|
|
1.IntroductionAt present, surgical treatment of cardiac valve disease occupies a considerable share of all cardiac treatment. In the last decade, the demand for heart valve transplants increased greatly. Even though quality, design, and properties of heart valve prostheses constantly improve, they are not comparable to the properties of native valves.1 After a transplant of a native implant, the recipient will be at risk of chronic immune rejection and lifelong immunosuppression therapy.2 That is why the overarching aim in the field of tissue engineering is the development and quality control of cell-free scaffolds suitable for the replacement of lesional tissues and organs. The large number of complications associated with implantation of an artificial cardiac valve necessitates high-quality processing of biomaterials. Decellularization is one of the helper methods of tissue engineering of heart valves and is aimed at the removal of cells from the tissue while maintaining the territorial matrix and the three-dimensional structure of the organ.2 The most popular method of decellularization of heart valves is chemical treatment of biomaterials.3 In particular, the detergent-enzymatic method is widely used with various detergent solutions and different duration of its exposure.4 To reduce the antigenic potential of tissue, the detergent-enzymatic technique (DET) decellularization process involves removal of cell constituents such as membranes and the related soluble proteins, bioplasts, the nucleus and nucleic acids (DNA and RNA) in it. In the meantime, the capacity for remodeling is retained. Nevertheless, the use of DET decellularization as a method of heart tissue cleaning can cause damage to such components as proteoglycans and glycosaminoglycans (GAGs), which are actively involved in cell growth, cell differentiation, and morphogenesis.4 The problem with preservation of the territorial matrix structure after exposure to detergent-enzymatic agents is still relevant and necessitates the use of sparing decellularization [biological, detergent-free (BIO)] methods.4 BIO decellularization effectively preserves GAGs, but it is difficult to ensure complete removal of cellular debris or unacceptable components of the territorial matrix using enzymatic treatment. Theoretically, in the extracellular matrix, researchers must preserve collagen, elastin, and GAGs, after which the main immunogenic components of cells, namely, lipid membranes, membrane-associated antigens, and soluble proteins, must be removed.5 Most of the existing methods of quality control of cardiac implants may be used only with limitations. Magnetic resonance imaging (MRI) does not allow researchers to evaluate the composition of a biological tissue or to achieve the necessary space resolution of cardiac tissue samples within a short scan time. Furthermore, MRI scanners are expensive and time consuming.6 Histology and scanning electron microscopy provide high-quality images of the microtexture of decellularization samples, and, as a result, they are the most popular methods for evaluation of a decellularization process, but they require destructive preparation of a sample.6 Limitations of these methods of quality control of implants can be overcome by Raman scattering spectroscopy. This method is a simple, noninvasive, and rapid approach to quality control of materials for tissue engineering.7–9 Thus, it was shown10 that Raman spectroscopy (RS) allows researchers to identify the composition of cardiac tissue and the proportions of matrix components. Some authors7 have conducted a comparative analysis of treated and control aortic valve cusps using Raman scattering spectroscopy. Comparing the Raman intensity of wave numbers in the region 400 to , those authors identified significant alterations in signal intensity for the extracellular matrix of the aorta. In one article,8 researchers showed an estimate of cytoreduction in decellularized bovine pericardia (during their lyophilization) using a Raman scattering method. Major changes in the spectrum were observed at wavenumbers 1004, 1240 to 1250, and corresponding to phenylalanine and type I and III amides. In one study,9 the RS method was used for assessing protein dissolution of cardiac tissue during decellularization. The intensity of the Raman bands corresponding to the vibration of amide I at wavenumber allows researchers to reveal differences in the total secondary structure of a protein among tissues. For this reason, RS is a promising, noninvasive, and noncontact way to assess the extracellular matrix. The aim of this study is to carry out an assessment of DET and BIO decellularization of cardiac implants by the Raman scattering spectroscopy method and to provide the typical spectra after the respective decellularization protocols. 2.Materials and MethodsWe used 50 decellularized aortic valves of adult pigs. The native valvular samples have been obtained from healthy pigs from a local slaughterhouse near the University of Duesseldorf (Germany). DET and BIO decellularization of valves with retrieval of extracellular matrices was carried out using technologies of decellularization of Heinrich-Heine University Duesseldorf (Germany): DET decellularization based on detergent with sodium dodecyl sulfate and sodium deoxycholate as the main agents; BIO decellularization means physiological, detergent-free decellularization based on latrunculin B, high ionic strength of a saline solution, and DNase. A detailed description of the protocol based on DET and BIO decellularization is presented elsewhere.5 The laboratory facility that was used in this work is described in Ref. 11 and includes a diode laser (LML-785.0RB-04), an optical Raman module (PBL 785), a spectrograph (Sharmrock SR-303i) with an integrated digital camera (ANDOR DV-420A-OE), and a computer. The use of this spectrograph provides a resolution of at a low level of intrinsic noise. To exclude the contribution of background to the Raman spectrum, we used the subtraction method of a background component of polynomial approximation with postfiltration of random noise effects. In this work, analysis of the Raman spectra was carried out in the range of 300 to . The laser radiation power of 400 mW within our exposure duration up to 200 s does not cause destructive changes in the samples. Registration of Raman spectra was carried out using an optical probe that was situated under the object at a distance of 7 mm. Parts of this work, i.e., the peak values for GAG, amide I, and amide III structures, have been published in a previous article.12 3.ResultsLet us consider typical normalized averaged Raman spectra for samples of the extracellular matrix of leaves and walls of cardiac implants obtained using DET and BIO decellularization (Fig. 1). Decoding of the Raman spectrum is shown in Table 1. Fig. 1Raman spectra, normalized to the average intensity value of the extracellular matrix of the heart valve: the leaves and walls, obtained using DET and BIO decellularization (spectra are shifted vertically for illustrative purposes). 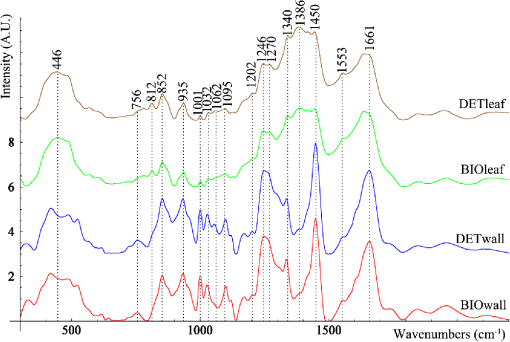 Table 1Decoding of Raman spectra.
As seen in Fig. 1, the samples of the extracellular matrix of the heart valves (leaf and wall) have spectral differences. It is noteworthy that during DET decellularization of the walls of cardiac implants, the intense peak is [deformation of proteins and nucleic acids (DNA)], less than that of the samples manufactured by a BIO decellularization protocol. At the same time, Fig. 1 shows that at wavenumber corresponding to amide III, corresponding to amide I (), and corresponding to (), proteins and lipids did not show spectral differences. This finding allows us to suggest that there were no significant differences between DET and BIO decellularization influence on proteins and lipids as well as the integrity of collagenous structures. Nevertheless, to confirm this finding, we must conduct a detailed analysis of the bands at 852 and , which correspond to () proline and oxyproline. The intensity of the band at corresponding to the symmetrical stretching of GAGs of chondroitin-6-sulfate is an important criterion for assessing DET and BIO decellularization. A reduction in peak intensity indicates leaching of sulfated GAGs during manufacture of the matrix. With decellularization procedures amide I is quite stable (), so this wavenumber served as a denominator in the coefficients below. Therefore, the ratio of the Raman bands may be used as lead criteria for evaluation of potential immunogenicity of extracellular matrices of heart valves where is the intensity values at wavenumbers for the components being analyzed.Recovery properties of implants are assessed by their structure and composition. Thus, preserved GAGs, proteins, proline, and hydroxyproline affect the ability to regenerate and elasticity. The residual nucleic acids (DNA) are capable of eliciting an immunological response in the recipient and rejection of the implants. The proposed coefficients allow us to assess these parameters and to evaluate these criteria. Figures 2 and 3 show two-dimensional (2-D) diagrams of the input optical coefficients , showing main changes in the composition of the walls and the cusp samples of the heart valves using DET and BIO treatment. For separation of overlapped peaks within the range of 700 to , we used MagicPlotPro 2.5.1 software with the representation of individual peaks of the Gaussian function. The position of the peaks on the wave numbers’ axis and peak width were preset. The spikes amplitude was determined by the iteration mechanism. The minimization of the residual amount or the achievement of an admissible number of iteration procedures (100) speak in capacity of stop-criteria. The method of PCA-analysis has proved itself to be good enough to identify the essential components during the data analysis, which includes Raman-scattering spectroscopy. Conversely, the component with the highest dispersion is not always the most informative, and the representation of components by linear combination of the original data is not always admissible. Fig. 22-D diagrams of the input optical coefficients for extracellular matrices of cardiac valves (leaves), obtained by DET and BIO decellularization. 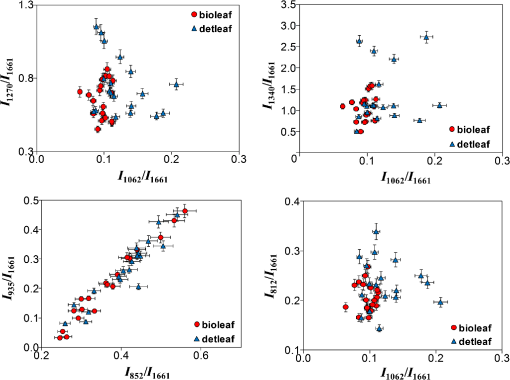 Fig. 32-D diagrams of the input optical coefficients for extracellular matrices of cardiac valves (walls), obtained by DET and BIO decellularization. 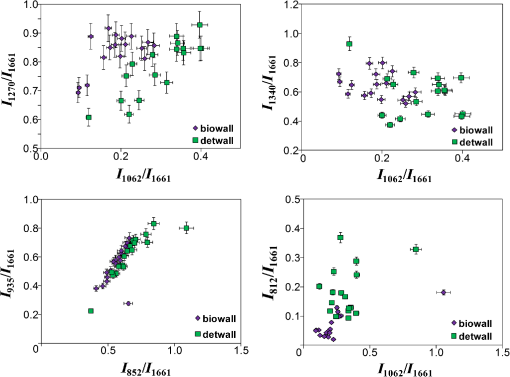 As shown in Figs. 2 and 3, in the extracellular matrices of leaves and walls during DET and BIO decellularization, there are no significant differences in impact on proteins and lipids, as well as the impact on the concentrations of proline, oxyproline, and basic amino acid residues of collagen, which are used as a status indicator of collagen in the tissue.18,19 There is a decrease in the intensity of band 812 during DET treatment of walls, which is responsible for the phosphodiester bond RNA (Fig. 3), but the difference is insignificant for leaves, probably because of the smaller thickness of the objects. In addition, analysis of 2-D diagrams revealed that DET decellularization yields better preservation of GAGs in comparison with BIO decellularization, as recently reported.12 On the other hand, during treatment of leaves by the protocol of DET decellularization, the amount of nucleic acids (DNA) is greater than in the samples subjected to BIO decellularization. 4.ConclusionInput optical coefficients enable a qualitative assessment of the protocols of DET and BIO decellularization in terms of the concentrations of GAG, proteins, amides, and DNA. Thus, the typical spectra of aortic and valvular tissues after DET and BIO decellularization are provided. It was found that the main spectral differences in extracellular matrices of cardiac walls between DET and BIO decellularization appear at wavenumber , corresponding to the deformation of proteins and nucleic acids (DNA). AcknowledgmentsThis work was supported by the Ministry of Education and Science. Resolution on Bioethics Committee received at the Samara State Medical University. ReferencesD. Schmidt et al.,
“Tissue engineering of heart valves using decellularized xenogeneic or polymeric starter matrices,”
Philos. Trans. R. Soc. B: Biol. Sci., 362 1505
–1512
(2007). http://dx.doi.org/10.1098/rstb.2007.2131 PTRBAE 0962-8436 Google Scholar
F. Moroni et al.,
“Decellularized matrices for cardiovascular tissue engineering,”
Am. J. Stem Cells, 3
(1), 1
–20
(2014). Google Scholar
A. V. Lavreshin et al.,
“Decellularization of aortic allografts and their morphological assessment,”
Geny i kletki (Genes and Cells), 9
(1), 64
–71
(2014). Google Scholar
P. M. Crapo et al.,
“An overview of tissue and whole organ decellularization processes,”
Biomaterials, 32
(12), 3233
–3243
(2011). http://dx.doi.org/10.1016/j.biomaterials.2011.01.057 BIMADU 0142-9612 Google Scholar
A. Lichtenberg et al.,
“In vitro re-endothelialization of detergent decellularized heart valves under simulated physiological dynamic conditions,”
Biomaterials, 27
(23), 4221
–4229
(2006). http://dx.doi.org/10.1016/j.biomaterials.2006.03.047 BIMADU 0142-9612 Google Scholar
C. K. Hagen et al.,
“High contrast microstructural visualization of natural acellular matrices by means of phase-based x-ray tomography,”
Sci. Rep., 5 18156
(2015). http://dx.doi.org/10.1038/srep18156 SRCEC3 2045-2322 Google Scholar
M. Votteler et al.,
“Non-invasive Raman spectroscopy of cardiovascular matrix,”
FASEB J., 25
(1), 681
–685
(2011). Google Scholar
R. Polak et al.,
“Care during freeze-drying of bovine pericardium tissue to be used as a biomaterial: a comparative study,”
Cryobiology, 63
(2), 61
–66
(2011). http://dx.doi.org/10.1016/j.cryobiol.2011.05.001 CRYBAS 0011-2240 Google Scholar
E. Kamilari et al.,
“Investigation of GAG-collagen interactions using spectroscopic techniques,”
Chem. Eng., 5 23
–25
(2013). BFECD9 Google Scholar
C. Krafft et al.,
“Raman and CARS microspectroscopy of cells and tissues,”
Analyst, 134 1046
–1057
(2009). http://dx.doi.org/10.1039/b822354h Google Scholar
E. V. Timchenko et al.,
“Using Raman spectroscopy to estimate the demineralization of bone transplants during preparation,”
J. Opt. Technol., 82
(3), 153
–157
(2015). http://dx.doi.org/10.1364/JOT.82.000153 JOTEE4 1070-9762 Google Scholar
A. Assmann et al.,
“Improvement of the in vivo cellular repopulation of decellularized cardiovascular tissues by a detergent-free, non-proteolytic, actin-disassembling regimen,”
J. Tissue Eng. Regener. Med.,
(2017). http://dx.doi.org/10.1002/term.2271 Google Scholar
E. Brauchle et al.,
“Raman spectroscopy as an analytical tool for melanoma research,”
Clin. Exp. Dermatol., 39 636
–645
(2014). http://dx.doi.org/10.1111/ced.12357 CEDEDE 1365-2230 Google Scholar
J. W. Chan et al.,
“Micro-Raman spectroscopy detects individual neoplastic and normal hematopoietic cells,”
Biophys. J., 90 648
–656
(2006). http://dx.doi.org/10.1529/biophysj.105.066761 BIOJAU 0006-3495 Google Scholar
T. T. Nguyen et al.,
“Characterization of type I and IV collagens by Raman microspectroscopy: identification of spectral markers of the dermo-epidermal junction,”
Spectrosc.: Int. J., 27
(5–6), 421
–427
(2012). http://dx.doi.org/10.1155/2012/686183 Google Scholar
A. Kunstar et al.,
“Label-free Raman monitoring of extracellular matrix formation in three-dimensional polymeric scaffolds,”
J. Royal Soc. Interface, 10
(86), 20130464
(2013). http://dx.doi.org/10.1098/rsif.2013.0464 Google Scholar
S. Tfaili et al.,
“Confocal Raman microspectroscopy for skin characterization: a comparative study between human skin and pig skin,”
Analyst, 137 3673
–3682
(2012). http://dx.doi.org/10.1039/c2an16292j ANLYAG 0365-4885 Google Scholar
A. Bonetti et al.,
“Carotenoids co-localize with hydroxyapatite, cholesterol, and other lipids in calcified stenotic aortic valves. Ex vivo Raman maps compared to histological patterns,”
Eur. J. Histochem., 59
(2), 2505
(2015). http://dx.doi.org/10.4081/ejh.2015.2505 Google Scholar
E. U. Otero et al.,
“Raman spectroscopy for diagnosis of calcification in human heart valves,”
Spectroscopy, 18 75
–84
(2004). http://dx.doi.org/10.1155/2004/975105 Google Scholar
|

