|
|
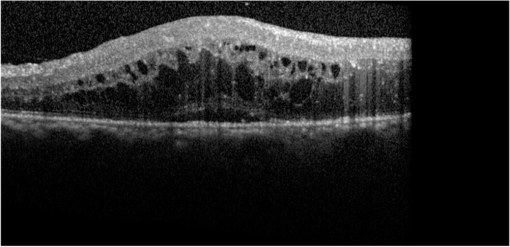
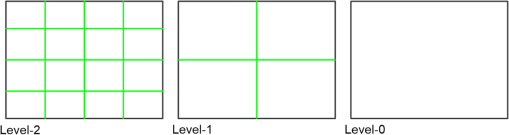
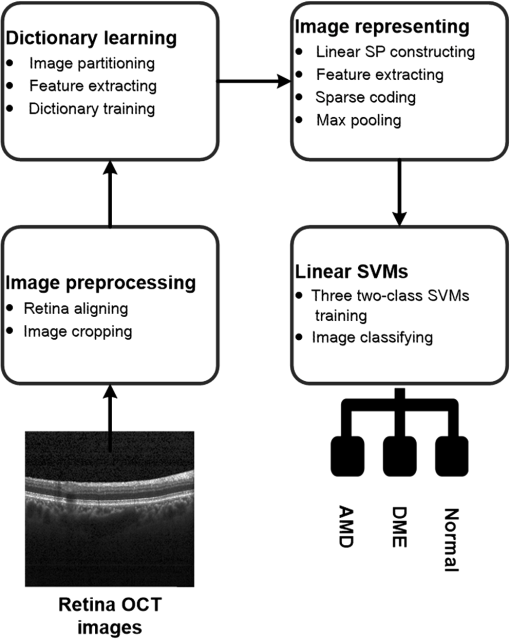
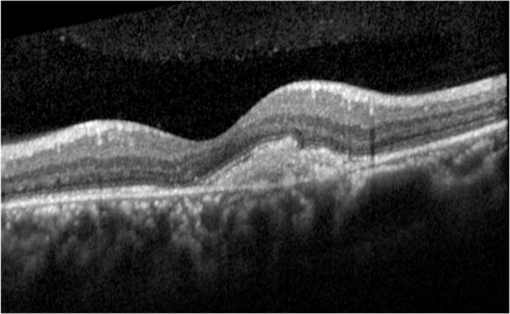
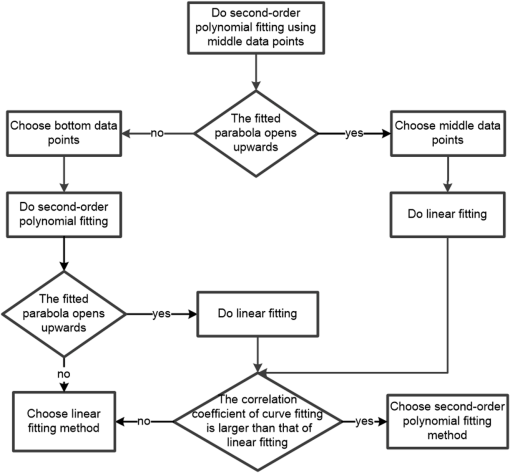
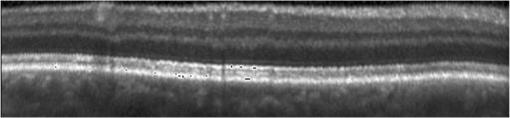
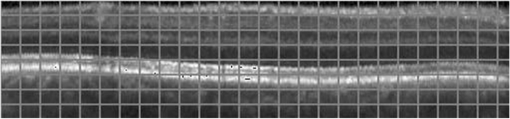
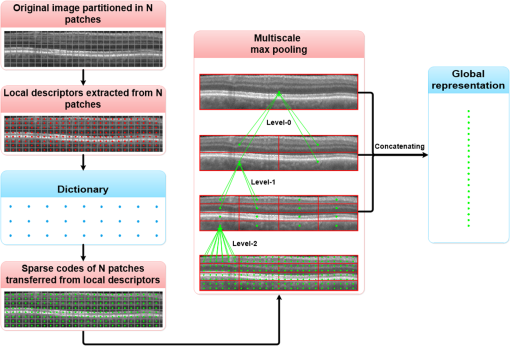
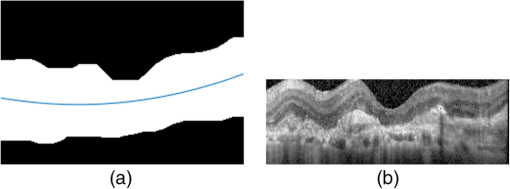
1.IntroductionOptical coherence tomography (OCT) has been widely adopted in ophthalmology as a clinical aid for identifying the presence of various ocular pathologies and their progression.1 The ability to visualize the internal structure of a retina makes it possible to diagnose diseases such as age-related macular degeneration (AMD) and diabetic macular edema (DME) (the leading cause of blindness of the elderly2 and the most common cause of irreversible vision loss in individuals with diabetes, respectively). Over the past two decades in the field of OCT image interpretation, a majority of the previous works on image processing and computer vision have been dedicated to methods of retinal layer segmentation,3–18 which we do not discuss in this paper. Many papers have also investigated OCT image classification.19–23 In addition, more methods have recently been proposed to address the problems about OCT image classification between patients without retinal pathologies and patients with retinal pathologies (especially AMD and DME). In 2011, Liu et al.24 proposed a methodology for detecting macular pathologies (including AMD and DME) in foveal slices of OCT images, in which they used local binary patterns and represented images using a multiscale spatial pyramid (SP) followed by a principal component analysis for dimension reduction. In 2012, Zheng et al.25 and Hijazi et al.26 proposed a method for representing images based on the graph. First, they decomposed images into a quad-tree collection; next, they employed the subgraph mining technology to analyze these quad-trees, and having the ability to distinguish subgraphs, they selected common subgraphs to generate the global vector for each image. Then they trained the classifier with the feature vectors. Finally, a binary classification over normal and AMD OCT images was performed. In 2014, Srinivasan et al.27 proposed a detection method to distinguish normal OCT volumes from DME and AMD volumes. In their work, a histogram of oriented gradients (HOGs) was extracted for each slice of a volume and fed to a linear support vector machine (SVM). All the aforementioned approaches are methods for classifying two-dimensional (2-D) OCT images, although the method in Ref. 27 could classify three-dimensional (3-D) OCT retina images. However, Albarrak et al.28 proposed a method that could directly deal with a 3-D OCT retinal image by first, decomposing the original 3-D OCT volume image into subvolume images and representing them with a tree structure; then representing the tree with high-frequency occurrence subgraphs obtained by using the subgraph mining technology; next, extracting the features of the subgraphs and concatenating them as the representation of the entire volume; finally, performing a binary classification with normal and AMD OCT images. In works that related to us,24,27 Srinivasan et al.’s preprocessing algorithm to align the retina region is suitable for the dataset that consists of OCT images with a clear or slightly distorted retinal pigment epithelium (RPE) layer [Fig. 1(a)] because the RPE layer estimation was used for alignment. Obviously, the RPE segmentation method is not accurate for estimating the RPE boundary with a severely distorted RPE layer as shown in Fig. 1(b). That is to say, the preprocessing algorithm in Ref. 27 is not suitable for more complex datasets that include OCT images of a retina with a severely distorted RPE layer. In the aspect of classification, the algorithm in Ref. 27 requires input cropped OCT images to have the same resolution, and it has a large memory requirement when the field of view, defined by the cropped area, is large. In Liu et al.’s work,24 the retina aligning method flattens the retina region by fitting to a parabola without segmenting the RPE layer, but our experiments show that this method does not deal well with some OCT images of retinas with severe diseases that make the lesion portion of the retina swell up as shown in Fig. 2. This means that in real applications the retina aligning algorithm in Ref. 24 could not perform well for complex datasets that include OCT images with aforementioned severe diseases, especially in a large amount. In addition, Liu et al.24 adopted a nonlinear SVM29 for classification, so that their algorithm had a computational complexity of in training and in testing, where is the training size, indicating that it is nontrivial to scale-up the algorithms to handle a large number of training images. Fig. 1OCT images of retina: (a) the one with clear RPE layer and (b) the one with a severely distorted RPE layer. 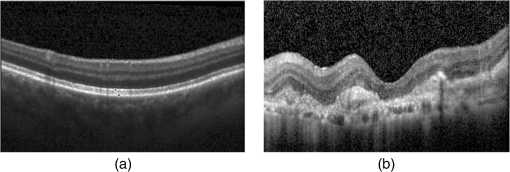 The rapid progress of image processing techniques brings an idea to the study of retinal OCT image processing and classification. To our knowledge, classification techniques based on image sparse representation are widely investigated and have been successfully applied in scene classification and face recognition fields. Linear SP matching (SPM) based on sparse coding (ScSPM)30 is a representative dictionary learning method for natural image classification. Its key idea is to replace -means vector quantization in Ref. 31 with sparse coding. In addition, the original SPM spatial pooling is obtained by calculating the histogram but ScSPM uses max pooling. The advantage of this method is the feasibility of employing a linear SVM model for classification, so that it not only has a better classification accuracy compared to the traditional method, but also remarkably reduces the complexity of SVMs to in training and a constant in testing, which provides a solution for large-scale image training and classifying tasks. In recent years, sparse representation techniques have been applied to OCT image processing, such as denoising and compression.32–34 Fang et al.32,33 applied the sparse representation to OCT image denoising and compressing. Kafieh et al.34 employed dual-tree complex wavelet transform instead of redundant discrete cosine transform as the initial dictionary to perform multiscale dictionary learning, whereby they proposed the complex wavelet dictionary learning method based on 2-D/3-D, which has been successfully applied in the course of OCT image noise reduction. To our knowledge, no works have been performed to use sparse coding and dictionary learning for detecting AMD and DME from OCT images. This paper proposes a general framework for distinguishing normal OCT images from DME and AMD scans based on sparse coding and dictionary learning. Here, a technique for preprocessing and alignment of a retina is proposed to address the deficiency of the previous methods, which cannot correctly classify a dataset that contains OCT images with a severely distorted retina region. Additionally, sparse coding and SP for preprocessing and an SVM for the classification are used to improve the automatic classification performance of the retina OCT images. The paper is organized as follows. Section 2 demonstrates our macular pathology classification method in detail. Section 3 presents our experimental results and analysis over two spectral domain OCT (SD-OCT) datasets, and Sec. 4 outlines conclusions. 2.ApproachSparse representation of signals has been widely investigated in recent years. Using an overcomplete dictionary that contains prototype signal atoms, signals are described by sparse linear combinations of these atoms. Here, the key problem is how to learn a dictionary to get the best sparse representation of signals. In practice, given a set of training signals, we search the dictionary that leads to the best representation for each member in this set under strict sparsity constraints. K-SVD is a representative dictionary learning method.35 It is an iterative method that alternates between sparse coding of examples based on the current dictionary and a process of updating the dictionary atoms to better fit the data. SPM is an SP image representation. We employ a three-level SPM in our work as shown in Fig. 3. The method partitions an image into blocks in three different scales (, 1, 2) and then computes the local image representation within each of the 21 blocks; finally, it concatenates all the local representations to form the vector representation of the image, i.e., the global representation of the image. ScSPM is an SPM based on sparse codes (SCs) of scale-invariant feature transform (SIFT) features.30 In this method, an image is partitioned into many patches; the SIFT feature descriptor of each patch is computed and its sparse representation is obtained for a learned dictionary; then the local representation of each block is obtained by max pooling all the SCs within the block. This representation is good enough for image classification by using a linear SVM classifier. In this paper, we use two techniques for image classification: SP with sparse coding as well as a multiclass linear SVM. Our classification approach consists of several steps that are illustrated in Fig. 4. First, we preprocess the OCT images to reduce morphological variations between OCT scans. Second, we fragment every cropped image in the training set into small patches and train a sparse dictionary with all SIFT descriptors extracted from the selected patches of the training images. Third, for each OCT image, we obtain its global representation by using an SP image representation, SCs, and max pooling. Fourth, three two-class linear SVMs are trained for image classification. 2.1.Image PreprocessingOCT images are usually rife with speckles and the position of the retina varies substantially among scans, which makes it nontrivial to align all the retina areas into a relatively unified position. Therefore, an aligning method for preprocessing is necessary. However, considering the previous works, there are three issues that we have taken into consideration. First, Srinivasan et al.27 aligned retina regions by fitting a second-order polynomial to the RPE layer and then flattened the retina. However, their method is invalidated once the RPE layer of the retina is too severely distorted to fit the curve of the RPE boundary of the retina [as depicted in Fig. 1(b)]. Second, Liu et al.24 aligned retina regions by fitting a second-order polynomial to the whole retina OCT image. Although their aligning method could deal with retina images that contain distorted RPE layers, the effectiveness of their method is seriously reduced when their algorithm is employed on a certain type of diseased retina where the lesion portion is swelling up as shown in Fig. 2. Third, there are many OCT images in our datasets where the RPE layer of the retina is straight but at a certain angle with the horizontal line as shown in Fig. 5. In that case, fitting the retina with a straight line might be better than with a parabola. To solve the three problems listed above and align retinas in a more robust way, we propose a fully automated aligning method possessing three salient characteristics. First, it perceives retinas without estimating their RPE boundary. Second, it extracts two sets of data points from a given image (for linear and curved fittings) and then automatically chooses one set of data points that is more representative of the retina morphology (in our paper, the two most common morphologies of retina are considered: curved and linear). Third, it automatically decides between a second-order polynomial and a straight line to fit the set of data points being chosen. Our aligning process is illustrated in Fig. 6, taking the most common case as an example (using one of the two sets of data points and the second-order polynomial fitting). Furthermore, in Sec. 3.1, we will present and discuss more cases that previous works24,27 have not taken into account. Fig. 6Description of the image aligning process: (a) original image, (b) BM3D denoising, (c) binarizing, (d) median filtering, (e) morphological closing, (f) morphological opening, (g) polynomial fitting, and (h) retina aligning. 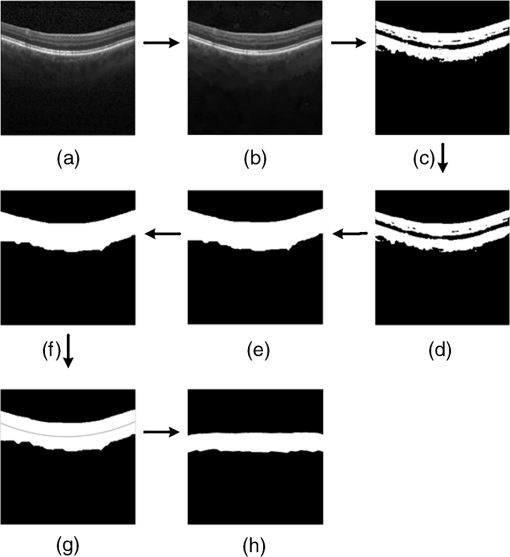 We separate our aligning method into three stages: perceiving stage, fitting stage, and normalizing stage. In the perceiving stage, our method detects the overall morphology of a retina in the following steps. First, the sparsity-based block matching and 3-D-filtering (BM3D) denoising method36 are used to reduce noises of the original image [Fig. 6(a)]. Second, the denoised image [Fig. 6(b)] is filtered by a threshold value to perceive the structure of the retina [Fig. 6(c)]. Third, a median filter is applied to remove detached black dots inside the retina [Fig. 6(d)], especially dots near the upper and lower edges of the white areas in Fig. 6(c), which could hamper the aligning effect in the following steps. Fourth, the morphological closing method is used to remove large black blobs inside the retina, which cannot be removed completely by the median filter [Fig. 6(e)]. Fifth, the morphological opening method is used to remove the detached white dots (caused by large noise speckles existing in some OCT scans, which are not presented in Fig. 6) outside the retina [Fig. 6(f)]. In the fitting stage, our method automatically chooses the set of data points and a fitting method, and the whole process for decision making in this stage is illustrated in Fig. 7. It could be described in two steps: Step 1: Choosing the set of data points for fitting. Our method extracts two sets of data points from Fig. 6(f): middle data points [i.e., each point is chosen from a unique column in the white area in Fig. 6(f), whose -coordinate is the index of the column and -coordinate is the arithmetic mean of -coordinates of all points in the column] and bottom data points [i.e., each point is chosen from a unique column in the white area in Fig. 6(f), whose -coordinate is the index of the column and -coordinate is the -coordinate of the corresponding bottom point in the column]. When choosing the sets of data points (the middle data points versus the bottom data points), our method performs the second-order polynomial fitting to the middle data points for judging, if the fitted parabola opens upward [Fig. 6(g)], then the middle data points are chosen; if the parabola opens downward, then the bottom data points are chosen. Step 2: Choosing the fitting method (linear fitting versus second-order polynomial fitting). In the case when the middle data points are chosen, our method performs the linear fitting to the data points and then calculates the correlation coefficients (calculated by using the MATLAB® command corrcoef) between the middle data points and the two sets of the fitted points (i.e., one is from the linear fitting and another is from the second-order polynomial fitting); then the fitting method corresponding to a larger correlation coefficient is chosen [i.e., a parabola is finally chosen to fit the middle data points in Fig. 6(g) because there is a larger correlation coefficient between the middle data points and the data points to the fitted parabola (as opposed to the fitted linear line)]. In the case when the bottom data points are chosen, our algorithm performs the second-order polynomial fitting to the bottom data points for judging, if the fitted parabola opens upward, then the linear fitting is done and the fitting method with a larger correlation coefficient is chosen; if the parabola opens downward, then the linear fitting method is chosen directly. In the normalizing stage, our method normalizes the retinas by aligning them to a relatively unified morphology and crops the images to trim out insignificant space. When the second-order polynomial fitting is chosen, the retina is flattened by moving each column of the image a certain distance according to the fitted curve [Fig. 6(h)]. In the case of the linear fitting, the retina is aligned by rotating the entire retina to an approximately horizontal position according to the angle between the fitted line and the horizontal line. When cropping the image, our method first detects the highest and lowest points of the white area when the retina region is flattened as shown in Fig. 6(h), and then, according to the points detected, it generates two horizontal lines to split the whole flattened image into three sections: upper, middle, and lower; finally, it vertically trims out the upper and lower sections (which are insignificant to the retinal characteristics) to get the middle section with no margins left (Fig. 8). In this way, our method could retain morphological structures of the retina that are useful for classification and leave out disturbances as much as possible. Our method performs the linear or second-order polynomial fitting of the middle data points in most common cases, which means that our algorithm employs all pixels within the retina [i.e., the white area in Fig. 6(f)] to compute and generate the input data points, whereby we perform the linear or curve fitting. We do not adopt the points of the bottom edge of the white areas [as shown in Fig. 6(f)] as our first choice for the fitting because the bottom line is relatively hard to fit because of its irregularity below the RPE layer. Moreover, based on our experience, the bottom edge of a retina [as shown in Fig. 6(f)] is roughly either straight or parabolic (opening upward)-shaped, which should not be fitted to a parabola opening downward. The cases in which the bottom data points are chosen and the fitted parabola opens downward (very rare in our experiments) are due to irregularities below the RPE layers, where the linear fitting performs much better in our experiments. Therefore, as we explain in Fig. 7, when the bottom data points are chosen and the fitted parabola opens downward, our algorithm directly chooses the linear fitting method (no longer considers the second-order polynomial fitting). 2.2.Dictionary LearningIn this phase, first we partition all the aligned and cropped images in the training dataset into small rectangular patches with a fixed size, as Fig. 9 demonstrates, and mix all patches from all the images in the training dataset randomly. Then we extract SIFT descriptors of every single random patch, each of which is a (where is 128) dimension vector ; next we build an SIFT descriptor set (), where is the number of patches selected for dictionary learning from a random collection of image patches partitioned from the training set attained in the previous step to solve following Eq. (1) iteratively by alternatingly optimizing over or (i.e., ) while fixing the other30,35 where is retained as the dictionary, is the number of bases chosen in the dictionary, i.e., the dictionary size. A unit -norm constraint on is typically applied to avoid trivial solutions. In the sparse coding phase, for an image represented as a set of SIFT descriptors , the sparse coding is obtained by optimizing Eq. (1) when the dictionary is fixed.In our experiments, we use 60,000 or more SIFT descriptors randomly extracted from the patches from all the images in our training set to train the dictionary by solving Eq. (1). Once we obtain the dictionary in this offline training, we can do online sparse coding efficiently on each descriptor of an image by solving Eq. (1) when the dictionary is fixed. 2.3.Image Feature RepresentationIn this phase, we build a global feature representation for each cropped OCT image. The constructing process is illustrated in Fig. 10. First, each cropped OCT image is partitioned into patches, and SIFT descriptors of all patches are extracted. Second, SCs of all the patches of the image are obtained by applying the prelearned and fixed dictionary. Third, we represent each image by employing a three-level SP with 21 blocks in total; for each block in level 2, we obtain its local representation by max pooling the SCs of all the patches in the block, and the local representation of each block in level 1 is obtained by max pooling the corresponding four block representations in level 2. In a similar way, the local representation of the block in level 0 can be acquired from level 1. Finally, the global feature representation of the image is obtained by concatenating all the local representations of all the blocks in the three levels. 2.4.Multiclass Linear Support Vector MachineIn this stage, we train three two-class linear SVMs as our classifiers, so that we can classify an input image into one of the three labels: AMD, DME, and normal. During the training phase, we train a single classifier per class, with the training samples of that class as positive samples and the others as negative samples. Let image be represented by , given the training data , , where (because in our experiment, DME, AMD, and normal OCT images are used in training and classification), we can obtain three two-class linear SVMs: AMD-against-all SVM, DME-against-all SVM, and normal-against-all SVM, each of which produces a real-valued confidence score for its decision, rather than just a class label. During the classification phase, for a single testing sample, we apply all the three classifiers (AMD, DME, and normal) to the sample and predict the label, for which the corresponding classifier reports the highest confidence score. 3.Experiments and ResultsIn this section, we introduce two different datasets we applied in our experiments, provide more cases in the image preprocessing step and the way we address them, describe several experiments, and present the experimental results over two datasets. In addition to the results from our own implementations, we also quote some results directly from the literature, especially those from Ref. 27. One of the datasets we applied for experiments is the Duke dataset published by Srinivasan et al., which was acquired in Institutional Review Board-approved protocols using Spectralis SD-OCT (Heidelberg Engineering Inc., Heidelberg, Germany) imaging at Duke University, Harvard University, and the University of Michigan. This dataset consists of 45 OCT volumes labeled as AMD (15 volumes), DME (15 volumes), and normal (15 volumes). The number of OCT scans in each volume varies from 36 to 97. We downloaded the full dataset from Ref. 37. The other dataset was obtained from clinics in Beijing, using CIRRUS TM (Heidelberg Engineering Inc., Heidelberg, Germany) SD-OCT device. The dataset consists of 168 AMD, 297 DME, and 213 normal OCT B-scans of a retina. All SD-OCT images are read and assessed by trained graders. For many images of a diseased retina with severely distorted RPE layers [which is similar to the case shown in Fig. 1(b)] in this dataset (roughly 27% for the AMD class and 6% for the DME class), it is not feasible to flatten the retina region by estimating the RPE boundaries. Our fully automated classification algorithm was coded in MATLAB® (The MathWorks, Natick, Massachusetts) and tested on a four-core PC with a Mac OS X EI Capitan 64-bit operating system, Core i7-3720QM CPU at 2.6 GHz (Intel, Santa Clara, California), and 8 GB of RAM. The ScSPM source code was downloaded from website38 and adopted in our own experiments. 3.1.Retina Region Aligning and FlatteningIn this section, we describe more cases that previous works24,27 did not consider. Figure 1 shows a severely distorted RPE layer of a retina that cannot be easily segmented and flattened by segmenting its RPE boundary as was done in Ref. 27. With our method outlined in Sec. 2.1, we can easily find the entire retina area and fit it with a second-order polynomial using its middle data points, as depicted in Fig. 11(a); then we flatten the whole retina by the fitted curve and trim out insignificant areas beyond the retina to acquire the cropped image in Fig. 11(b). Figure 12 demonstrated the retina flattening process with linear fitting by using the middle data points of the retina region. Figure 12(a) shows the original retina OCT image. In this case, first the middle data points were employed owing to the fitted parabola opening upward; then the linear fitting method was selected [Fig. 12(b)] via correlation comparison, which means that the linear fitting method had a better representation of the overall morphology of the retina. Next, the fitted line was used to align the retina regions [as shown in Fig. 12(c)]. Finally, the cropped image is obtained [Fig. 12(d)]. The following example is the case when our algorithm automatically choses the lower edge of the retina to perform the linear fitting, as illustrated in Fig. 13. Figure 13(a) is the original image sharing the same feature with Fig. 2, in which the retina suffers local swelling in the lesion position. According to our algorithm in Sec. 2.1 (Fig. 7), the middle data points were first used to perform the second-order polynomial fitting. Since the fitted parabola opened downward, the bottom data points were chosen to perform the second-order polynomial fitting, and the linear fitting method was chosen because the parabola opened upward and the correlation coefficient corresponding to the second-order polynomial fitting was smaller than that to the linear fitting [Fig. 13(b)]. Finally, the fitted linear line was used to align the whole retina to a relatively horizontal direction [Fig. 13(c)]. Then we cropped the image to obtain the final image [Fig. 13(d)]. In this example, when we used Liu et al.’s method24 to align the same images [Fig. 13(a)], which means employing the middle data points and flattening the retina region according to the downward parabola, we finally obtained Fig. 13(e), in which the retina was flattened in a reverse way and the curvature of the retina became larger. There are many cases like this one in our datasets, which Liu et al.’s preprocessing method could not deal with well. Fig. 13Case of preprocessing, in which the bottom data points of the retina were employed to fit with a straight line. (a) The original OCT image. (b) An intermediate result in our preprocessing process when the bottom data points were fitted with a straight line. (c) An intermediate result in our preprocessing process when the retina was aligned according to the fitted line. (d) The preprocessed result with our method. (e) The preprocessed result with Liu et al.’s method. 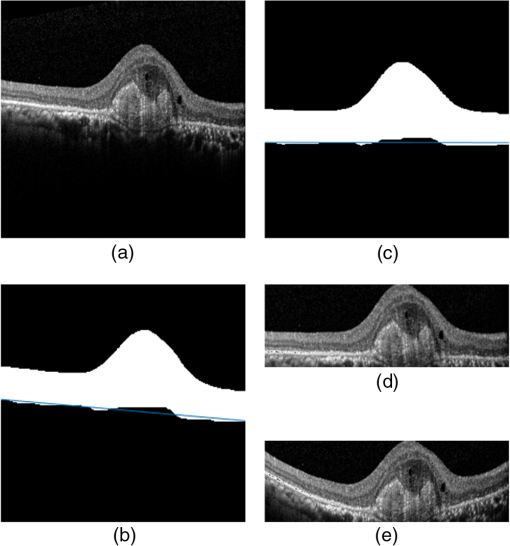 3.2.Classification PerformanceWe validated our classification algorithm by testing on the two datasets mentioned above. In the experiments, we first obtained the cropped retina images. The parameters selected for preprocessing are listed below: 45 and 35 for sigma for the Duke dataset and the Beijing clinical dataset, respectively, while denoising images with the BM3D method; binary threshold selected from Ostu’s algorithm39 for each image automatically; median filter, disk-shaped structure element with size 40 for morphological closing and size 5 for opening. We observed that our aligning method with these parameters could roughly align all the retinas to a horizontal and unifying state with a little morphological variance and insignificant area. We chose different parameters for sigma for two datasets because their average noise levels of OCT images are different, and we chose these parameters by constantly fine tuning them to make the aligning effect best for each dataset. In real applications, we would suggest choosing a sigma parameter that is suitable for the images in the corresponding dataset as much as possible. The average preprocessing time per image was about 9.2 s. In the dictionary learning phase, the SIFT descriptors (with dimensions for each of them) were extracted from patches, which were densely sampled from each training image on a grid with step-size eight pixels; 60,000 SIFT descriptors extracted from random patches in the training images were used to train dictionary; the dictionary size was set to be 1024, where each visual element in the dictionary was of size 128; 0.3 for one free parameter in Eq. (1) was set when sparse coding. The number of iterations (1, 5, 10, and 15) and their influences on the experimental results were tested. For each training set, dictionary learning was just done once and was done offline. These parameters may be not optimized, but the following experimental results show them to be excellent. We first conducted our test on the Duke dataset (experiment 1). To compare our algorithm with that in Ref. 27 fairly, we used leave-three-out cross-validation for 45 times as was done in Ref. 27. For each time, the multiclass linear classifier was trained on 42 volumes, excluding one volume from each class, and tested on the three volumes that were excluded from training. This process resulted in each of the 45 SD-OCT volumes being classified once, each using 42 of the other volumes as the training data. Since a volume contains many OCT images of a specific person, we appoint a volume to a class (AMD, DME, or normal), into which the most images in the volume have been classified. We used our proposed preprocessing method without RPE layer boundary segmentation, and the iteration times in dictionary learning were set to be 1 for saving the dictionary learning time. The actual dictionary learning time is approximately 763 s for each leave-three-out cross-validation. The cross-validation results are shown in Table 1. As can be seen from Table 1, 100% of 30 OCT volumes under the AMD and DME classes were correctly classified, being equal to that in Duke; and 93.33% of 15 OCT volumes under the normal class were correctly classified with our method, which is higher than that with the method proposed in Duke27 (86.67%). This shows that our proposed classification method using sparse coding and dictionary learning for retina OCT images performs much better than that in Ref. 27, which adopts HOGs as feature descriptors and a linear SVM for classification. Table 1Fraction of volumes correctly classified with the two methods on the Duke dataset.
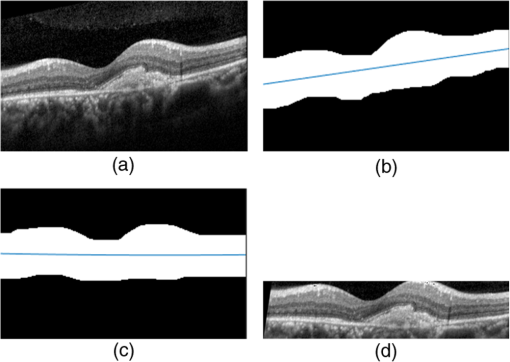
By analyzing our experimental results in detail, we found that, among the 15 OCT volumes under the normal class, 14 volumes were correctly classified and 1 volume (i.e., normal volume 6) was misclassified into the DME class. Next, we try to find reasons why normal volume 6 was misclassified. Figure 14(a) is a typical case of images, not from normal volume 6, being correctly classified, from which we can see that the cropped image retains only the areas between the upper layer and the RPE layer of the retina, which contains only the most useful information of image classification. In contrast, Fig. 14(b) is a typical case of images, from normal volume 6, being misclassified, from which we can see that its large portion of insignificant area below the RPE layer visually resembling the pathological structures presented in the DME cases was retained. Since these insignificant areas widely exist in normal volume 6 but not in other volumes after our preprocessing, we speculate that the insignificant areas below the RPE layer of the retina led to normal volume 6 being misclassified into the DME class. To further prove our speculation, we excluded 45 rows starting from the bottom of each aligned and cropped image in normal volume 6 to roughly trim out the insignificant areas below RPE layers; then we conducted experiment 1 again with the slightly modified dataset, and the fraction of volumes for each category (AMD, DME, and normal) correctly classified is 100%. This justified our speculation and showed a good performance of our algorithm. Fig. 14Retina OCT images under the normal class in the Duke dataset flattened and cropped by the aligning method proposed in this paper. (a) A typical case from normal volumes except from normal volume 6, which was correctly classified. (b) A case from normal volume 6, which was misclassified. 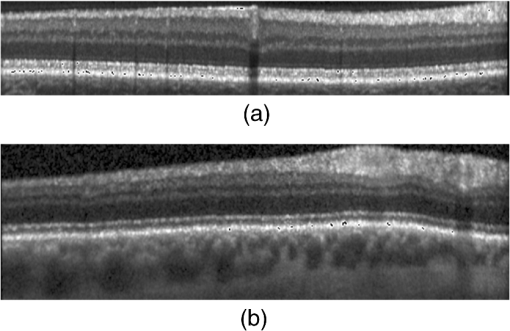 We further performed an experiment (experiment 2) on the clinic dataset. Here, we employed our preprocessing method to obtain the aligned and cropped OCT images and classify them. We used a leave-three-out cross-validation on images, where the multiclass linear classifier was trained on 675 OCT B-scans, excluding one OCT B-scans randomly selected from each class, and tested on the three scans that were excluded from training. We did 300 times cross-validations, so that this process resulted in each of the 678 OCT B-scans being roughly classified once. The correct classification rates of normal, AMD, and DME subjects are presented in Table 2. In this test, the iteration times in dictionary learning were also set to be 1 for saving the dictionary learning time. Table 2Correct classification rate (%) during cross-validation on the clinic dataset.
As shown in Table 2, the correct classification rates for the AMD and DME classes are 99.67% () and 100% () for the normal class. To fully evaluate our aligning method, we considered the clinic dataset preprocessed by Liu et al.’s preprocessing method and conducted the exact same experiment (300 times leave-three-out cross-validations), and the results are as follows: 100% () for both normal and DME classes and 97.67% () for the AMD class. In experiment 2, one DME scan was misclassified during leave-three-out cross-validations performed 300 times as shown in Table 2; however, the misclassified DME scan was coincidently not chosen during leave-three-out cross-validations performed 300 times when we perform the classification with Liu et al.’s aligning method (i.e., in each cross-validation loop, one OCT scan of each of three classes is randomly chosen for testing). Thus, we conducted one more classification on the clinic dataset preprocessed by Liu et al.’s aligning method, and this time we chose the misclassified DME scan on purpose, and it turned out that the DME scan was misclassified (into the normal class) anyway. Hence, our aligning method promises a better performance during classification. In the above-mentioned experiments, the iteration times in dictionary learning were set to be 1, and the dictionaries were trained with 60,000 SIFT descriptors. It can be seen from Tables 1 and 2 that the experimental results are excellent for the two datasets. However, the learned dictionary is obviously not optimal for one iteration, the more iterations, the better the representation of the dictionary. In addition, the total number of SIFT descriptors selected from the training set should be determined by the image dimension and the total number of images fed for training; for a given dataset, the more descriptors selected, the better the representation of the dictionary. More iteration times and employed descriptors in the dictionary-learning phase mean higher computational cost. In the following, we conducted more experiments (experiment 3) on the clinic dataset with different training sets and iteration times. To obtain reliable results, we repeated the experimental process by 10 times with different randomly selected images in the clinic dataset for training and the rest for testing. The correct classification rates of normal, AMD, and DME subjects were recorded for every time. We reported our final results by the mean and the standard deviation of the classification rates of AMD, DME, and normal, respectively. Here, we first chose half of the AMD, DME, and normal images (84 AMD images, 148 DME images, and 106 normal images) for training (simply called dataset training) and the rest (84 AMD images, 149 DME images, and 107 normal images) for testing. Then we chose two-thirds of AMD, DME, and normal images (112 AMD images, 198 DME images, and 142 normal images) for training (simply called dataset training) and the rest (56 AMD images, 99 DME images, and 71 normal images) for testing. In this experiment, the dictionaries are trained for 5, 10, and 15 iteration times, where the number of SIFT descriptors is fixed to be 60,000. The experimental results are given in Table 3. Table 3Correct classification rate (%) comparison of different proportion images for training on the clinic dataset.
Several characteristics can be concluded here:
The computational performances in experiment 3 are as follows: the dictionary learning times are approximately 3400, 6700, and 10,000 s for 5, 10, and 15 iteration times, respectively. For the dataset training, the average SVM training on 338 examples is 2.3 s, and the average classification time for each cropped image is 3.0 s. For the dataset training, the average SVM training on 452 examples is 2.8 s, and the average classification time for each cropped image is 3.1 s. To evaluate the influence of the number of SIFT descriptors on the learned dictionary and the correct classification rate when the iteration times are fixed, we have conducted more experiments using the aforementioned dataset training under the condition on 80,000 SIFT descriptors randomly extracted from the training image with 5 iteration times. The final correct classification rates (%) for normal, AMD, and DME are , , and , respectively. The result is the best in all the experimental results that is presented in Table 3. This shows that the dictionary learned from 80,000 descriptors is better than that trained from 60,000 descriptors. We did not conduct experiments with bigger iteration times due to the relative high computational cost of dictionary learning. 4.ConclusionIn this paper, we propose a method for OCT image classification between AMD, DME, and normal retina OCT scans. This method was successfully tested in different datasets for the detection of AMD and DME. The proposed method does not rely on the segmentation of retina layers. This is a significantly important feature when dealing with retina diseases that alter retinal layers and thus complicate the layer boundary segmentation task. Moreover, a multiclass linear SVM classifier based on SCs and dictionary learning of the preprocessed OCT images are used to detect AMD and DME diseases. This method has a much better performance in the Duke dataset than the conventional method in Ref. 27 and possesses an excellent classification performance on our clinical dataset. Our algorithm is a potentially impactful tool for the computer-aided diagnosis and screening of ophthalmic diseases. The preprocessing method for retina OCT images in our algorithm has many advantages, including (1) input OCT images could have different resolutions, which means that our input data could be captured with different scanning protocols, which is often the case in real-world clinical practice; (2) it does not rely on any retina layer segmentation algorithms, hence, it is suitable for OCT images with severe retina diseases; (3) it provides linear or second-order polynomial fitting method to flatten retina region, therefore, variations between OCT images in morphologies could be reduced greatly; and (4) the cropped OCT images obtained are data relative, which could have different image sizes. The latter three features are different from those in Ref. 27. The classification method based on dictionary learning is introduced to successfully classify the above-processed OCT images. Its classifier training time complexity is when the dictionary has been prelearned and its image classification time is a constant for every image, where is the number of training samples of OCT images. Compared to the algorithm in Ref. 27, which has a large memory requirement when the field-of-view defined by the cropped area is large, the memory requirement of our algorithm is independent of the field of view. In our experiments, algorithm parameters for preprocessing are given. Some parameters for dictionary learning and classifier training are used according to ScSPM’s code implementation for natural image classification. Although they are not optimal for OCT images, their performance is excellent for our experiments. The limitation of our algorithm is the efficiency of the dictionary-learning phase, which prevents us from doing further validation during cross-validation with a larger dictionary, more iteration times, and a larger number of training patches. To our knowledge, the computational time in the learning dictionary by solving Eq. (1) increases rapidly with the iteration times and the number of the training patches selected to train the dictionary. In our experiments, for a fixed number of SIFT descriptors, different iteration times were set and their influence on the computational cost and classification rate were tested. To solve the problem of the computational efficiency of dictionary learning for large training examples, some processing techniques, such as double sparse model40 and online dictionary learning,41 have been developed. These methods could solve our problem to some extent, and the development of such methods for OCT image classification is part of our ongoing work. In addition, deep learning has been proven to be a good method in medical imaging,42 so the development of the deep learning method for OCT image classification is another part of our ongoing work. AcknowledgmentsThis work was supported by the National Natural Science Foundation of China under Grant No. 61671272. We would like to thank the two anonymous reviewers for their careful reading of our paper and their many insightful comments and suggestions that greatly improved the paper. ReferencesJ. S. Schuman et al., Optical Coherence Tomography of Ocular Diseases, Slack, Thorofare, New Jersey
(2004). Google Scholar
C. B. Rickman et al.,
“Dry age-related macular degeneration: mechanisms, therapeutic targets, and imagingdry AMD mechanisms, targets, and imaging,”
Invest. Ophthalmol. Visual Sci., 54
(14), ORSF68
–ORSF80
(2013). http://dx.doi.org/10.1167/iovs.13-12757 IOVSDA 0146-0404 Google Scholar
B. J. Antony et al.,
“A combined machine-learning and graph-based framework for the segmentation of retinal surfaces in SD-OCT volumes,”
Biomed. Opt. Express, 4
(12), 2712
–2728
(2013). http://dx.doi.org/10.1364/BOE.4.002712 BOEICL 2156-7085 Google Scholar
A. Carass et al.,
“Multiple-object geometric deformable model for segmentation of macular OCT,”
Biomed. Opt. Express, 5
(4), 1062
–1074
(2014). http://dx.doi.org/10.1364/BOE.5.001062 BOEICL 2156-7085 Google Scholar
S. J. Chiu et al.,
“Validated automatic segmentation of AMD pathology including drusen and geographic atrophy in SD-OCT images,”
Invest. Ophthalmol. Visual Sci., 53
(1), 53
–61
(2012). http://dx.doi.org/10.1167/iovs.11-7640 IOVSDA 0146-0404 Google Scholar
S. J. Chiu et al.,
“Automatic segmentation of seven retinal layers in SDOCT images congruent with expert manual segmentation,”
Opt. Express, 18
(18), 19413
–19428
(2010). http://dx.doi.org/10.1364/OE.18.019413 OPEXFF 1094-4087 Google Scholar
D. C. DeBuc et al.,
“Reliability and reproducibility of macular segmentation using a custom-built optical coherence tomography retinal image analysis software,”
J. Biomed. Opt., 14
(6), 064023
(2009). http://dx.doi.org/10.1117/1.3268773 JBOPFO 1083-3668 Google Scholar
D. C. Fernández, H. M. Salinas and C. A. Puliafito,
“Automated detection of retinal layer structures on optical coherence tomography images,”
Opt. Express, 13
(25), 10200
–10216
(2005). http://dx.doi.org/10.1364/OPEX.13.010200 OPEXFF 1094-4087 Google Scholar
H. Ishikawa et al.,
“Macular segmentation with optical coherence tomography,”
Invest. Ophthalmol. Visual Sci., 46
(6), 2012
–2017
(2005). http://dx.doi.org/10.1167/iovs.04-0335 IOVSDA 0146-0404 Google Scholar
A. Lang et al.,
“Retinal layer segmentation of macular OCT images using boundary classification,”
Biomed. Opt. Express, 4
(7), 1133
–1152
(2013). http://dx.doi.org/10.1364/BOE.4.001133 BOEICL 2156-7085 Google Scholar
M. A. Mayer et al.,
“Retinal nerve fiber layer segmentation on FD-OCT scans of normal subjects and glaucoma patients,”
Biomed. Opt. Express, 1
(5), 1358
–1383
(2010). http://dx.doi.org/10.1364/BOE.1.001358 BOEICL 2156-7085 Google Scholar
A. Mishra et al.,
“Intra-retinal layer segmentation in optical coherence tomography images,”
Opt. Express, 17
(26), 23719
–23728
(2009). http://dx.doi.org/10.1364/OE.17.023719 OPEXFF 1094-4087 Google Scholar
M. Mujat et al.,
“Retinal nerve fiber layer thickness map determined from optical coherence tomography images,”
Opt. Express, 13
(23), 9480
–9491
(2005). http://dx.doi.org/10.1364/OPEX.13.009480 OPEXFF 1094-4087 Google Scholar
L. A. Paunescu et al.,
“Reproducibility of nerve fiber thickness, macular thickness, and optic nerve head measurements using StratusOCT,”
Invest. Ophthalmol. Visual Sci., 45
(6), 1716
–1724
(2004). http://dx.doi.org/10.1167/iovs.03-0514 IOVSDA 0146-0404 Google Scholar
M. Shahidi, Z. Wang and R. Zelkha,
“Quantitative thickness measurement of retinal layers imaged by optical coherence tomography,”
Am. J. Ophthalmol., 139
(6), 1056
–1061
(2005). http://dx.doi.org/10.1016/j.ajo.2005.01.012 AJOPAA 0002-9394 Google Scholar
Y. Sun et al.,
“3D automatic segmentation method for retinal optical coherence tomography volume data using boundary surface enhancement,”
J. Innovative Opt. Health Sci., 9
(2), 1650008
(2016). http://dx.doi.org/10.1142/S1793545816500085 Google Scholar
K. Vermeer et al.,
“Automated segmentation by pixel classification of retinal layers in ophthalmic OCT images,”
Biomed. Opt. Express, 2
(6), 1743
–1756
(2011). http://dx.doi.org/10.1364/BOE.2.001743 BOEICL 2156-7085 Google Scholar
Q. Yang et al.,
“Automated segmentation of outer retinal layers in macular OCT images of patients with retinitis pigmentosa,”
Biomed. Opt. Express, 2
(9), 2493
–2503
(2011). http://dx.doi.org/10.1364/BOE.2.002493 BOEICL 2156-7085 Google Scholar
P. B. Garcia-Allende et al.,
“Morphological analysis of optical coherence tomography images for automated classification of gastrointestinal tissues,”
Biomed. Opt. Express, 2
(10), 2821
–2836
(2011). http://dx.doi.org/10.1364/BOE.2.002821 BOEICL 2156-7085 Google Scholar
K. W. Gossage et al.,
“Texture analysis of optical coherence tomography images: feasibility for tissue classification,”
J. Biomed. Opt., 8
(3), 570
–575
(2003). http://dx.doi.org/10.1117/1.1577575 JBOPFO 1083-3668 Google Scholar
C. A. Lingley-Papadopoulos et al.,
“Computer recognition of cancer in the urinary bladder using optical coherence tomography and texture analysis,”
J. Biomed. Opt., 13
(2), 024003
(2008). http://dx.doi.org/10.1117/1.2904987 JBOPFO 1083-3668 Google Scholar
P. Pande et al.,
“Automated classification of optical coherence tomography images for the diagnosis of oral malignancy in the hamster cheek pouch,”
J. Biomed. Opt., 19
(8), 086022
(2014). http://dx.doi.org/10.1117/1.JBO.19.8.086022 JBOPFO 1083-3668 Google Scholar
Y. Sun and M. Lei,
“Method for optical coherence tomography image classification using local features and earth mover’s distance,”
J. Biomed. Opt., 14
(5), 054037
(2009). http://dx.doi.org/10.1117/1.3251059 JBOPFO 1083-3668 Google Scholar
Y.-Y. Liu et al.,
“Automated macular pathology diagnosis in retinal OCT images using multi-scale spatial pyramid and local binary patterns in texture and shape encoding,”
Med. Image Anal., 15
(5), 748
–759
(2011). http://dx.doi.org/10.1016/j.media.2011.06.005 Google Scholar
Y. Zheng, M. H. A. Hijazi and F. Coenen,
“Automated ‘disease/no disease’ grading of age-related macular degeneration by an image mining approach,”
Invest. Ophthalmol. Visual Sci., 53
(13), 8310
–8318
(2012). http://dx.doi.org/10.1167/iovs.12-9576 IOVSDA 0146-0404 Google Scholar
M. H. A. Hijazi, F. Coenen and Y. Zheng,
“Data mining techniques for the screening of age-related macular degeneration,”
Knowledge-Based Syst., 29 83
–92
(2012). http://dx.doi.org/10.1016/j.knosys.2011.07.002 KNSYET 0950-7051 Google Scholar
P. P. Srinivasan et al.,
“Fully automated detection of diabetic macular edema and dry age-related macular degeneration from optical coherence tomography images,”
Biomed. Opt. Express, 5
(10), 3568
–3577
(2014). http://dx.doi.org/10.1364/BOE.5.003568 BOEICL 2156-7085 Google Scholar
A. Albarrak et al.,
“Volumetric image mining based on decomposition and graph analysis: an application to retinal optical coherence tomography,”
in 13th Int. Symp. on Computational Intelligence and Informatics (CINTI 2012),,
263
–268
(2012). Google Scholar
C. Cortes and V. Vapnik,
“Support-vector networks,”
Mach. Learn., 20
(3), 273
–297
(1995). http://dx.doi.org/10.1007/BF00994018 MALEEZ 0885-6125 Google Scholar
J. Yang et al.,
“Linear spatial pyramid matching using sparse coding for image classification,”
in IEEE Conf. on Computer Vision and Pattern Recognition,
1794
–1801
(2009). Google Scholar
S. Lazebnik, C. Schmid and J. Ponce,
“Beyond bags of features: spatial pyramid matching for recognizing natural scene categories,”
in IEEE Computer Society Conf. on Computer Vision and Pattern Recognition (CVPR 2006),
2169
–2178
(2006). http://dx.doi.org/10.1109/CVPR.2006.68 Google Scholar
L. Fang et al.,
“3-D adaptive sparsity based image compression with applications to optical coherence tomography,”
IEEE Trans. Med. Imaging, 34
(6), 1306
–1320
(2015). http://dx.doi.org/10.1109/TMI.2014.2387336 ITMID4 0278-0062 Google Scholar
L. Fang et al.,
“Fast acquisition and reconstruction of optical coherence tomography images via sparse representation,”
IEEE Trans. Med. Imaging, 32
(11), 2034
–2049
(2013). http://dx.doi.org/10.1109/TMI.2013.2271904 ITMID4 0278-0062 Google Scholar
R. Kafieh, H. Rabbani and I. Selesnick,
“Three dimensional data-driven multi scale atomic representation of optical coherence tomography,”
IEEE Trans. Med. Imaging, 34
(5), 1042
–1062
(2015). http://dx.doi.org/10.1109/TMI.2014.2374354 ITMID4 0278-0062 Google Scholar
M. Elad and M. Aharon,
“Image denoising via sparse and redundant representations over learned dictionaries,”
IEEE Trans. Image Process., 15
(12), 3736
–3745
(2006). http://dx.doi.org/10.1109/TIP.2006.881969 IIPRE4 1057-7149 Google Scholar
K. Dabov et al.,
“Image denoising by sparse 3-D transform-domain collaborative filtering,”
IEEE Trans. Image Process., 16
(8), 2080
–2095
(2007). http://dx.doi.org/10.1109/TIP.2007.901238 IIPRE4 1057-7149 Google Scholar
P. P. Srinivasan et al.,
“Srinivasan_BOE_2014,”
http://www.duke.edu/~sf59/Srinivasan_BOE_2014_dataset.htm Google Scholar
N. Otsu,
“A threshold selection method from gray-level histograms,”
Automatica, 11
(285–296), 23
–27
(1975). ATCAA9 0005-1098 Google Scholar
R. Rubinstein, M. Zibulevsky and M. Elad,
“Double sparsity: learning sparse dictionaries for sparse signal approximation,”
IEEE Trans. Signal Process., 58
(3), 1553
–1564
(2010). http://dx.doi.org/10.1109/TSP.2009.2036477 ITPRED 1053-587X Google Scholar
J. Sulam et al.,
“Trainlets: dictionary learning in high dimensions,”
IEEE Trans. Signal Process., 64
(12), 3180
–3193
(2016). http://dx.doi.org/10.1109/TSP.2016.2540599 ITPRED 1053-587X Google Scholar
H. Greenspan, B. van Ginneken and R. M. Summers,
“Guest editorial deep learning in medical imaging: overview and future promise of an exciting new technique,”
IEEE Trans. Med. Imaging, 35
(5), 1153
–1159
(2016). http://dx.doi.org/10.1109/TMI.2016.2553401 ITMID4 0278-0062 Google Scholar
BiographyYankui Sun is an associate professor in the Department of Computer Science and Technology, Tsinghua University, Beijing, China. He received his PhD in manufacturing engineering of aeronautics and astronautics from Beihang University in 1999. He is the author of more than 80 journal and conference papers and has written two books. His current research interests include OCT image processing and so on. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||