|
|
1.IntroductionColorectal cancer is the third most common form of malignancy in the United States, and approximately 149,900 new cases and 50,000 deaths occur each year, resulting in a major cause of morbidity and mortality.1 Screening for this disease is commonly done with endoscopy, in which the mucosal surface of the colon is examined with reflected white light, to look for raised lesions called polyps. Such polyps can be either benign (hyperplastic) or premalignant (dysplastic, termed adenomas), but definitive pathological diagnosis cannot be made unless a biopsy is performed. Hyperplastic polyps have no risk of evolving into malignancy, while dysplastic lesions have a predictable risk that increases with the size of the lesion.2 Dysplastic mucosa can persist in a latent phase for approximately before transforming; thus, a window of opportunity exists to prevent cancer if adequate techniques for early detection can be developed. In addition, numerous benign inflammatory polyps can be found in patients with inflammatory bowel disease that cannot be distinguished endoscopically from dysplastic polyps.3 Polyp removal during endoscopy incurs an increased risk of bleeding or perforation. In addition, it takes time to excise these lesions, thus prolonging the duration of sedation, and additional cost is incurred for histopathological examination of tissue. Thus, a technique that can identify dysplasia during endoscopy could have tremendous diagnostic value in medical care. The absorption of infrared light by the interatomic bonds in tissue macromolecules can be measured by techniques of Fourier transform infrared (FTIR) spectroscopy. This method has demonstrated promise for detection of cancer in a number of tissues including skin,4 cervix,5, 6, 7, 8 esophagus,9, 10 stomach,11, 12 lung,13 ovary,14 prostate,15 and colon.16, 17, 18, 19 FTIR spectra can reveal the molecular composition of the tissues without use of exogenous contrast agents.20, 21 Instead, a number of endogenous biomolecules can provide a characteristic spectrum of infrared absorption peaks constituting a molecular “fingerprint,” revealing the biochemical composition of the tissue. The use of FTIR to detect the presence and to measure the concentration of these specific molecules can provide important diagnostic information. Previously, we have measured FTIR spectra with attenuated total reflectance (ATR) by placing the tissue specimen in contact with a zinc selenide (ZnSe) crystal.10 In this mode, the infrared beam reflects at the interface between the crystal and the tissue below the critical angle and undergoes total internal reflection generating an evanescent wave with a depth of 2 to 3 microns into the tissue. This was used to determine the relative concentrations of the key endogenous biomolecules. Recently, flexible optical fibers have been developed with polycrystalline silver halide materials for collecting FTIR spectra that may be configured to be endoscope-compatible and can enable interrogation of the molecular composition of tissues during endoscopic procedures.22, 23, 24, 25, 26, 27, 28, 29, 30, 31 Fibers that transmit infrared light can be made with a variety of materials; however, silver halides, such as , have advantages for in vivo applications that include broad spectral transmission, good mechanical flexibility, low optical attenuation, long-term stability, nonhygroscopicity, and nontoxicity.24, 26, 28 Silver halide transmits infrared radiation from , depending on the fiber composition, with low attenuation ( at ),24 and can be fabricated into optical fibers that are several meters in length. Here, we demonstrate the use of an endoscope-compatible silver halide optical fiber to collect FTIR spectra from freshly excised specimens of colonic mucosa, including normal, hyperplastic, and dysplastic mucosa. Because dysplasia is one of the early steps in the transformation process toward cancer, the molecular changes at this stage are expected to have subtle spectral differences compared to that of normal colonic mucosa and hyperplasia. Thus, we will need sophisticated spectral processing algorithms to accurately classify these specimens. The mid-infrared (MIR) spectral range is evaluated because this regime contains numerous absorption maxima of fundamental rotational and vibrational modes of biochemical bonds.22 2.Methods2.1.Recruitment and Specimen ProcessingA total of 37 subjects undergoing routine screening colonoscopy with a mean age of (range ) were recruited for this study. The protocol was approved by the human subjects committee (IRB) at Stanford University and the Veterans Affairs Palo Alto Health Care System (VAPAHCS). After informed consent, routine screening was performed using a standard video colonoscope in the Endoscopy Unit at the VAPAHCS Hospital. When a polyp was identified, a pinch biopsy with jumbo forceps was taken from the lesion and an adjacent site of normal-appearing mucosa. The endoscopic appearance and anatomic location of each lesion biopsied was recorded. Immediately after resection, half of the biopsy specimen was placed on moist gauze over ice in a container for transport to the FTIR spectrometer. The other half of each specimen was sent for routine histopathological evaluation. Water was removed from the mucosal surface of the research specimen by blowing cool air for . FTIR spectra were then obtained, as described in the following. Following the collection of spectra, each piece of tissue was then rehydrated with normal saline (0.9%) and placed in 10% buffered formalin. The tissues were processed and cut in sections, stained with hematoxylin and eosin (H&E), and analyzed by two gastrointestinal pathologists (MRA, JMC). Only specimens whose morphology were consistent with the endoscopic findings (polypoid versus normal appearing) were included in the analysis. This resulted in spectra from of the original 37 subjects, including , 17, and 24 specimens of normal, hyperplasia, and dysplasia, respectively. 2.2.FTIR Spectra Collection from Colon SpecimensAn FTIR spectrometer (Nexus 470, Thermo Electron, Madison, Wisconsin) was used to measure the spectra and was continuously purged with air to remove water vapor and . A Multi-Loop-MIR silver halide optical fiber (Harrick Scientific, Pleasantville, New York) is coupled to a FiberMate module via SMA connectors. This module contains a set of elliptical (EM) and flat mirrors (FM), as shown in Fig. 1, that define the optical path, and is inserted into the signal arm of the Michelson interferometer used by the spectrometer. The input and output fibers are each one meter long and have a diameter of . The distal end of the fibers terminates into a handpiece that has an overall diameter of with a -diam replaceable fiber tip. The diameter of the handpiece is determined by the minimum bend radius of the silver halide fiber. In the attenuated total reflectance (ATR) configuration, the fiber tip is formed into a loop so that there is no physical interruption of the light path between the light delivery (input) and collection (output) fibers. The specimen is placed on a linear translational stage, which is adjusted until the distal tip of the fiber is in contact for spectral collection. A mercury cadmium telluride (MCT) detector cooled with liquid nitrogen and set at an autogain of 4 was used to record the spectra with a resolution of using 36 co-added scans for both the background and tissue. The extent of hydration was determined using a ratio between the Amide I and Amide II bands at 1650 and , respectively. This process was used to adjust the drying time to provide a consistent peak at . Fig. 1The optical fiber is coupled to the spectrometer via SMA connectors, and the optical path is defined by a set of elliptical (EM) and flat mirrors (FM). The input and output arms of the fiber are one meter long each with a diameter of , and the distal end terminates into a handpiece that has a diameter of and attaches to a replaceable loop tip. The photo shows the handpiece next to a quarter for size comparison. 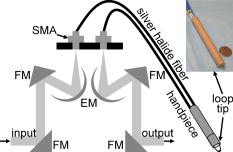 2.3.Spectral AnalysisThe FTIR spectra were evaluated using partial least squares (PLS) discriminant analysis,32, 33, 34 and the algorithms were implemented using the SIMPLS algorithm in the PLS̱Toolbox, a commercially available software package (Eigenvector Research, Inc., Wenatchee, Washington).35 The spectra were first preprocessed to remove variance that is not relevant for predicting class membership. The simplest preprocessing method is interval PLS, where a subset of wavelengths (wave numbers) is identified that provides a superior predictive value in comparison to use of all the wavelengths collected.35 Interval PLS discriminant analysis is performed by conducting an exhaustive search for the best combination of wavelengths. In this study, the fingerprint regime was divided into six subranges, and all possible combinations of subranges were tested. The two other basic categories of preprocessing are sample-wise and variable-wise methods. Sample-wise methods act on each spectrum one at a time to remove unwanted variance. Sample-wise pre-processing methods evaluated in this analysis include along-spectrum derivation, baseline subtraction, and normalization. Derivation was performed by applying the Savitsky-Golay algorithm,36 where a second-order polynomial was fit to a window around each value of the spectrum, and the value was then replaced with the second-order coefficient of the polynomial. Multiple window widths were also tested. When the derivation was not applied, a baseline spectrum, calculated as a low-order polynomial fit to the end regions of each spectrum, was subtracted. The relative values of spectral measurements often have greater predictive value than that of the absolute measurements. In this case, normalization is useful and helps all spectra to have an equal impact in the model. Three different types of metric were evaluated, with either the area under the curve of a spectrum, the vector length of a spectrum, or the maximum value of a spectrum undergoing normalization. Variable-wise preprocessing methods act on each spectral wavelength independently but require multiple samples for parameter estimation before they are implemented. The variable-wise preprocessing method tested in this study was unit-variance scaling. In this method, the value of a spectrum (or preprocessed spectrum, as unit-variance scaling always occurs as the last preprocessing step) at each wavelength was centered by subtracting the mean value across all samples and then was scaled to unit variance by dividing by the standard deviation. Each subrange was independently normalized to have the same area under the curve. The normalized values were then scaled to unit-variance by subtracting the mean and dividing by the standard deviation at each wavelength. By normalizing and scaling to unit-variance, the mixture of units due to a combination of baseline-subtraction in some subranges and second derivative pre-processing in others are eliminated. 2.4.Classification Algorithm DesignWe sought to identify diagnostic information from the subtle differences observed in the mid-infrared absorbance spectra collected from colonic mucosa, including normal, hyperplasia, and dysplasia. We developed a double binary detection algorithm that consists of one model to discriminate hyperplasia from dysplasia and another to distinguish normal from dysplasia. These distinctions are clinically relevant to patients that are found to have a very large number of polyps , where time limitations become a factor for lesion removal. The hyperplasia-versus-dysplasia and normal-versus-dysplasia models use spectra from respective tissue specimens for training. The total number of PLS discriminant analysis models tested for each is determined by the number of unique preprocessing combinations and exceeded . The accuracy of each component of the double binary model is assessed using leave-one-subject-out cross-validation to provide prospective analysis. The 30 most accurate hyperplasia-versus-dysplasia and normal-versus-dysplasia models were then selected for each. All 900 combinations of these two models were then tested in the double binary algorithm, again using leave-one-subject-out where spectra from each subject are rotated out for cross-validation. After completing the rotation, each spectrum has classification values for both hyperplasia-versus-dysplasia and normal-versus-dysplasia regardless of the true tissue class. Classification thresholds are varied in two dimensions, and the double binary algorithm is scored using histopathology as the “gold standard.” The final algorithm reported consists of the best hyperplasia-versus-dysplasia and normal-versus-dysplasia combination in terms of sensitivity and specificity for dysplasia. 3.ResultsThe average FTIR spectra collected with the silver halide optical fiber from specimens of colonic mucosa, including normal, hyperplasia, and dysplasia, are shown in Fig. 2 . An average ratio of (SEM) was found between the Amide I and Amide II peaks from the spectra and is used as an internal control. The spectra reveal subtle differences among the three histological classes in terms of location and height of the absorbance peaks. The spectra are divided into the optimal six subranges found from the algorithm described in Sec. 2 and are indicated by the dashed vertical lines. These subranges correspond to the groups of vibrational modes listed in Table 1 (spectral ranges 1 to 6). Representative histology (H&E) for the specimens of colonic mucosa studied are shown in Fig. 3 . Although some dessication artifact was observed, it did not prevent reliable classification of the mucosal samples into normal, hyperplastic, or dysplastic categories. Because of the possibility that the two halves of each biopsy specimen might differ histologically, the results reported herein are based on interpretation of the half-specimen from which the FTIR spectra were obtained. Fig. 2The mean mid-infrared absorbance spectra from biopsy specimens of colonic mucosa, including normal , hyperplasia , and dysplasia , collected with the silver halide optical fiber show subtle differences in average location and height of the absorbance peaks. 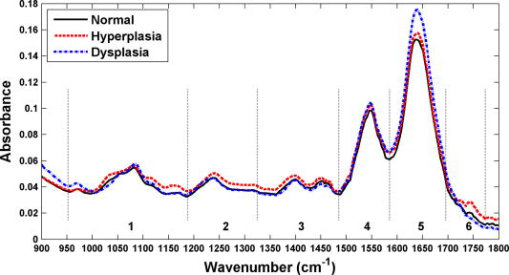 Fig. 3Histology of specimens examined after FTIR evaluation. (a) Normal—In contrast to mucosal specimens placed directly into formalin fixative, following FTIR evaluation, there is some disruption of surface epithelial cells as a result of dessication. In most instances, this did not preclude diagnostic assessment on the specific mucosal samples examined by FTIR. (b) Hyperplasia—The characteristic serrated profile of crypt and surface epithelial cells is present. (c) Dysplasia—Both adenomatous crypt and villiform architecture are present. Original magnification, . 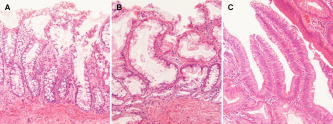 Table 1Primary mid-infrared absorption band assignments to tissue biomolecules in the 950cm−1to1800cm−1 spectral regime. The references cited are within 5cm−1 of those found in this study.10, 16, 17, 18, 37, 38, 39
The classification of each specimen using the best double binary algorithm is shown in the scatter plot in Fig. 4a, with the optimal thresholds for distinguishing hyperplasia-versus-dysplasia and normal-versus-dysplasia indicated by the solid vertical and dashed horizontal lines, respectively. Dysplasia was classified with sensitivity and specificity of 96% and 92%, respectively, and the accuracy and positive predictive value was 93% and 82%, respectively. In Fig. 4a, the four data points for normal (black crosses) in the lower-left corner represent false positives on the dysplasia-versus-hyperplasia decision line, and the data point for dysplasia (red circle) in the lower-right corner represents a false negative on the hyperplasia-versus-dysplasia decision line. The samples in the bottom-right quadrant of Fig. 4a are classified as nondysplasia by the hyperplasia-versus-dysplasia model. The specificity for the hyperplasia specimens alone was 93%. The receiver operator characteristic (ROC) curve for the double binary algorithm applied to all samples is shown in Fig. 4b, and reveals an optimal trade-off between sensitivity and specificity of 96% and 92%, respectively, for dysplasia and nondysplasia, shown by the black dot in the upper-left corner. The area under the curve was 0.953. Fig. 4Results of best two-part algorithm for classification of tissue. (a) Scatter plot shows true positives for dysplasia as red circles in shaded area, true negatives for nonadenoma as black crosses (normal) and blue squares (hyperplasia) in the unshaded area. Optimal thresholds for distinguishing hyperplasia-versus-dysplasia and normal-versus-dysplasia are indicated by the solid vertical and dashed horizontal lines, respectively. (b) ROC curve for best two-part algorithm shows optimal trade-off between sensitivity and specificity of 96% and 92% (black dot). The area under the curve is 0.953. 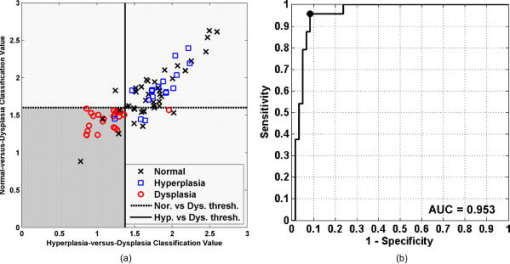 The spectral data after wavelength selection and preprocessing for input to the PLS discriminant analysis regression are shown by class average in Fig. 5a for the normal-versus-dysplasia model and in Fig. 5b for the hyperplasia-versus-dysplasia model. Note that the optimal model for normal-versus-dysplasia includes subrange 5, where Amide I and water are the predominant contributors to the absorbance peaks. This model used subranges 4 and 5 with baseline-removal only, while Savitzky-Golay second-derivative preprocessing was applied to subranges 2 and 6, respectively. The hyperplasia-versus-dysplasia model excluded subrange 5 only. Subrange 4 was used with baseline-removal, while the Savitzky-Golay second-derivative preprocessing was applied to subranges 1, 2, 3, and 6. The hyperplasia-versus-dysplasia model used the same normalization scheme as the normal-versus-dysplasia model and was also scaled to unit-variance. The normal-versus-dysplasia and hyperplasia-versus-dysplasia models both employed only two latent variables, suggesting that the scaling process removes extraneous variance that would otherwise be captured by additional latent variables. Fig. 5Preprocessed spectra and variable importance for projection (VIP) curves. The mean preprocessed spectra for (a) the normal-versus-dysplasia model and (b) the hyperplasia-versus-dysplasia model are shown. The spectra are divided into six optimal subranges as shown by the dotted vertical lines, and the peaks are assigned in Table 1. (c) VIP scores for the normal-versus-dysplasia and hyperplasia-versus-dysplasia models reflect the relative importance of each wave number to the model. 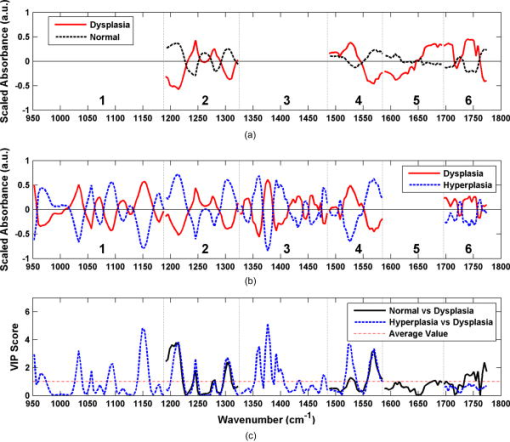 The variable importance for projection (VIP) curves for the normal-versus-dysplasia and hyperplasia-versus-dysplasia models are shown in Fig. 5c. This score summarizes the importance of each wave number for modeling both the variance of the predictors (spectra) and that of the response (classes).40 By definition, the average VIP score is one, and variables with values much greater than one are the most important for the model, while those with values much less than one make a negligible contribution. The VIP results show that the glycogen peaks for hyperplasia at 1026 and (Table 1) in sub-range 1 are important for distinguishing them from dysplasia and that the Amide I and lipid peaks of subranges 5 and 6 are relatively unimportant. While these spectral differences can be clearly discerned in the average spectra of Fig. 2, the differences in protein, lipid, and phosphate peaks of subranges 2, 3, and 4 are much more subtle in the average spectra. However after preprocessing, these differences are amplified, and these peaks are clearly valuable for classification. 4.DiscussionHere, we demonstrate the use of an optical fiber to collect FTIR spectra remotely from freshly excised colonic mucosa and find that adequate signal could be obtained with use of a silver halide fiber that is transparent in the mid-infrared regime, a sensitive liquid nitrogen cooled MCT detector, and an efficient FTIR spectrometer. While the fiber used in this study is not sufficiently long or thin enough to pass through a standard medical colonoscope, there are specialty instruments available, including therapeutic sigmoidoscopes, that have comparable lengths and channel diameters. The fiber is sealed within a protective jacket, and the exposed loop tip was replaced after several measurements to prevent degradation of signal. Because the specimens were studied immediately after excision, we expect that these results will be representative of that found in viable colonic mucosa. We observed subtle differences in the average location and magnitude of the absorbance peaks among normal, hyperplasia, and dysplasia in the fingerprint region between 950 and . Regression of these datasets is challenging because they are underdetermined and collinear. That is, there are more spectral peaks that exist than there are spectra in each class, and a high correlation occurs between absorbance maxima for unique biomolecules. Nonetheless, we were able to tease out these subtle spectral differences by performing an exhaustive search for the optimal subrange intervals to be used in the PLS discriminant analysis with rigorous spectral preprocessing techniques. In the end, we were able to develop a double binary algorithm that could distinguish normal from dysplasia and hyperplasia from dysplasia with high sensitivity and specificity. We are able to simplify this complex spectral dataset into a handful of latent variables that could then be evaluated by PLS discriminant analysis. The underlying assumption of this analysis is that the observations are generated by a process driven by a small number of latent (not directly observed) variables, and the data measured can be projected onto these latent variables by maximizing the covariance of predictor variables (spectral absorbances) and responses (class membership). The number of latent variables is limited to four in this study to avoid overfitting the spectra. Residual water can be a serious interference with FTIR spectra. However, with attenuated total reflectance (ATR), we need to remove water only within the most superficial 2- to 3-micron layer of the tissue. This depth is much less than the dimension of a single cell, so the removal of water over this layer is feasible. As a consequence, we were able to address the spectral peak associated with water, a major source of absorbance in tissue. Our model for distinguishing hyperplasia from dysplasia could completely eliminate this effect by avoiding use of subrange 5 [Fig. 5b]. In addition, we can provide physiological explanation to interpret the spectral changes observed among the three classes of colonic mucosa with use of Table 1. Our spectra provides a unique “snapshot” of the biochemical composition of the mucosa at different steps along the cancer transformation process.20 The absorbance peak for RNA rises relative to that of DNA. Some of these spectral differences are apparent in Fig. 2, while others are not visually obvious. We also found that in addition to the well-known glycogen, nucleic acid, and amide protein peaks, there is useful diagnostic information in the lipids and weaker protein vibrations. In particular, it is notable that subranges 1 (glycogen and nucleic acids) and 3 (proteins and lipids) are not included in the normal-versus-dysplasia model at all, while they are important components of the hyperplasia-versus-dysplasia model, suggesting that these biomolecules have an important role in cellular proliferation. For future development of this technology, the use of a classification algorithm that minimizes the amount of information required for calibration transfer between the optical fiber instruments is desirable. With the use of unit-variance scaling, a mean and standard deviation at each wavelength must be included as part of the calibration transfer, in addition to the regression coefficients. However, unit-variance scaling was found to provide clearly better performance. If the additional burden upon calibration transfer presented by unit-variance scaling is not significant even in the production of a large number of optical fiber instruments, then unit-variance scaling should be employed as a preprocessing component in the algorithm utilized by this technique. 5.ConclusionsThese results demonstrate that collection and use of FTIR spectra with an endoscope-compatible optical fiber is feasible for distinguishing premalignant colonic mucosa and that the relatively simple spectral biomarkers used to identify dysplasia can potentially be measured in vivo with continued development of this technology. These exciting findings suggest that continued development of an endoscope compatible instrument for collecting FTIR spectra to identify premalignant mucosa in the colon is feasible and has implications for evaluating histopathology in situ as a novel approach for early detection of cancer in the colon that may be generalized to hollow organs. AcknowledgmentsThis work was funded in part by grants from the National Institutes of Health, including K08 DK67618, P50 CA93990, and U54 CA136429 (TDW), and the Department of Defense USAMRCMC W81XWH-05-C-0086 (RW), by the Stanford Infrared Science and FEL Center as a grant from the U.S. Air Force Office of Sponsored Research (F9550-04-1-0075; CHC) and U54 CA105296 (JMC and CHC). References, Cancer Facts and Figures 2008,
(2008) Google Scholar
C. Liu and J. M. Crawford, Robbins and Cotran Pathologic Basis of Disease, 7th ed.Saunders, Philadelphia
(2005). Google Scholar
D. T. Rubin, J. A. Rothe, J. T. Hetzel, R. D. Cohen, and S. B. Hanauer,
“Are dysplasia and colorectal cancer endoscopically visible in patients with ulcerative colitis?,”
Gastrointest. Endosc., 65 998
–1004
(2007). https://doi.org/10.1016/j.gie.2006.09.025 0016-5107 Google Scholar
S. F. Chew, B. R. Wood, C. Kanaan, J. Browning, D. MacGregor, I. D. Davis, J. Cebon, B. D. Tait, and D. McNaughton,
“Fourier transform infrared imaging as a method for detection of HLA class I expression in melanoma without the use of antibody,”
Tissue Antigens, 69 252
–258
(2007). https://doi.org/10.1111/j.1399-0039.2006.00775.x 0001-2815 Google Scholar
S. Mark, R. K. Sahu, K. Kantarovich, A. Podshyvalov, H. Guterman, J. Goldstein, R. Jagannathan, S. Argov, and S. Mordechai,
“Fourier transform infrared microspectroscopy as a quantitative diagnostic tool for assignment of premalignancy grading in cervical neoplasia,”
J. Biomed. Opt., 9 558
–567
(2004). https://doi.org/10.1117/1.1699041 1083-3668 Google Scholar
A. Podshyvalov, R. K. Sahu, S. Mark, K. Kantarovich, H. Guterman, J. Goldstein, R. Jagannathan, S. Argov, and S. Mordechai,
“Distinction of cervical cancer biopsies by use of infrared microspectroscopy and probabilistic neural networks,”
Appl. Opt., 44 3725
–3734
(2005). https://doi.org/10.1364/AO.44.003725 0003-6935 Google Scholar
M. J. Romeo, B. R. Wood, M. A. Quinn, and D. McNaughton,
“Removal of blood components from cervical smears: implications for cancer diagnosis using FTIR spectroscopy,”
Biopolymers, 72 69
–76
(2003). https://doi.org/10.1002/bip.10284 0006-3525 Google Scholar
B. R. Wood, M. A. Quinn, F. R. Burden, and D. McNaughton,
“An investigation into FTIR spectroscopy as a biodiagnostic tool for cervical cancer,”
Biospectroscopy, 2 143
–153
(1996). https://doi.org/10.1002/(SICI)1520-6343(1996)2:3<143::AID-BSPY1>3.0.CO;2-9 1075-4261 Google Scholar
J. S. Wang, J. S. Shi, Y. Z. Xu, X. Y. Duan, L. Zhang, J. Wang, L. M. Yang, S. F. Weng, and J. G. Wu,
“FT-IR spectroscopic analysis of normal and cancerous tissues of esophagus,”
Aquat. Microb. Ecol., 9 1897
–1899
(2003). 0948-3055 Google Scholar
T. D. Wang, G. Triadafilopoulos, J. M. Crawford, L. R. Dixon, T. Bhandari, P. Sahbaie, S. Friediand, R. Soetikno, and C. H. Contag,
“Detection of endogenous biomolecules in Barrett’s esophagus by Fourier transform infrared spectroscopy,”
Proc. Natl. Acad. Sci. U.S.A., 104 15864
–15869
(2007). https://doi.org/10.1073/pnas.0707567104 0027-8424 Google Scholar
N. Fujioka, Y. Morimoto, T. Arai, and M. Kikuchi,
“Discrimination between normal and malignant human gastric tissues by Fourier transform infrared spectroscopy,”
Cancer Detect. Prev., 28 32
–36
(2004). https://doi.org/10.1016/j.cdp.2003.11.004 0361-090X Google Scholar
Q. B. Li, X. J. Sun, Y. Z. Xu, L. M. Yang, Y. F. Zhang, S. F. Weng, J. S. Shi, and J. G. Wu,
“Diagnosis of gastric inflammation and malignancy in endoscopic biopsies based on Fourier transform infrared spectroscopy,”
Clin. Chem., 51 346
–350
(2005). https://doi.org/10.1373/clinchem.2004.037986 0009-9147 Google Scholar
K. Yano, S. Ohoshima, Y. Gotou, K. Kumaido, T. Moriguchi, and H. Katayama,
“Direct measurement of human lung cancerous and noncancerous tissues by Fourier transform infrared microscopy: can an infrared microscope be used as a clinical tool?,”
Anal. Biochem., 287 218
–225
(2000). https://doi.org/10.1006/abio.2000.4872 0003-2697 Google Scholar
C. M. Krishna, G. D. Sockalingum, R. A. Bhat, L. Venteo, P. Kushtagi, M. Pluot, and M. Manfait,
“FTIR and Raman microspectroscopy of normal, benign, and malignant formalin-fixed ovarian tissues,”
Anal. Bioanal. Chem., 387 1649
–1656
(2007). https://doi.org/10.1007/s00216-006-0827-1 1618-2642 Google Scholar
E. Gazi, J. Dwyer, P. Gardner, A. Ghanbari-Siahkali, A. P. Wade, J. Miyan, N. P. Lockyer, J. C. Vickerman, N. W. Clarke, J. H. Shanks, L. J. Scott, C. A. Hart, and M. Brown,
“Applications of Fourier transform infrared microspectroscopy in studies of benign prostate and prostate cancer. A pilot study,”
J. Pathol., 201 99
–108
(2003). https://doi.org/10.1002/path.1421 0022-3417 Google Scholar
S. Argov, J. Ramesh, A. Salman, I. Sinelnikov, J. Goldstein, H. Guterman, and S. Mordechai,
“Diagnostic potential of Fourier-transform infrared microspectroscopy and advanced computational methods in colon cancer patients,”
J. Biomed. Opt., 7 248
–254
(2002). https://doi.org/10.1117/1.1463051 1083-3668 Google Scholar
S. Argov, R. K. Sahu, E. Bernshtain, A. Salman, G. Shohat, U. Zelig, and S. Mordechai,
“Inflammatory bowel diseases as an intermediate stage between normal and cancer: an FTIR-microspectroscopy approach,”
Biopolymers, 75 384
–392
(2004). https://doi.org/10.1002/bip.20154 0006-3525 Google Scholar
B. Rigas, S. Morgello, I. S. Goldman, and P. T. T. Wong,
“Human colorectal cancers display abnormal Fourier-transform infrared-spectra,”
Proc. Natl. Acad. Sci. U.S.A., 87 8140
–8144
(1990). https://doi.org/10.1073/pnas.87.20.8140 0027-8424 Google Scholar
S. Takahashi, A. Satomi, K. Yano, H. Kawase, T. Tanimizu, Y. Tuji, S. Murakami, and R. Hirayama,
“Estimation of glycogen levels in human colorectal cancer tissue: relationship with cell cycle and tumor outgrowth,”
World J. Gastroenterol., 34 474
–480
(1999). 1007-9327 Google Scholar
P. G. L. Andrus and R. D. Strickland,
“Cancer grading by Fourier transform infrared spectroscopy,”
Biospectroscopy, 4 37
–46
(1998). https://doi.org/10.1002/(SICI)1520-6343(1998)4:1<37::AID-BSPY4>3.0.CO;2-P 1075-4261 Google Scholar
R. K. Sahu, S. Argov, A. Salman, U. Zelig, M. Huleihel, N. Grossman, J. Gopas, J. Kapelushnik, and S. Mordechai,
“Can Fourier transform infrared spectroscopy at higher wavenumbers (mid IR) shed light on biomarkers for carcinogenesis in tissues?,”
J. Biomed. Opt., 10 054017
(2005). https://doi.org/10.1117/1.2080368 1083-3668 Google Scholar
U. Bindig, I. Gersonde, M. Meinke, Y. Becker, and G. Muller,
“Fiber-optic IR-spectroscopy for biomedical diagnostics,”
Spectroscopy Int. J., 17 323
–344
(2003). Google Scholar
U. Bindig, M. Meinke, I. Gersonde, O. Spector, I. Vasserman, A. Katzir, and G. Muller,
“IR-biosensor: flat silver halide fiber for bio-medical sensing?,”
Sens. Actuators B, 74 37
–46
(2001). https://doi.org/10.1016/S0925-4005(00)00709-7 0925-4005 Google Scholar
U. Bindig, and G. Muller,
“Fiber-optic laser-assisted infrared tumour diagnostics (FLAIR),”
J. Phys. D, 38 2716
–2731
(2005). https://doi.org/10.1088/0022-3727/38/15/026 0022-3727 Google Scholar
U. Bindig, H. Winter, W. Wasche, K. Zelianeos, and G. Muller,
“Fiber-optical and microscopic detection of malignant tissue by use of infrared spectrometry,”
J. Biomed. Opt., 7 100
–108
(2002). https://doi.org/10.1117/1.1412439 1083-3668 Google Scholar
C. Charlton, A. Katzir, and B. Mizaikoff,
“Infrared evanescent field sensing with quantum cascade lasers and planar silver halide waveguides,”
Anal. Chem., 77 4398
–4403
(2005). https://doi.org/10.1021/ac048189a 0003-2700 Google Scholar
S. M. Chiacchiera, and E. M. Kosower,
“Deuterium-exchange on micrograms of proteins by attenuated total reflection fourier-transform infrared-spectroscopy on silver-halide fiber,”
Anal. Biochem., 201 43
–47
(1992). https://doi.org/10.1016/0003-2697(92)90171-3 0003-2697 Google Scholar
Z. Hammody, M. Huleihel, A. Salman, S. Argov, R. Moreh, A. Katzir, and S. Mordechai,
“Potential of ‘flat’ fibre evanescent wave spectroscopy to discriminate between normal and malignant cells in vitro,”
J. Microsc., 228 200
–210
(2007). https://doi.org/10.1111/j.1365-2818.2007.01840.x 0022-2720 Google Scholar
Y. Matsuura, S. Kino, E. Yokoyama, T. Katagiri, H. Sato, and H. Tashiro,
“Flexible fiber-optics probes for Raman and FT-IR remote spectroscopy,”
IEEE J. Sel. Top. Quantum Electron., 13 1704
–1708
(2007). https://doi.org/10.1109/JSTQE.2007.910111 1077-260X Google Scholar
A. Ring, V. Schreiner, H. Wenck, K. P. Wittern, L. Kupper, and R. Keyhani,
“Mid-infrared spectroscopy on skin using a silver halide fiber probe in vivo,”
Skin Res. Technol., 12 18
–23
(2006). https://doi.org/10.1111/j.0909-725X.2006.00130.x 0909-752X Google Scholar
S. Simhony, E. M. Kosower, and A. Katzir,
“Fourier-transform infrared-spectra of aqueous protein mixtures using a novel attenuated total internal reflectance cell with infrared fibers,”
Biochem. Biophys. Res. Commun., 142 1059
–1063
(1987). https://doi.org/10.1016/0006-291X(87)91522-1 0006-291X Google Scholar
S. Dejong,
“SIMPLS—an alternative approach to partial least-squares regression,”
Chemom. Intell. Lab. Syst., 18 251
–263
(1993). https://doi.org/10.1016/0169-7439(93)85002-X 0169-7439 Google Scholar
P. Geladi, and B. R. Kowalski,
“Partial least-squares regression—a tutorial,”
Anal. Chim. Acta, 185 1
–17
(1986). https://doi.org/10.1016/0003-2670(86)80028-9 0003-2670 Google Scholar
S. Wold, A. Ruhe, H. Wold, and W. J. Dunn,
“The collinearity problem in linear-regression—the partial least-squares (PLS) approach to generalized inverses,”
SIAM (Soc. Ind. Appl. Math.) J. Sci. Stat. Comput., 5 735
–743
(1984). 0196-5204 Google Scholar
B. Wise, N. Gallagher, R. Bro, J. Shaver, W. Windig, and R. Koch, PLS̱Toolbox Version 4.0 Manual, 2006). Google Scholar
A. Savitzky and M. J. E. Golay,
“Smoothing and differentiation of data by simplified least squares procedures,”
Anal. Chem., 36 1627
–1639
(1964). https://doi.org/10.1021/ac60214a047 0003-2700 Google Scholar
E. Bogomolny, M. Huleihel, Y. Suproun, R. K. Sahu, and S. Mordechai,
“Early spectral changes of cellular malignant transformation using Fourier transform infrared microspectroscopy,”
J. Biomed. Opt., 12 024003
(2007). https://doi.org/10.1117/1.2717186 1083-3668 Google Scholar
M. J. German, A. Hammiche, N. Ragavan, M. J. Tobin, L. J. Cooper, S. S. Matanhelia, A. C. Hindley, C. M. Nicholson, N. J. Fullwood, H. M. Pollock, and F. L. Martin,
“Infrared spectroscopy with multivariate analysis potentially facilitates the segregation of different types of prostate cell,”
Biophys. J., 90 3783
–3795
(2006). https://doi.org/10.1529/biophysj.105.077255 0006-3495 Google Scholar
G. W. Lucassen, G. N. A. van Veen, and J. A. J. Jansen,
“Band analysis of hydrated human skin stratum corneum attenuated total reflectance Fourier transform infrared spectra in vivo,”
J. Biomed. Opt., 3 267
–280
(1998). https://doi.org/10.1117/1.429890 1083-3668 Google Scholar
I. G. Chong and J. Chi-Hyuck,
“Performance of some variable selection methods when multicollinearity is present,”
Chemom. Intell. Lab. Syst., 78 103
–112
(2005). https://doi.org/10.1016/j.chemolab.2004.12.011 0169-7439 Google Scholar
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||

