|
|
1.IntroductionImaging has become an indispensable tool in both the biological sciences and medicine. In the past two decades, there has been a huge increase in the number of imaging technologies and their applications. In particular, fluorescent imaging has been most rapidly adapted for in vitro and in vivo analysis of biological processes.1 Visualization of processes occurring in the complex environment of the cell and/or tissue needs an appropriate cellular marking procedure, and fluorescent dye has often been used as a straightforward technique. However, since fluorescent intensity may decrease during in vivo cellular proliferation, the use of fluorescent dye is not always suitable for in vivo imaging.2 Therefore, genetically encoded biological probes serve as stable and high-performance tools to visualize cellular fate in living animals. The development of genetic molecular tags such as green fluorescent protein (GFP) from the jellyfish (Aequorea victoria) has mostly accelerated the revolution occurring in this field over the past decade. In fact, GFP as the most popular biological light source has offered important opportunities for the investigation of a wide variety of biological processes in living cells and animals.3, 4 To obtain a stable optic cellular source, we developed GFP-transgenic (Tg) animals, including rats and rabbits.2, 5 These GFP-Tg animals have been employed as valuable cellular and organ sources for cell therapy and transplantation studies.6, 7, 8, 9 For instance, the cultured stem/progenitor cells of GFP-Tg rats transplanted into the spinal cord survived for a long time after transplantation (around 50 days), demonstrating a stable in vivo GFP expression.6 Moreover, application to oligodendrocyte replacement in models of white matter insult and disease also demonstrated the engraftment and survival of GFP-positive oligodendrocytes in the host white matter and cerebral cortex.9 Thus, the evidence suggests that GFP-Tg animals provide stable cell sources even after cell proliferation and differentiation. Recent advances in gene manipulation allowed the development of a variety of transgenic animals,10, 11, 12 and the procedure used for the microinjection of animal zygotes has continuously improved.13, 14 However, since expression of an injected expression vector depends on the integration site of the genome and the copy number, it is not always easy to obtain transgenics that ubiquitously express a particular cDNA, even under the general promoter.15 In fact, our previous results demonstrated that the tissue/organ expression profile depended on the line of established Tg animals.2 Nonetheless, the characteristics of established Tg animals led to high-demand animal resources for new biomedical translational research fields such as tissue engineering and regeneration medicine. Since the use of authentic transgenic technology through microinjection into zygotes is not always suitable with large transgenic animals, we aimed to generate cloned pigs on the basis of the somatic cell nuclear transfer method16, 17 and the supportive method for reconstructed embryos.18 In this study we generate new GFP-Tg Jinhua pigs, determine the expression profile of GFP emission light in the Tg pig, and demonstrate a stable reproductive performance. The GFP Tg pig may represent a valuable large-animal resource. 2.Experimental Materials and Methods2.1.Plasmid Construction and Polymerase Chain ReactionThe pEGFPneo expression vector [Fig. 1a ] was generated by insertion of the PGKneo polyA cassette from the pGKNeoPolyA/pUC19 plasmid into the pCX-GFP vector. The expression plasmids, pCX-GFP and pGKNeo poly-A, were kindly provided by Kashiwazaki (University of Tsukuba, Tsukuba, Japan). GFP cDNA was driven under the chicken β-actin promoter and cytomegalovirus immediate-early 1 gene enhancer.19 The liner HindIII fragment of the pEGFPneo expression plasmid was transferred into primary fibroblasts of Jinhua pigs (see next). Fig. 1Establishment of GFP-expressing Jinhua pigs. Representative scheme of the transgene composition. (a) The neomycin-resistant gene is driven under a mouse phosphoglycerate kinase 1 promoter (PGK), and the enhanced GFP cDNA is driven under the chicken β-actin promoter and cytomegalovirus immediate-early 1 gene enhancer (CAG).19 (b) Representative scheme of the creation of a cloned pig. (c) GFP expression in cells for nuclear transfer. A G418-resistant single fibroblast was grown in culture. Left panel, visible light; right panel, excitation light. Original magnification 100×. (d) Genotype inspection of a cloned pig. GFP-specific sequences were detected by PCR analysis. Tg, transgenic pig; WT, wild-type pig. (e) Representative image of the generated cloned pig under an excitation light. Arrows indicate parts with strong GFP expression (skin, oral and nasal mucosa, hoof wall).  For confirmation of GFP transduction into the cloned pig, polymerase chain reaction (PCR) was performed using AmpliTaq Gold polymerase (Applied Biosystems Incorporated, Foster City, California). The EGFP sequence was amplified using the following primers: forward, -TGA ACC GCA TCG AGC TGA AGG G- ; reverse, -TCC AGC AGG ACC ATG TGA TCG C- . PCR conditions for each set of primers included initial treatment at 94°C for 2 min, followed by 35 cycles consisting of denaturation at 94°C for 30 sec, annealing at 65°C for 30 sec, followed by extension at 72°C for 2 min. PCR products (307 bp) were analyzed on a 1.5% agarose gel. 2.2.Animals and Preparation of Transgenic Donor CellsChinese Jinhua pigs were maintained under an experimental protocol approved by the Judging Committee of Transgenic Experiments of Shizuoka Prefectural Swine and Poultry Research Center and Experimental Animal Ethics of Jichi Medical University. Primary fibroblasts from the skin of a 4-day-old female Jinhua pig were grown to confluency in a 100-mm tissue culture dish. Cells ( to ) were trypsinized and transduced with the liner pEGFPneo (10 μg) using an electroporation system [Gene Pulser II; Bio-Rad Company, Limited, Hercules, California; at 0.240 kV, 500 μF in 900 μl of phosphate buffered saline (PBS) without and ]. Electroporated cells were then cultured in a 100-mm-diam culture dish and maintained with Dulbecco modified Eagle medium (DMEM) (11965-092; Gibco, Carlsbad, California) containing 10% fetal bovine serum (FBS) and 150 μg/ml G418 geneticin (Gibco) at 37°C in a humidified atmosphere of 5% in air for 2 weeks. The cells were trypsinized and moved onto 48-well culture plates with one cell per well. GFP-expressing cells were cultured until confluency in 35-mm-diam culture dishes, and thereafter in 60-mm dishes over the course of 6 months. The cells were frozen before nuclear transfer. Donor cells for nuclear transfer were cultured until confluency in a 35-mm dish and produced a synchronized cell cycle by serum starvation (0.5% FBS-DMEM) for 6 days. 2.3.Somatic Cell Nuclear TransferMature oocytes and parthenogenotes were produced by methods previously described.18 Immature oocytes of ovaries collected from a local abattoir were cultured for 40 h, and the maturity of the oocytes was assessed under a stereoscopic microscope. Only oocytes that possessed a distinct first polar body were classified as reaching metaphase 2 and used for nuclear transfer. Nuclear transfer was performed using the microinjection method.16, 17 The nuclei were each introduced into a single enucleated oocyte by piezo-actuated microinjection. Electrostimulation was performed 48 h after the start of maturation (2 to 4 h after nucleus microinjection) in an activation medium containing 280-mM D-mannitol, 0.05-mM , 0.1-mM , and 0.01% (w/v) polyvinyl alcohol. Pulses were delivered to cells placed between two wire electrodes (1 mm apart) in a fusion dish (CUY5000P1, Nepa Gene Company, Limited, Ichikawa, Japan) by applying a single direct-current pulse of 150 kV/cm for a duration of 99 μsec. The stimulated oocytes were transferred to porcine zygote medium (PZM)20 supplemented with cytochalasin B (4 μg/ml) for 2 h to prevent cytokinesis, after which the culture was continued in PZM containing 0.3% bovine serum albumin at 38.5°C under 5% and 5% for 110 h. After this period, reconstructed embryos that developed into morula-blastocysts were transferred to the uteri of surrogate sows with parthenogenetic embryos developed at the same stage.18 2.4.Isolation and Culture of Mesenchymal Stromal Cells In Vitro Differentiation Assay, and Hepatocyte IsolationBone marrow cells (BMCs) from cloned pigs were harvested by flushing femurs with ice-cold PBS. Cells were filtered through a 70-μm nylon mesh and plated in or flasks with DMEM/F-12 (Gibco, Grand Island, New York) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. The cultures were kept in a humidified atmosphere containing 5% and 95% air at 37°C. Nonadherent cells were removed after 24 h. Adherent cells were trypsinized with 0.25% trypsin-EDTA (Gibco), harvested and then plated into new flasks at every 90% confluency. Adherent mesenchymal stromal cells (MSCs) from passage 2 were frozen in liquid nitrogen for future use. Early passage cells were examined for their capacity to differentiate in culture. For the in vitro differentiation assay, passage 4 MSCs were tested for their ability to differentiate into osteocytes and adipocytes.21 For adipocyte differentiation, cells were cultured with Differentiation Media Bullet Kit-Adipogenic (Lonza, Basel, Switzerland) according to the manufacturer’s instructions. MSCs were cultured in 6-well plates with MSC culture medium until they reached confluency. Cells were then exposed to three cycles of adipogenic induction medium alternating with adipogenic maintenance medium. Following three complete cycles of induction/maintenance, the MSCs were cultured for 7 more days in supplemented adipogenic maintenance medium. Cell differentiation to adipocytes was confirmed by Oil Red O (Muto Chemicals, Tokyo, Japan) staining. For osteogenic differentiation, cells were plated and the culture medium was replaced with Differentiation Media Bullet Kit-Osteogenic (Lonza) until confluence. Cells were stained with alizarin red S (Wako Pure Chemicals). Primary hepatocytes from swine liver were prepared by the conventional perfusion method with collagenase digestion. Hepatocytes were seeded at cells per dish in 10-cm-diam plastic dishes and cultured in William’s E complete medium (Gibco, Grand Island, New York) supplemented with 10% FBS, 1-nM insulin, and 1-nM dexamethasone in a humidified atmosphere of 5% air at 37°C. 2.5.Flow CytometryPeripheral blood cells were isolated from 10 ml of freshly drawn heparinized blood of pigs, and red cells were lysed in a buffer containing 155-mM , 10-mM and 0.1-mM EDTA. Precipitated white cells were resuspended in 0.1% FBS-PBS, and cells were analyzed using FACSCalibur (Beckton Dickinson, Mountain View, California) and FlowJo analysis software (Tree Star, San Carlos, California). 2.6.Histological Analysis, Nissl Staining, and Hematoxylin and Eosin stainingThree male GFP-Tg Jinhua cloned pigs were used for histological observations. Animals (8 to 36 months, weighing approximately 70 to 120 kg) were deeply anesthetized by inhalation of isoflurane (4% 1 L/min, Dinippon Pharmaceutical Company, Osaka, Japan) and intramuscular injection of domitor (0.6 mL, Meiji seika, Tokyo, Japan) and dormicum (0.6 mL, Astellas Pharma, Tokyo, Japan). Tissues (brain, pancreas, skeletal muscle, cardiac muscle, small intestine, stomach, liver, colon, and testis) were isolated from anesthetized Tg animals and fixed in 4% paraformaldehyde (Merck KGaA, Darmstadt, Germany) with 0.1-M phosphate buffer (pH 7.4). Tissue samples were soaked in 20% sucrose in PBS at 4°C for 2 to 4 days, frozen in OCT compound (Tissue-Tek, Sakura Finetechnical Company, Tokyo, Japan), and then sectioned at a thickness of 10 to 40 μm using a cryostat (Leica CM 1850, Leica Incorporated, Nussloch, Germany). Sections were then mounted onto silane-coated slides. To identify the cell architecture of the brain, delipidated brain sections were immersed in 0.1% solution of Cresylecht violet (Chroma-Gesellschaft, Munster, Germany) overnight at 37°C (Nissl staining). Slides were dehydrated through graded ethanol baths, delipidated in xylene, and then mounted with Mount-Quick (Daidosangyo Company, Tokyo, Japan). For hematoxylin and eosin (HE) staining, all samples except for brain were embedded in paraffin (Wako Pure Chemicals) and then sectioned at a thickness of 4 μm using a sliding microtome (REM-700, Yamato Kohki Industrial Company, Saitama, Japan). Sections were deparaffinized, rehydrated to water, and then stained with hematoxylin (Wako Pure Chemicals) and eosin (Wako Pure Chemicals). All frozen sections except for brain were counterstained with DAPI (Sigma Chemical, Saint Louis, Missouri). GFP fluorescence was examined using a fluorescent microscope Keyence BZ-9000 (Keyence, Tokyo, Japan). Micrographs were taken using a digital camera attached to the same microscope. Digital images were processed with Adobe Photoshop CS2 to adjust the final plates. 2.7.Survey of Reproductive and Growth Performance in Green Fluorescent Protein PigsA primary GFP Jinhua pig mated with wild-type Jinhua boars and farrowed the second generation of GFP-Tg piglets (three litters). Boars of the second generation mated with five wild-type Jinhua sows and the sows farrowed the third generation. For surveillance of reproductive and growth performance in GFP-Tg Jinhua pigs, pregnant pigs that conceived GFP pigs entered the farrowing unit 1 week before the expected date of parturition and were housed in an individual section (1.2 × 2.5 m) of a slated barn until weaning. Piglets were weaned at 30 days of age. After weaning, piglets were reared in pens (2.4 × 2.5 m) for each litter. Each pen had a feeder and water cup that allowed free access to feed (18% crude protein, 3080 kcal/kg digestible energy) and water throughout the experiment. The body weight of piglets was recorded every week until they reached two months of age. 2.8.Statistical AnalysesAll statistical analyses were performed with StatView software (Windows©version 5; SAS Institute Incorporated, Cary, North Carolina). Data concerning born-alive rate (number born alive/number of total born) and weaning rate (number of weaning piglets/number born alive) between GFP-positive and GFP-negative piglets were analyzed by a Fisher’s exact test. Data regarding body weight of piglets were analyzed using two-way factorial ANOVA and the Tukey-Kramer multiple range test. 3.Results3.1.Generation of Green Fluorescent Protein Expressing Cloned PigsPrimary fibroblasts from the skin (4-day-old female Jinhua pig) were isolated and grown for transgenic donor cells. The liner pEGFPneo plasmid DNA (10 μg) was transduced with primary fibroblasts using an electroporation system. After G418 selection for 2 weeks, 212 colonies (18.0%) of 1177-sorted cells were grown to confluence in 35-mm culture dishes. One of the cell lines that expressed a strong GFP signal was used for nuclear transfer [Fig. 1c]. 23 (9.2%) of 249 somatic cell nuclear transferred oocytes developed to the morula-blastocyst stage 110 h after nuclear transfer. These morula-blastocysts (average 7.7/recipient) were cotransferred with parthenogenetic embryos (average 17.3/recipient) into three recipient gilts, two of which became pregnant and each farrowed a total of two piglets (8.7%: ) at day 113 after nuclear transfer. Both of the two cloned Jinhua pigs possessed GFP-specific sequences according to PCR analysis [Fig. 1d]. Visual inspection under an excitation light demonstrated that GFP-derived fluorescence was clearly expressed in the skin, oral and nasal mucosa, and hoof wall of the transgenic pigs [Fig. 1e]. 3.2.Green Fluorescent Protein Expression Pattern in the Newly Created Cloned PigIn an effort to examine the expression pattern of newly generated cloned pigs, various organs were removed from the transgenic pigs and their macroscopic expression pattern was determined (Fig. 2 ). While skeletal muscle and pancreas showed strong GFP expression in the cloned pigs, expression in gastrointestinal tracts and eyes was weak in comparison. GFP signals in the kidney and liver of cloned pigs appeared moderate and heterogeneous. Fig. 2Macroscopic GFP expression profile in various organs from cloned pigs. Representative organs (skeletal muscle, heart, kidney, eye, liver, pancreas, small intestine, and colon) were removed from GFP-expressing pigs and macroscopically examined under a visible (left) or 489-nm excitation (right) light. Results derived from one of two independent experiments showing similar results. 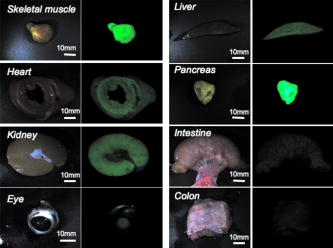 We further analyzed microscopic GFP expression patterns in various organs (Table 1 and Figs. 3, 4, 5 ). GFP expression sites in the central nervous system (CNS) are summarized in Fig. 3. Nisslstaining was also performed to identify the cell architecture of the brain [Figs. 4a, 4b, 4c, 4d, 4e, 4f, 4g, 4h, 4i, 4j, 4k]. Small-sized and round-shaped GFP-positive cell bodies were found in the olfactory bulb [Figs. 3a and 4d], lateral ventricle [Figs. 3b, 3c, 4g, 4h], and hippocampus [Figs. 3c and 4f]. In contrast, large-sized cell bodies were observed in the cerebellum [Fig. 4j]. Both large- and small-sized cell bodies were found in the medula oblongata [Fig. 4l]. GFP-positive fibers were found in the olfactory bulb [Figs. 4b and 4d], cerebellum [Fig. 4j], and medula oblongata [Fig. 4l]. Fig. 3Schematic illustration of GFP expression sites in the brain of cloned pigs. Coronal sections are illustrated from (a) rostral to (e) caudal. The green circle represents GFP-positive cell bodies and the red line indicates GFP-positive fibers. (Color online only.) 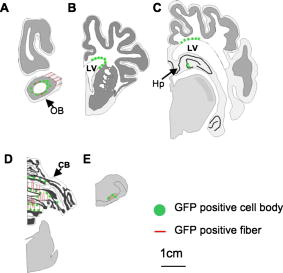 Fig. 4Photomicrographs of GFP fluorescence and Nissl-staining in the brain of cloned pigs. (a) to (d) olfactory bulb; (e) and (f) hippocampus; (g) and (h) lateral ventricle; (i) and (j) cerebellum; (k) and (l) medula oblongata. (a), (c), (e), (g), (i), and (k): Nissl staining. (b), (d), (f), (h), (j), and (k): 489-nm excitation light. 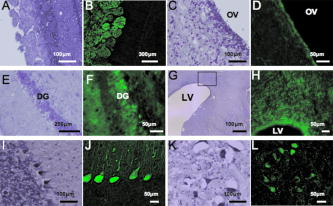 Fig. 5Microscopic GFP expression in representative tissue sections from cloned pigs other than brain. (a), (c), (e), (g), (i), (k), (m), (o), and (q) Representative tissue sections (skin, skeletal muscle, cardiac muscle, liver, pancreas, stomach, small intestine, colon, and testis) from GFP-expressing pigs were inspected under 489-nm excitation light HE staining: (b), (d), (f), (h), (j), (l), (n), (p), and (r). (s) and (t) Hemogram of peripheral blood from a GFP-Tg pig. Leukocytes (granulocytes and mononuclear cells) expressed GFP, although erythrocytes showed no expression. (s) 489-nm excitation light; (t) visible light. (u) Representative flow-cytometrical GFP-expression pattern in peripheral blood leukocytes. More than 90% of leukocytes were GFP-positive. WT, leukocytes from a wild-type pig; Tg, leukocytes from a GFP-transgenic pig. Results derived from one of two independent experiments showing similar results.  Table 1Expression profile of GFP-expressing Jinhua pigs. GFP expression was determined microscopically under a 489-nm excitation light.
GFP expression of the skin was predominant in the epidermis (the granular layer and stratum spinosum) and the hair follicle, but less so in the dermis [Figs. 5a and 5b]. Expression in skeletal [Figs. 5c and 5d] and cardiac muscle [Figs. 5e and 5f] appeared modestly heterogeneous, but most of the muscle fibers were GFP-positive. Regarding expression in the liver, parenchymal cells appeared GFP-positive and interstitial cells were GFP-negative [Figs. 5g and 5h]. Acinus cells in the pancreas were strongly GFP-positive [Figs. 5i and 5j]. In the gastrointestinal tract, GFP was heterogeneously expressed in the epithelium of the stomach, intestine, and colon [Figs. 5k, 5l, 5m, 5n, 5o, 5p]. Primary and secondary spermatocytes in the testis weakly expressed GFP, although sperms were almost totally GFP-negative [Figs. 5q and 5r]. Since it is important to know the fate of leukocytes in many biomedical studies, we examined GFP expression of peripheral blood cells from cloned pigs [Figs. 5s and 5t]. Leukocytes were GFP-positive, granulocytes exhibited a particularly strong expression of GFP, and mononuclear cells were also moderately GFP-positive. Notably, GFP expression of erythrocytes was definitely negative. FACS analysis also revealed strong GFP expression in peripheral leukocytes [Fig. 5u]. 3.3.Green Fluorescent Protein Expression in Potential Cell SourcesTo restore form and function to damaged tissues, a cell transplantation strategy has emerged as a potential therapeutic approach involving the use of autologous cells. Therefore, we examined GFP expression in the processed cells for transplantation (Fig. 6 ). Since mesenchymal stem cells (MSCs) possess a high expansion potential and genetic stability22 and can be easily isolated and transferred from the laboratory to the bedside, we first examined GFP expression in bone-marrow-derived MSCs. As shown in Fig. 6a, sufficient levels of GFP expression were observed in isolated MSCs. The cells rapidly proliferated and formed colonies, and GFP expression levels were not altered, even within several cell passages (data not shown). These cells were capable of differentiating successfully into osteocytes [Fig. 6c], but poorly into adipocytes [Fig. 6b]. These results demonstrate that GFP expression was stable in MSCs from cloned pigs and preferentially differentiated into osteocytes. We next addressed GFP expression in cultured parenchymal hepatocytes from the cloned pigs [Figs. 6d, 6e, 6f]. Sufficient levels of GFP expression were observed in proliferating hepatocytes, but not in adherent hepatocytes with the contact inhibition. A similar phenomenon was observed in Langerhans islets isolated from cloned pig pancreas [Figs. 6g, 6h, 6i]. Fig. 6GFP expression in potential cellular sources. (a) Strong GFP expression in bone-marrow-derived MSCs from the GFP-transgenic pig. Under appropriate differentiation conditions, MSCs were capable of differentiating into (c) adipocytes (stained with Oil red O for lipid droplets) and (d) osteocytes (stained with alizarin red for mineral deposition). Original magnification 20×. (d), (e), and (f) GFP expression in cultured parenchymal hepatocytes from cloned pigs. (d) Visible light, (e) excitation light. (f) Representative merged image of GFP-expressing cultured hepatocytes. (g) Visual inspection of the pancreas of cloned pigs (visible light). Langerhans islets isolated from the cloned pig pancreas were cultured and GFP expression was examined under (h) a visible and (i) 489-nm excitation light (original magnification 200×). (Color online only.) 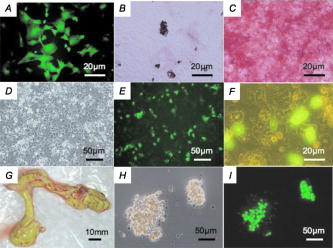 3.4.Reproductive Performance of Green Fluorescent Protein Jinhua PigsA piglet of GFP-cloned piglets died 2 days after birth, but the remainder grew up normally and expressed estrus. The primary GFP-cloned female mated with a wild-type boar by artificial insemination and farrowed the second generation of GFP pigs. The average litter size and average number of weaned piglets in three parities were 11 and 9.7, respectively [Fig. 7a ]. To examine the GFP expression rate in the second generation, PCR analysis and inspection by excitation light were also performed. The results showed that the GFP expression rate was 51.5% . Two boars of second-generation GFP Jinhua pigs mated with five females of wild-type pigs. The third generations (total 52 piglets) were born and 22 (42.3%) of these individuals were GFP-positive in genotype and phenotype. These results indicate that reproductive activity is successfully maintained in the established GFP-cloned Jinhua pigs, and that the introduced GFP transgene can be stably transmitted to pigs in subsequent generations. Fig. 7Reproductive and growth performance of GFP-expressing pigs. (a) Summary of average litter size and average number of weaned piglets in three parities. (b) Growth performance of GFP cloned pigs. GFP, GFP cloned pigs. Wild, wild-type pigs; ♀, female; ♂, male. The female is heavier than the male after reaching 4 months of age (4, 6, and 7 weeks: *, p < 0.05; 8 weeks: **, p < 0.01). 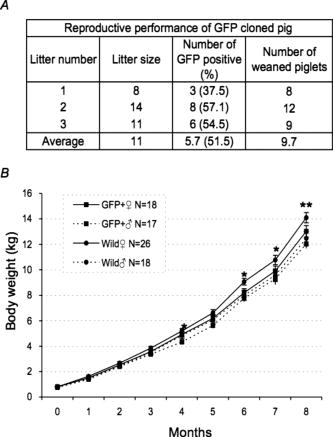 3.5.Growth Performance of Green Fluorescent Protein Transgenic PigsWe further evaluated the influence of the GFP transgene on growth performance of GFP-Tg pigs. A total of 85 piglets (33 of second-generation and 52 of third-generation piglets) were examined using body weight. The born-alive rate in GFP-positive and -negative pigs was 89.7% and 95.7% , respectively. The weaning rate in GFP positive and negative piglets was 100% and 97.7% , respectively. There were no differences between GFP-positive and -negative piglets for born-alive and weaning rates. The body weights of 79 born-alive piglets did not differ between GFP-positive and -negative piglets, while the female was heavier than the male after 4 months of age (4, 6, and 7 weeks: P < 0.05; 8 weeks: P < 0.01) [Fig. 7b]. As with wild-type Jinhua pigs, the body size of GFP-positive piglets reached plateau levels around 24 months (male: 96.0 ± 4.7 kg [n = 8]; female: 107.6 ± 13.3 kg [n = 12]), and the miniature size was maintained. These results demonstrate that the GFP transgene has less effect on growth performance of Tg pigs. 4.DiscussionsWe created new GFP-expressing pigs using a somatic cell cloning technique. The remarkable features presented as an imaging source include: 1. the born GFP-Tg pig demonstrated an organ/tissue-dependent expression pattern; 2. it displayed normal growth and fertility; and 3. the introduced GFP gene was transmitted to pigs in subsequent generations. The new GFP-expressing Jinhua pigs could provide new cellular/tissue light sources for biological imaging. The Jinhua pig is a kind of indigenous Chinese pig.23 The growth and reproductive traits of this pig were evaluated using microsatellite markers.24, 25 It has been reported that the mean litter size is 11 piglets, and that the pig shows premature growth and high multiplication.26, 27 The adult body weight of Jinhua pigs ranges from 90 to 110 kg (data not shown). Although Jinhua pigs have sufficient reproductive ability, the pigs have not been used as a commercial base resource [due to the low carcass lean content (29 to 30%) compared with large white pigs (53 to 54%).27 However, since Jinhua pigs exhibit a middle body size similar to the human body size, organs from this breed may represent an appropriate organ resource of xeno-transplantation for humans. GFP expression was observed in various organs in this study, although expression levels differed between tissues/organs. It is very important for researchers to know the expression profile for various tissues/organs, because it is impossible to regulate the integration site and the copy number of transgenes into the genome in transgenic animals.2, 22 Since our previous results demonstrated that naked GFP-expression plasmid DNA was successfully expressed in the pig liver,28 this general promoter and enhancer could be driven in various tissues/organs of cloned pigs. In particular, GFP expression was strong in the skeletal muscle, pancreas, heart, and kidney. These organs are potentially available for organ transplantation experiments as in the case of rats.2 In terms of experimental cell therapy, it may also be possible to use neural progenitor cells. However, the culture system for pig neural progenitor cells may be required for future in vivo animal experiments, whereas the system for cell culture and large-animal experiments remains to be established. At the very least, MSCs, hepatocytes, and islet cells of the pancreas seem to be available for cell transplantation studies. The present study demonstrated that an introduced GFP gene was very stable in Jinhua pigs, and that it was transmitted to the second and third generations. The first sow transmitted the gene to the second generation (both male and female pigs), and the second-generation male pig transmitted it to the third generation (both male and female pigs), suggesting a stable genotype and phenotype transmission. This also allows us to preserve this animal source as a fertilized egg and/or sperm. The cells used for the nuclear transfer in this study were grown in culture from one cell of fibroblasts, and they had eventually divided around 20 times. Nevertheless, the evidence that normal individual pigs were successfully generated suggests that somatic cells of the pig may be resistant to gene alterations, including epigenetic mutations, in comparison with other animals such as cows.29, 30, 31, 32 This may represent a species-specific characteristic. As indicated in Fig. 7a, the GFP-transmission rate was around 50% from the sow to the second generation. This suggests the transduced gene was integrated into one portion of a chromosome. It was thought that gene transmission by this somatic cell cloning technique was almost totally equivalent to the case of intracytoplasmic sperm injection-mediated DNA transfer.33 This study also demonstrated that the GFP gene did not influence the growth or reproduction of the cloned pig greatly. Since the GFP-Tg pigs were maintained in hemizygous conditions in the present study, any influence under homozygous conditions remains to be elucidated. In conclusion, new GFP-expressing pigs showed normal growth and stable reproductive activity. Taking advantage of stable light sources, the GFP expression profile in these pigs may provide useful imaging information in research fields such as tissue engineering, experimental regenerative medicine, and transplantation. AcknowledgmentsWe would like to thank S. Kashiwazaki (University of Tsukuba, Japan) for donating pCX-GFP and pGKNeoPolyA/pUC19 plasmids. This study was supported by a research grant to author Kawarasaki from Shizuoka Prefecture, a grant to Kobayashi from the Japan Society for the Promotion of Science (project number 18300143; 2006-2008), and Health and Labour Science Research grants from the Ministry of Health, Labor and Welfare, and by a grant from the “Strategic Research Platform” Project for Private Universities: matching fund subsidy from the Ministry of Education, Culture, Sports, Science and Technology of Japan (2008). ReferencesR. Weissleder and M. J. Pittet,
“Imaging in the era of molecular oncology,”
Nature (London), 452
(7187), 580
–589
(2008). https://doi.org/10.1038/nature06917 0028-0836 Google Scholar
T. Murakami and E. Kobayashi,
“Color-engineered rats and luminescent LacZ imaging: a new platform to visualize biological processes,”
J. Biomed. Opt., 10
(4), 41204
(2005). https://doi.org/10.1117/1.2007947 1083-3668 Google Scholar
J. Lippincott-Schwartz and G. H. Patterson,
“Development and use of fluorescent protein markers in living cells,”
Science, 300
(5616), 87
–91
(2003). https://doi.org/10.1126/science.1082520 0036-8075 Google Scholar
R. M. Hoffman,
“The multiple uses of fluorescent proteins to visualize cancer in vivo,”
Nat. Rev. Cancer, 10
(10), 796
–806
(2005). https://doi.org/10.1038/nrc1717 1474-175X Google Scholar
R. Takahashi, T. Kuramochi, K. Aoyagi, S. Hashimoto, I. Miyoshi, N. Kasai, Y. Hakamata, E. Kobayashi, and M. Ueda,
“Establishment and characterization of CAG/EGFP transgenic rabbit line,”
Transgenic Res., 16
(1), 115
–120
(2007). https://doi.org/10.1007/s11248-006-9043-1 0962-8819 Google Scholar
A. K. Mothe, I. Kulbatski, R. L. van Bendegem, L. Lee, E. Kobayashi, A. Keating, and C. H. Tator,
“Analysis of green fluorescent protein expression in transgenic rats for tracking transplanted neural stem/progenitor cells,”
J. Histochem. Cytochem., 53
(10), 1215
–1226
(2005). https://doi.org/10.1369/jhc.5A6639.2005 0022-1554 Google Scholar
Y. Yamaguchi, T. Kubo, T. Murakami, M. Takahashi, Y. Hakamata, E. Kobayashi, S. Yoshida, K. Hosokawa, K. Yoshikawa, and S. Itami,
“Bone marrow cells differentiate into wound myofibroblasts and accelerate the healing of wounds with exposed bones when combined with an occlusive dressing,”
Breast Cancer Res. Treat., 152
(4), 616
–622
(2005). 0167-6806 Google Scholar
T. Shimizu, H. Sekine, J. Yang, Y. Isoi, M. Yamato, A. Kikuchi, E. Kobayashi, and T. Okano,
“Polysurgery of cell sheet grafts overcomes diffusion limits to produce thick, vascularized myocardial tissues,”
FASEB J., 20
(6), 708
–710
(2006). 0892-6638 Google Scholar
J. S. Francis, A. Olariu, E. Kobayashi, and P. Leone,
“GFP-transgenic Lewis rats as a cell source for oligodendrocyte replacement,”
Exp. Neurol., 205
(1), 177
–189
(2007). https://doi.org/10.1016/j.expneurol.2007.01.039 0014-4886 Google Scholar
C. V. Hunter, L. S. Tiley, and H. M. Sang,
“Developments in transgenic technology: applications for medicine,”
Trends Mol. Med., 11
(6), 293
–298
(2005). https://doi.org/10.1016/j.molmed.2005.04.001 Google Scholar
L. M. Houdebine,
“Transgenic animal models in biomedical research,”
Methods Mol. Biol., 360 163
–202
(2007). 1064-3745 Google Scholar
H. Niemann, X. C. Tian, W. A. King, and R. S. Lee,
“Epigenetic reprogramming in embryonic and foetal development upon somatic cell nuclear transfer cloning,”
Reproduction, 135
(2), 151
–163
(2008). https://doi.org/10.1530/REP-07-0397 Google Scholar
L. Tesson, J. Cozzi, S. Menoret, S. Remy, C. Usal, A. Fraichard, and I. Anegon,
“Transgenic modification of the rat genome,”
Transgenic Res., 14
(5), 531
–546
(2005). https://doi.org/10.1007/s11248-005-5077-z 0962-8819 Google Scholar
C. T. Dann and D. L. Garbers,
“Production of knockdown rats by lentiviral transduction of embryos with short hairpin RNA transgenes,”
Methods Mol. Biol., 450 193
–209
(2008). https://doi.org/10.1007/978-1-60327-214-8_14 1064-3745 Google Scholar
T. Nakanishi, A. Kuroiwa, S. Yamada, A. Isotani, A. Yamashita, A. Tairaka, T. Hayashi, T. Takagi, M. Ikawa, Y. Matsuda, and M. Okabe,
“FISH analysis of 142 EGFP transgene integration sites into the mouse genome,”
Genomics, 80
(6), 564
–574
(2002). https://doi.org/10.1006/geno.2002.7008 0888-7543 Google Scholar
A. Onishi, M. Iwamoto, T. Akita, S. Mikawa, K. Takeda, T. Awata, H. Hanada, and A. C. F. Perry,
“Pig cloning by microinjection of fetal fibroblast nuclei,”
Science, 289
(5482), 1188
–1190
(2000). https://doi.org/10.1126/science.289.5482.1188 0036-8075 Google Scholar
T. Wakai, S. Sugimura, K. Yamanaka, M. Kawahara, H. Sasada, H. Tanaka, A. Ando, E. Kobayashi, and E. Sato,
“Production of viable cloned miniature pig embryos using oocytes derived from domestic pig ovaries,”
Cloning Stem Cells, 10
(2), 249
–262
(2008). https://doi.org/10.1089/clo.2007.0045 Google Scholar
T. Kawarasaki, M. Otake, S. Tsuchiya, M. Shibata, K. Matsumoto, and N. Isobe,
“Co-transfer of parthenogenotes and single porcine embryos leads to full-term development of the embryos,”
Anim. Reprod. Sci., 112
(1–2), 8
–21
(2009). https://doi.org/10.1016/j.anireprosci.2008.03.022 0378-4320 Google Scholar
H. Niwa, K. Yamamura, and J. Miyazaki,
“Efficient selection for high-expression transfectants with a novel eukaryotic vector,”
Gene, 108
(2), 193
–199
(1991). https://doi.org/10.1016/0378-1119(91)90434-D 0378-1119 Google Scholar
K. Yoshioka, C. Suzuki, A. Tanaka, I. M. Anas, and S. Iwamura,
“Birth of piglets derived from porcine zygotes cultured in a chemically defined medium,”
Biol. Reprod., 66
(1), 112
–119
(2002). https://doi.org/10.1095/biolreprod66.1.112 0006-3363 Google Scholar
H. Inoue, T. Murakami, T. Ajiki, M. Hara, Y. Hoshino, and E. Kobayashi,
“Bioimaging assessment and effect of skin wound healing using bone marrow-derived mesenchymal stromal cells with the artificial dermis in diabetic rats,”
J. Biomed. Opt., 13
(6), 064036
(2008). https://doi.org/10.1117/1.3042266 1083-3668 Google Scholar
M. Hara, T. Murakami, and E. Kobayashi,
“In vivo bioimaging using photogenic rats: fate of injected bone marrow-derived mesenchymal stromal cells,”
J. Autoimmun, 30
(3), 163
–171
(2008). https://doi.org/10.1016/j.jaut.2007.12.007 0896-8411 Google Scholar
S. J. Li, S. L. Yang, S. H. Zhao, B. Fan, M. Yu, H. S. Wang, M. H. Li, B. Liu, T. A. Xiong, and K. Li,
“Genetic diversity analyses of 10 indigenous Chinese pig populations based on 20 microsatellites,”
J. Anim. Sci. (Savoy, Ill.), 82
(2), 368
–374
(2004). 0021-8812 Google Scholar
X. F. Zhao, N. Y. Xu, X. X. Hu, and N. Li,
“Effects of microsatellite in the regulatory region of IGF 1 on growth traits in Jinhua swine,”
Yi Chuan., 29
(2), 206
–210
(2007). Google Scholar
J. Zhao, G. J. Nie, J. Z. Zhang, X. L. Guo, and N. Y. Xu,
“Influence of MyoG gene on reproductive trails in Jinhua pig,”
Yi Chuan., 27
(6), 893
–8978
(2005). Google Scholar
M. Shibata, M. Otake, S. Tsuchiya, M. Chikyu, A. Horiuchi, and T. Kawarazaki,
“Reproductive and growth performance in Jinhua pigs cloned from somatic cell nuclei and the meat quality of their offspring,”
J. Reprod. Dev., 52
(5), 583
–590
(2006). https://doi.org/10.1262/jrd.18004 Google Scholar
J. P. Bidanel, J. C. Caritez, and C. Legault,
“Ten years experiments with Chinese pig in France.1. breed evaluation,”
Pig News Info., 11 345
–348
(1990). Google Scholar
H. Yoshino, K. Hashizume, and E. Kobayashi,
“Naked plasmid DNA transfer to the porcine liver using rapid injection with large volume,”
Gene Ther., 13
(24), 1696
–1702
(2006). https://doi.org/10.1038/sj.gt.3302833 0969-7128 Google Scholar
Y. K. Kang, D. B. Koo, J. S. Park, Y. H. Choi, H. N. Kim, W. K. Chang, K. K. Lee, and Y. M. Han,
“Typical demethylation events in cloned pig embryos. Clues on species-specific differences in epigenetic reprogramming of a cloned donor genome,”
J. Biol. Chem., 276
(43), 39980
–39984
(2001). https://doi.org/10.1074/jbc.M106516200 0021-9258 Google Scholar
K. Inoue, T. Kohda, J. Lee, N. Ogonuki, K. Mochida, Y. Noguchi, K. Tanemura, T. Kaneko-Ishino, F. Ishino, and A. Ogura,
“Faithful expression of imprinted genes in cloned mice,”
Science, 295
(5553), 297
(2002). https://doi.org/10.1126/science.295.5553.297 0036-8075 Google Scholar
N. Ogonuki, K. Inoue, Y. Yamamoto, Y. Noguchi, K. Tanemura, O. Suzuki, H. Nakayama, K. Doi, Y. Ohtomo, M. Satoh, A. Nishida, and A. Ogura,
“Early death of mice cloned from somatic cells,”
Nat. Genet., 30
(3), 253
–254
(2002). https://doi.org/10.1038/ng841 1061-4036 Google Scholar
I. Wilmut, N. Beaujean, P. A. de Sousa, A. Dinnyes, T. J. King, L. A. Paterson, D. N. Wells, and Young Le,
“Somatic cell nuclear transfer,”
Nature (London), 419
(6907), 583
–586
(2002). https://doi.org/10.1038/nature01079 0028-0836 Google Scholar
M. Kurome, H. Ueda, R. Tomii, K. Naruse, and H. Nagashima,
“Production of transgenic-clone pigs by the combination of ICSI-mediated gene transfer with somatic cell nuclear transfer,”
Transgenic Res., 15
(2), 229
–240
(2006). https://doi.org/10.1007/s11248-006-0004-5 0962-8819 Google Scholar
|

