http://dx.doi.org/10.1038/nrc3080 NRCAC4 1474-175X Google Scholar
T. Risler,
“Focus on the physics of cancer,”
New J. Phys., 17
(5), 055011
(2015). http://dx.doi.org/10.1088/1367-2630/17/5/055011 NJOPFM 1367-2630 Google Scholar
B. D. Hoffman and J. C. Crocker,
“Cell mechanics: dissecting the physical responses of cells to force,”
Annu. Rev. Biomed. Eng., 11 259
–288
(2009). http://dx.doi.org/10.1146/annurev.bioeng.10.061807.160511 ARBEF7 1523-9829 Google Scholar
G. Cheng et al.,
“Micro-environmental mechanical stress controls tumor spheroid size and morphology by suppressing proliferation and inducing apoptosis in cancer cells,”
PLoS One, 4
(2), e4632
(2009). http://dx.doi.org/10.1371/journal.pone.0004632 POLNCL 1932-6203 Google Scholar
B. Ladoux and A. Nicolas,
“Physically based principles of cell adhesion mechanosensitivity in tissues,”
Rep. Prog. Phys., 75
(11), 116601
(2012). http://dx.doi.org/10.1088/0034-4885/75/11/116601 RPPHAG 0034-4885 Google Scholar
J. Fu et al.,
“Mechanical regulation of cell function with geometrically modulated elastomeric substrates,”
Nat. Methods, 7
(9), 733
–736
(2010). http://dx.doi.org/10.1038/nmeth.1487 1548-7091 Google Scholar
R. S. Gieni and M. J. Hendzel,
“Mechanotransduction from the ECM to the genome: are the pieces now in place?,”
J. Cell. Biochem., 104
(6), 1964
–1987
(2008). http://dx.doi.org/10.1002/jcb.21364 JCEBD5 0730-2312 Google Scholar
D. E. Jaalouk and J. Lammerding,
“Mechanotransduction gone awry,”
Nat. Rev. Mol. Cell Biol., 10
(1), 63
–73
(2009). http://dx.doi.org/10.1038/nrm2597 NRMCBP 1471-0072 Google Scholar
L. P. Ivers et al.,
“Dynamic and influential interaction of cancer cells with normal epithelial cells in 3D culture,”
Cancer Cell Int., 14
(1), 108
(2014). http://dx.doi.org/10.1186/s12935-014-0108-6 Google Scholar
S. Hu et al.,
“Manipulating cell adhesions with optical tweezers for study of cell-to-cell interactions,”
J. Biomed. Nanotechnol., 9
(2), 281
–285
(2013). http://dx.doi.org/10.1166/jbn.2013.1528 Google Scholar
X. Gou et al.,
“Applying combined optical tweezers and fluorescence microscopy technologies to manipulate cell adhesions for cell-to-cell interaction study,”
IEEE Trans. Biomed. Eng., 60
(8), 2308
–2315
(2013). http://dx.doi.org/10.1109/TBME.2013.2255287 IEBEAX 0018-9294 Google Scholar
X. Guo et al.,
“The effect of neighboring cells on the stiffness of cancerous and non-cancerous human mammary epithelial cells,”
New J. Phys., 16
(10), 105002
(2014). http://dx.doi.org/10.1088/1367-2630/16/10/105002 NJOPFM 1367-2630 Google Scholar
K. Kamińska et al.,
“The role of the cell–cell interactions in cancer progression,”
J. Cell. Mol. Med., 19
(2), 283
–296
(2015). http://dx.doi.org/10.1111/jcmm.2015.19.issue-2 Google Scholar
S. Nawaz et al.,
“Cell visco-elasticity measured with AFM and optical trapping at sub-micrometer deformations,”
PLoS One, 7
(9), e45297
(2012). http://dx.doi.org/10.1371/journal.pone.0045297 POLNCL 1932-6203 Google Scholar
G. Coceano et al.,
“Investigation into local cell mechanics by atomic force microscopy mapping and optical tweezer vertical indentation,”
Nanotechnology, 27
(6), 065102
(2015). http://dx.doi.org/10.1088/0957-4484/27/6/065102 NNOTER 0957-4484 Google Scholar
A. Calzado-Martin et al.,
“Effect of actin organization on the stiffness of living breast cancer cells revealed by peak-force modulation atomic force microscopy,”
ACS Nano, 10
(3), 3365
–3374
(2016). http://dx.doi.org/10.1021/acsnano.5b07162 ANCAC3 1936-0851 Google Scholar
C. C. De Fromentel et al.,
“Epithelial HBL-100 cell line derived from milk of an apparently healthy woman harbours SV40 genetic information,”
Exp. Cell Res., 160
(1), 83
–94
(1985). http://dx.doi.org/10.1016/0014-4827(85)90238-1 ECREAL 0014-4827 Google Scholar
M. S. Yousafzai et al.,
“Substrate-dependent cell elasticity measured by optical tweezers indentation,”
Opt. Lasers Eng., 76 27
–33
(2016). http://dx.doi.org/10.1016/j.optlaseng.2015.02.008 OLENDN 0143-8166 Google Scholar
F. Gittes and C. F. Schmidt,
“Interference model for back-focal-plane displacement detection in optical tweezers,”
Opt. Lett., 23
(1), 7
–9
(1998). http://dx.doi.org/10.1364/OL.23.000007 OPLEDP 0146-9592 Google Scholar
M.-H. Lee et al.,
“Mismatch in mechanical and adhesive properties induces pulsating cancer cell migration in epithelial monolayer,”
Biophys. J., 102
(12), 2731
–2741
(2012). http://dx.doi.org/10.1016/j.bpj.2012.05.005 BIOJAU 0006-3495 Google Scholar
J. Guck et al.,
“Optical deformability as an inherent cell marker for testing malignant transformation and metastatic competence,”
Biophys. J., 88
(5), 3689
–3698
(2005). http://dx.doi.org/10.1529/biophysj.104.045476 BIOJAU 0006-3495 Google Scholar
F. Guilak, J. R. Tedrow and R. Burgkart,
“Viscoelastic properties of the cell nucleus,”
Biochem. Biophys. Res. Commun., 269
(3), 781
–786
(2000). http://dx.doi.org/10.1006/bbrc.2000.2360 BBRCA9 0006-291X Google Scholar
R. Kristal-Muscal, L. Dvir and D. Weihs,
“Metastatic cancer cells tenaciously indent impenetrable, soft substrates,”
New J. Phys., 15
(3), 035022
(2013). http://dx.doi.org/10.1088/1367-2630/15/3/035022 NJOPFM 1367-2630 Google Scholar
P. Katira, R. T. Bonnecaze and M. H. Zaman,
“Modeling the mechanics of cancer: effect of changes in cellular and extra-cellular mechanical properties,”
Front. Oncol., 3 145
(2013). http://dx.doi.org/10.3389/fonc.2013.00145 Google Scholar
E. Moeendarbary et al.,
“The cytoplasm of living cells behaves as a poroelastic material,”
Nat. Mater., 12
(3), 253
–261
(2013). http://dx.doi.org/10.1038/nmat3517 NMAACR 1476-1122 Google Scholar
J. W. Haycock,
“3D cell culture: a review of current approaches and techniques,”
3D Cell Culture: Methods and Protocols, Methods in Molecular Biology, 1
–15 Humana Press, New York
(2011). Google Scholar
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||