|
|
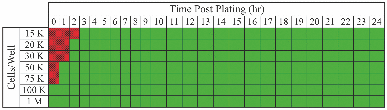
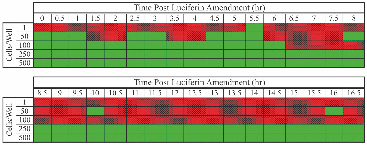
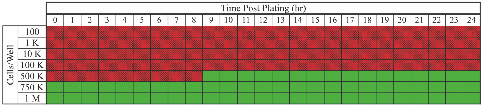
1.IntroductionWe have recently demonstrated that autonomous bioluminescent production from a mammalian cell line expressing human-optimized (ho) bacterial luciferase (lux) cassette genes can be used as a target for cell culture and small animal bioluminescent imaging.1 Here we compare the bioluminescent expression of a mammalian HEK293 cell line transfected with the holux genes with the bioluminescent expression of the same cell line expressing a commercially available, ho-firefly luciferase gene (luc) and the fluorescent expression of a commercially available, improved green fluorescent protein (GFP). luc and gfp are two of the most widely known and used reporter genes for optical imaging2 and therefore provide excellent points of comparison for determining if holux expression would be beneficial in a given experiment. The three systems are intrinsically different, and as such, have the potential to fulfill alternative niches within the needs of the bioimaging community. The holux system is unique among bioluminescent systems because of its ability to autonomously synthesize and/or scavenge all required substrates from the host cell in order to produce bioluminescence in a fully autonomous fashion. The system itself is composed of five genes with the luxA and luxB gene products forming the heterodimeric luciferase enzyme and the luxD, luxC , and luxE gene products forming a transferase, a synthase, and a reductase, respectively, that work together to produce and regenerate the required myristyl aldehyde co-substrate from endogenous myristyl groups. A sixth gene, frp, encodes an NAD(P)H:flavin reductase that helps to cycle endogenous flavin mononucleotide (FMN) into the required FMNH2 co-substrate. Along with molecular oxygen, these components supply the enzyme with all the required products to produce a bioluminescent signal at 490 nm.3 The Luc system catalyzes the oxidation of reduced luciferin in the presence of ATP-Mg2+ and oxygen to generate CO2, AMP, PPi, oxyluciferin, and yellow-green light at a wavelength of 562 nm. This reaction was originally reported to occur with a quantum yield of 0.88,4 but has since been shown to actually achieve a quantum yield closer to only 0.41.5 The Luc system used in these experiments utilizes a commercially available holuc gene from the Promega Corporation (luc2). This gene encodes for an altered protein that improves translational efficiency in the mammalian cellular background and has also been destabilized to promote lower background and increased induction levels.6 The GFP system is perhaps one of the most well characterized and longest studied of any reporter system available.7 It differs from the holux and Luc systems in that it is a fluorescent reporter. The oxidized GFP protein, upon excitation at 395 or 478 nm, produces a fluorescent emission signal at 507 nm, allowing for detection. These experiments use an improved gfp gene that is commercially available from Invitrogen. This version of gfp has been optimized for higher levels of solubility and greater than 40-fold increase in fluorescent yield over the wild-type GFP protein.8 Despite these engineered improvements, the requisite excitation signal for this, like the majority of GFP-variants, elicits high levels of background fluorescence under small animal imaging conditions.2, 9 In this respect, it will be tested only under cell culture conditions, since its use in small animal imaging is becoming increasingly supplanted by proteins or dyes that emit light in the near infrared range where autofluorescence and absorption levels are lower.10 The Luc and GFP imaging targets are representative of the types of reporter systems commonly employed in the optical imaging community11 and provide well known benchmarks against which to compare the bioluminescent expression of the new holux system. Here we compare the luminescent profile and intensity of holux-expressing HEK293 cells with the luminescent and fluorescent profiles and intensities of HEK293 cells expressing the human optimized Luc and improved GFP systems in cell culture and the holux and Luc bioluminescent systems under small animal imaging conditions. The shape and duration of the resulting light signals over time are compared, as are respective signal intensities and minimum detectable reporter cell numbers to establish which type of reporter system may be most appropriate under a given set of imaging conditions. 2.Experimental2.1.Transfection and Selection of Cell LinesTransfection was carried out in six-well Falcon tissue culture plates (Thermo-Fisher, Pittsburgh, Pennsylvania). HEK293 cells were passaged into each well at a concentration of ∼4 × 105 cells/well in complete medium the day before transfection. Plasmid vectors were purified from 100 ml overnight cultures of E. coli using the Wizard Purefection plasmid purification system (Promega, Madison, Wisconsin). On the day of transfection, cell medium was removed and replaced and vector DNA was introduced using Lipofectamine 2000 (Invitrogen, Carlsbad, California). Twenty-four hours post-transfection, the medium was removed and replaced with complete medium supplemented with the appropriate antibiotic. Selection of successfully transfected clones was performed by refreshing selective medium every 4 to 5 days until all untransfected cells had died. At this time, colonies of transfected cells were removed by scraping, transferred to individual 25 cm2 cell culture flasks, and grown in complete medium supplemented with the appropriate antibiotics. 2.2.In Vitro ImagingActively growing HEK293 cells expressing either pLuxCDEfrp:CO/pLuxAB (holux), pGL4.50[luc2/CMV/Hygro] (Luc), or pCDNA3.1-CT-GFP (GFP) were trypsinized and harvested from 75 cm2 tissue culture flasks and viable cell counts were determined as the average of two counts using a hemocytometer. Serial dilutions of cells ranging from ∼1 × 106 cells per well to ∼100 cells per well were plated in each of three wells in opaque 24-well tissue culture plates in DMEM without phenol red and supplemented with 10% fetal bovine serum, 0.01 millimolar (mM) nonessential amino acids, and 0.01 mM sodium pyruvate. Because of the autofluorescent nature of mammalian tissues, HEK293 cells expressing GFP were subjected to alternate conditions to better elucidate the GFP-based fluorescent signal from tissue autofluorescent background. Along with all surveyed cell population sizes, an equal number of untransfected HEK293 cells were plated to determine autofluorescent levels from each population size. In addition, cells were plated in phosphate buffered saline (PBS) to reduce background fluorescent detection from the medium. All values were reported as background corrected averages, which were obtained by subtracting the measured autofluorescent background value from the average fluorescent flux of each cell population size. For Luc-expressing cell lines, all wells were spiked with 0.07 mg D-luciferin/ml (Caliper Life Sciences, Hopkinton, MA) immediately before imaging. In all assays a sample of medium without cells present was included to determine the level of background detection. Photon counts were recorded using an IVIS Lumina in vivo imaging system and analyzed with Living Image 3.0 software (Caliper Life Sciences, Hopkinton, Massachusetts). To determine the change in light output over time, average radiance for population sizes of ∼1 × 106 cells was determined in photons (p)/s/cm2/steridian (sr) for each well using integration times of 10 min (holux), 10 s (Luc), or 1 s (GFP) and reported as the average of three runs with the standard error of the mean. Following initial analysis, the minimal detectable cell number was determined by performing a second assay using cell concentrations ranging between the lowest detectable number of the initial assay and the highest undetectable number of cells plated and comparing the average radiance of each population to the level of background light detected over cell-free medium. For all measurements, statistical differences were determined by using student's t tests with a p value cutoff of p = 0.05. 2.3.In Vivo Bioluminescent ImagingAll animal work was performed in adherence to the institutional guidelines put forth by the animal care and use committee of the University of Tennessee. All animal research procedures were approved by the University of Tennessee Animal Care and Use Committee (protocol number 1411) and were in accordance with National Institutes of Health guidelines. Five week old nu/nu (nude) mice (NCRNU-M, Taconic Farms Inc., New York) were anesthetized via isoflurane inhalation until unconscious. Subcutaneous injections were performed in both the shoulder and hip of each subject (n = 3) for a total of n = 6 subcutaneous injections for each cell line tested. Subjects receiving holux cells were injected with ∼5 × 106 cells. Subjects receiving Luc cells were injected with ∼5 × 105 cells. Cell counts were determined as the average of two counts using a hemocytometer. All injections were performed in a 100 μl volume of PBS. Due to the of the lack of endogenous bioluminescent processes in mammalian tissue, and to control for changes in overall animal size and dispersion of reporter-tagged cells following injection, readings were gathered as total flux values and presented in photons (p)/second (s). Luc treated subjects were immediately imaged following intraperitoneal injection of 150 mg D-luciferin/kg using a 1 s integration time, whereas holux treated subjects were immediately imaged following the injections using 1 min integration times. Total flux from each injection site was determined by drawing regions of interest (ROI) of identical size over each location. Readings were recorded over a 60 min period to determine the change in flux over time. To measure bioluminescent flux following intraperitoneal injection of the cell lines, each subject (n = 2) received a single injection of ∼1 × 106 cells (Luc) or ∼1 × 107 cells (holux). All injections were performed in a 100 μl volume of PBS. Luc treated subjects were immediately imaged following intraperitoneal injection of 150 mg D-luciferin/kg using a 10 s integration time and holux treated subjects were immediately imaged following injection using 1 min integration times. Total flux from each subject was determined by drawing an ROI of identical size over the animal. Readings were recorded over a 60 min period to determine the change in flux over time. To determine the minimal detectable number of cells in vivo, a subject was subcutaneously injected at three locations – the scruff of the neck, the mid back, and hip – with the relevant range of cells as determined by the previously described in vitro minimum detectable cell number assays in a 100 μl volume of PBS. Subjects were then imaged using integration times of up to 10 min to determine if a luminescent signal could be detected above background at the injected concentration of cells. For all measurements, statistical differences were determined by using student's t tests with a p value cutoff of p = 0.05. 3.Results3.1.In Vitro Holux DetectionCells expressing the holux cassette genes produced a visible light signal over a range from approximately 1 × 106 to 1.5 × 104 cells/well using a 10 min integration time [Fig. 1a]. A detectable light signal was inconsistently observed at a concentration of ∼1 × 104 cells/well, however, this was determined not to be significantly distinguishable from background (p = 0.72) in the given experiment (Fig. 2). In general, detection of lower cell populations as significantly different than background was more feasible at time points further from the initial plating (Table 1). The luminescent profile of holux expression demonstrated a consistent increase in average radiance from an initial post-plating value of 1800 p/s/cm2/sr to a peak of 6400 p/s/cm2/sr 16 hr post-plating [Fig. 1b]. Following peak bioluminescence, the cells expressed a slow decrease in average radiance over the remainder of the 24 hr assay, averaging a reduction of 135 (± 16) p/s/cm2/sr per hr. Expression was consistent over the course of the assay with the standard error of the mean averaging 58 (± 4) p/s/cm2/sr at each time point surveyed. Fig. 1Pseudocolor representation of the bioluminescent or fluorescent flux from cell concentrations ranging from 1 million (1M) to several thousand (K) to approximately single cell levels (NEG = negative control wells) stably transfected with (a) holux, (c) Luc, or (e) GFP. Red lines indicate the combination of two separate runs, each represented by the corresponding color scale on the right or left side of the figure. The yellow box in (e) indicates wells containing equal numbers of untransfected HEK293 cells to determine levels of background autofluorescence. Note that autoscaling of the pseudocolor image assigns brighter colors and larger areas to the larger population sizes of low level detection experiments although their scale indicates overall lower levels of flux compared to larger population sizes. Average bioluminescent or fluorescent flux dynamics for the (b) holux, (d) Luc, and (f) GFP-containing cell populations of ∼1 × 106 cells over a 24 hr period demonstrate the differences in signal intensity over time. 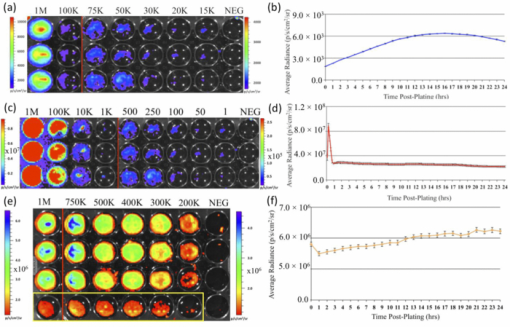 Fig. 2Despite presenting an intermittently detectable pseudocolor image, a population of ∼10000 holux-expressing cells could not be statistically differentiated from background light detection. Boxes represent the mean values of three trials, reported with overlapping standard error of the means. 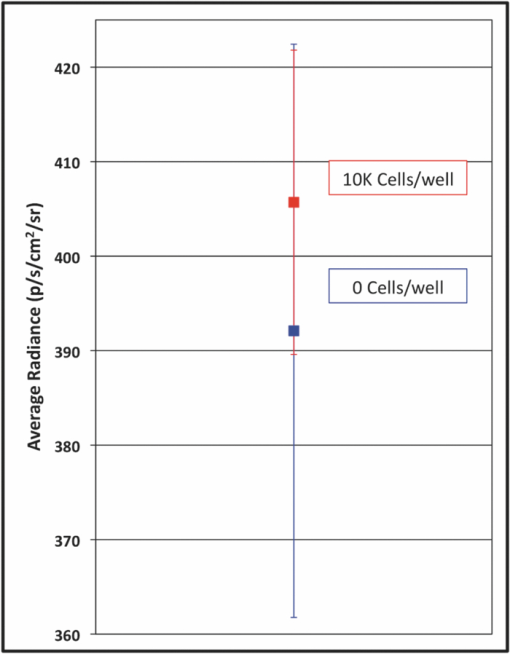 Fig. 6Comparison of pseudocolor images of subcutaneously and intraperitonealy injected holux and Luc Cells. Subcutaneously injected (a) holux- or (b) Luc-expressing cells are capable of presenting relatively similar images despite the large differences in total flux from each reporter system if the integration time is increased from 1 s (Luc) to 60 s (holux). Similar increases must be made to maintain uniform representative detection following intraperitoneal injection of the (c) holux and (d) Luc cells as well, with the holux system requiring a 60 s integration time to achieve similar pseudocolor patterning as a 10 s integration of the Luc system. 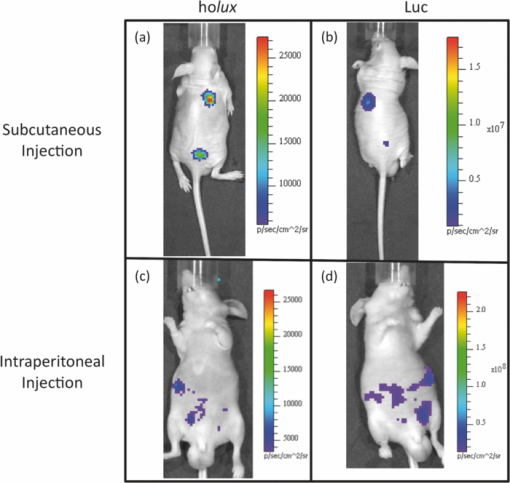 Table 1Larger population sizes of lux-expressing cells were visible sooner following plating. Green boxes represent time points where the indicated cell population was significantly distinguishable from the background. Red hatched boxes represent time points where the indicated cell population was not significantly distinguishable from background light detection. 3.2.In Vitro Luc DetectionCells expressing the human-optimized luc2 gene displayed a significantly greater average radiance (p < 0.01) than those expressing the human-optimized lux genes and as a result were visible at lower cell concentrations. Luc-expressing cells produced a visible signal over a range from ∼1 × 106 down to 2.5 × 102 cells/well at an integration time of 1 s [Figs. 1c and 3a]. Although concentrations as low as 50 cells/well could be differentiated from background if the integration time was extended to 10 s [Fig. 3b], this concentration of cells was not determined to be statistically greater than background light detection (p = 0.65) while using the 1 s integration time required to prevent saturation of the camera at the higher cell concentrations [Fig. 3a]. Detection of the Luc-tagged cell populations showed the opposite trend of those expressing the holux genes and was generally easier to differentiate from background at time points closer to luciferin addition (Table 2). The luminescent profile of the Luc-expressing cells displayed a large initial intensity, with a peak average radiance of 8.9 (± 0.4) × 107 p/s/cm2/sr 10 min following addition of 0.07 mg D-luciferin/ml. This level of radiance was not maintained, however, and had decreased to 3.0 (± 0.3) × 107 p/s/cm2/sr by 40 min post-addition. The decrease in radiance occurred during the period 10 to 30 min post-addition of substrate, after which the signal remained steady (± 9.3 × 105 p/s/cm2/sr) for the remainder of the assay. Concurrent with the higher bioluminescent output of the Luc-expressing cells compared to holux was a larger standard error. The average error over the course of the Luc luminescence assay was 2.9 (± 0.6) × 106 p/s/cm2/sr [Fig. 1d]. Fig. 3Short integration times (∼1 s) are required to prevent saturation of the CCD camera when using a Luc-based reporter system due to its high levels of bioluminescent flux following D-luciferin amendment. (a) However, at integration times of 1 s it is not possible to differentiate Luc-expressing cell populations below ∼250 cells from background light detection. (b) Increasing the integration time to ∼10 s in the absence of larger population sizes to prevent camera saturation allows for detection down to ∼50 cells. Boxes represent the mean values of three trials, reported with the standard error of the mean. 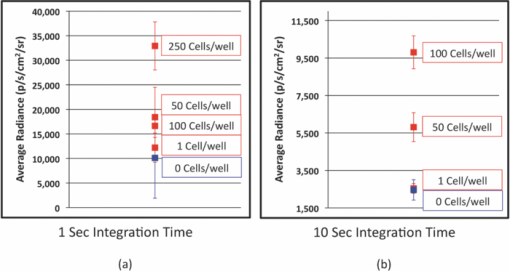 Table 2Larger populations of Luc-expressing cells were visible over longer periods of time following the addition of D-luciferin. Due to the highly dynamic nature of Luc expression, readings are reported at 30 min intervals. Green boxes represent time points where the indicated cell population was significantly distinguishable from background. Red hatched boxes represent time points where the indicated cell population was not significantly distinguishable from background light detection. 3.3.In Vitro GFP DetectionFluorescent detection from GFP emission presented the least sensitive lower limits of detection for any of the three reporter systems tested when PBS was used as the assay medium. Under these conditions, detection ranged from ∼1 × 106 down to 5 × 105 cells/well [Fig. 1e]. Although wells of less than ∼5 × 105 cells/well clearly show fluorescent signals, they were not significantly different from background following subtraction of background tissue autofluorescence (Fig. 4). Similar to the holux-expressing cells, detection ability increased for the smaller population sizes over the course of the assay (Table 3). Average radiance increased slightly but not significantly (p = 0.08) over the course of the assay from an initial value of 6.0 (± 0.06) × 106 p/s/cm2/sr to a peak of 6.6 (± 0.07) × 106 p/s/cm2/sr by 22 hr after the initial plating. Over the full course of the assay the average radiance remained relatively steady at 6.2 (± 0.05) × 106 p/s/cm2/sr with an average error of 7.3 (± 0.3) × 104 p/s/cm2/sr [Fig. 1f]. A full comparison of the pertinent expression data for all three reporter systems is detailed in Table 4. Fig. 4Cells expressing GFP were visible down to population sizes of ∼5 × 105 cells. Boxes represent the mean values of three trials, reported with the standard error of the mean. 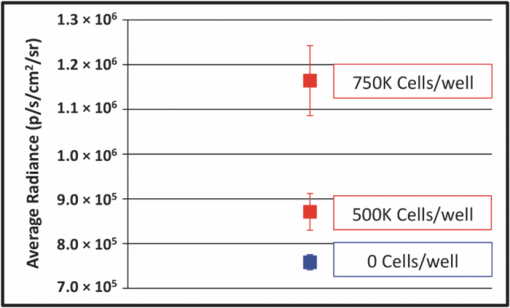 Table 3GFP-expressing cells could be significantly differentiated from background fluorescence detection at all time points following plating when greater than ∼5 × 105 cells were present. Detection of ∼5 × 105 cells became possible nine hours after plating, while detection of less than ∼5 × 105 cells was not possible at any of the time points surveyed. Green boxes represent time points where the indicated cell population was able to be significantly distinguishable from background. Red hatched boxes represent time points where the indicated cell population was not significantly distinguishable from background light detection. Table 4Summary of comparisons between the holux, Luc, and GFP reporter systems under in vitro and in vivo imaging conditions.
3.4.In Vivo Holux DetectionAverage flux from subcutaneous injection of ∼5 × 106 holux-expressing cells was 1.5 (± 0.2) × 105 p/s and remained relatively constant over the full course of the 60 min assay, displaying a minimum flux of 1.3 (± 0.1) × 105 p/s and a maximum of 1.5 (± 0.2) × 105 p/s. The standard errors of the readings were relatively low, averaging 1.6 (± 0.3) × 104 p/s and therefore provided readings with increased resolution compared to the Luc reporter system. Over the full course of the assay, the bioluminescent profile remained relatively flat, displaying a range of 2.8 × 104 p/s between the lowest and highest recorded values [Fig. 5a]. To obtain a representative pseudocolor image during acquisition, integration times of 1 min were used, however, we have previously demonstrated detection following subcutaneous injection of ∼5 × 106 holux-expressing cells is possible using ∼30 s integration times.1 It was also demonstrated that following subcutaneous injection the lower level for detection was 25000 cells when using increased integration times (∼10 min) [Fig. 5b]. Fig. 5Comparison of in vivo bioluminescence for holux and Luc cells. (a) The bioluminescent signal following subcutaneous injection of holux-expressing cells remains relatively stable following injection and (b) is detectable down to a minimum of ∼25000 cells. (c) Signal dynamics are significantly altered, but of approximately the same strength following intraperitoneal injection. (d) Total flux from subcutaneous injection of Luc-expressing cells is significantly higher, and (e) as such is detectable down to ∼2500 cells. (f) Bioluminescent output from intraperitoneal injected Luc-expressing cells expressed peak flux immediately following D-luciferin injection, but then quickly diminished over the remainder of the assay. 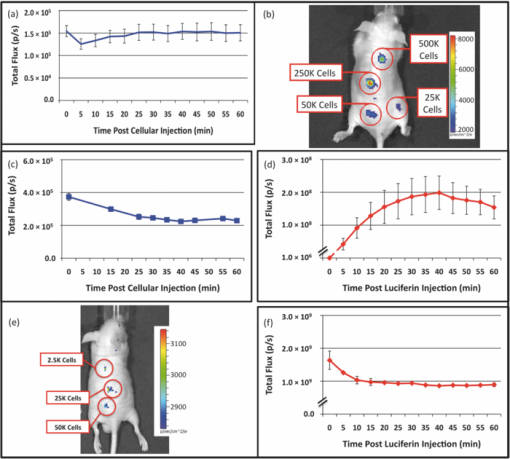 Intraperitoneal injections of ∼1 × 107 holux-expressing cells yielded a disparate bioluminescent profile from that of the subcutaneous injections. The largest total flux was immediately measured following substrate injection at a rate of 3.6 (± 0.2) × 105 p/s. Following this initial light output, the total flux continued to trend downward over the remainder of the assay [Fig. 5c]. The greatest decrease, presumably from dispersion of the cells following injection, occurred during the first 15 min, during which the total flux decreased from the maxima to 2.4 (± 0.2) × 105 p/s. After this time, the rate of bioluminescent production remained relatively flat, decreasing ∼67000 p/s by the final time point of the 60 min assay. Due to the diffusion of cells within the intraperitoneal cavity following injection and the increased amount of scattering and absorption associated with intraperitoneal imaging, pseudocolor images obtained using a 60 s integration time were not as well defined as those from the subcutaneous injections despite the injection of a higher number of cells [Figs. 6a and 6c]. The expression value differences (in p/s) that lead to these changes in pseudocolor representation are presented in Table 4. 3.5.In Vivo Luc DetectionSubcutaneous injection of ∼5 × 105 Luc-containing cells produced a bell curve of bioluminescent production. Immediately following intraperitoneal injection of 150 mg D-luciferin/kg, the average total flux from each injection site was 1.0 (± 0.2) × 106 p/s. Total flux then rapidly increased over the next 40 min to a maximum of 2.0 (± 0.5) × 108 p/s before declining for the remainder of the 60 min assay. Along with the increased flux values were increased error ranges at each time point as compared to the holux-expressing cell line. Standard error of each reading averaged 4.0 (± 0.5) × 107 p/s [Fig. 5d]. Visual detection of a signal was never problematic, with a 1 s integration providing ample exposure for facile visual representation of the subcutaneous injection site [Fig. 6b]. With the system under the control of the CMV promoter, the minimum detectable cell number was determined to be 2500 under subcutaneous imaging conditions [Fig. 5e]. Intraperitoneal injections of ∼1 × 106 Luc-expressing cells produced a much different time dependent bioluminescent expression profile than that obtained following subcutaneous injections [compare Fig. 5f to Fig. 5d]. The magnitude of bioluminescent flux notwithstanding, the time dependent bioluminescent profile following intraperitoneal injection of Luc-expressing cells yielded a profile similar to that obtained following intraperitoneal injections of holux-expressing cells [compare Fig. 5f to Fig. 5c]. The highest total flux occurred immediately after intraperitoneal injection of 150 mg D-luciferin/kg at 1.6 (± 0.3) × 109 p/s. The bioluminescent flux then quickly decreased to 1.0 (± 0.1) × 109 p/s by 10 min post-luciferin injection. For the remaining 50 min of the assay the total flux remained relatively constant, averaging 9.2 (± 0.2) × 108 p/s. As with the holux-expressing cells, integration time had to be extended to obtain a representative visual image of the intraperitoneal injection site. Intraperitoneal injection of ∼1 × 106 Luc-expressing cells, followed by immediate imaging post-D-luciferin injection using a 10 s integration time, produced a pseudocolor visual representation similar to the pseudocolor images obtained using a 60 s integration time following injection of ∼1 × 107 holux-expressing cells, but did not produce images that were as well defined as those following subcutaneous injection [Figs. 6b and 6d]. This is presumably due to the increases in absorbance and scattering associated with injection into the intraperitoneal cavity. A summary of the differences between Luc expression in vivo or in vitro following either a subcutaneous or intraperitoneal injection can be found in Table 4. 4.DiscussionThere have been numerous demonstrations of the bioluminescent and fluorescent profiles obtained in culture or small animal imaging when employing the Luc or GFP proteins as targets. The variety and scope of published literature utilizing these or versions of these reporters is testament to their usefulness, as well as the expression strategies to which they can be adapted within the confines of a particular experimental design. To aid in the comparison of the three different systems under conditions that are as uniform and comparable as could be achieved, each was expressed in the same cellular background (HEK293) and placed under the control of identical cytomegalovirus (CMV) promoters. The use of identical promoters should encourage similar levels of expression when each construct is expressed in the HEK293 cell line.12 However, in the holux cell line, although luxAB is driven by the CMV promoter, the luxC and luxE genes are instead under the control of the human elongation factor 1 alpha (Ef1α) promoter. Because the previously published demonstration of holux function was designed in this manner,1 it was not subjected to any modification prior to expression in order to allow for consistent comparison with the previously published results. As expected, bioluminescence from the Luc system was detectable at lower cell concentrations and displayed a significantly larger total flux than equal numbers of holux-containing cells in the mouse imaging experiments and its detection level was lower than both the lux and GFP reporters in the cell culture imaging scenarios. Under conditions where only small populations of Luc-expressing cells were assayed in cell culture, as few as 50 Luc cells/well were visible [Figs. 1c and 3b], compared with a minimum of 15000 cells/well for the holux system [Fig. 1a], and 5.0 × 105 cells/well in the GFP system when cells were imaged in PBS [Figs. 1e and 4]. The need to use PBS as a liquid medium to detect lower GFP-expressing cell numbers due to the autofluorescence from the cell culture medium represents a crucial problem with using fluorescent systems for prolonged cell culture imaging. The lack of medium components such as serum and nutrients required for low-level fluorescent detection does not promote continued cellular growth, thereby preventing potential autonomous fluorescent monitoring without regular medium changes. The inclusion of these compounds can prevent this, but increases the minimum detectable cell number beyond 1 million cells/well, and therefore could not be detected under our imaging conditions. Another approach to overcome the poor sensitivity of GFP in culture is to use an alternate cell line capable of more efficiently expressing the reporter. It has previously been demonstrated that GFP expression under the control of the CMV promoter in the MCF-7 breast cancer cell line is capable of being detected at lower numbers of GFP-expressing cells/well.13 However, these experiments were conducted in wells of significantly smaller surface area (0.32 cm2 as compared to 1.9 cm2) than used in these experiments. When the results from both experiments are normalized to media volume, this corresponds to a lower detection level of ∼250 cells/μl using MCF-7 cells compared to ∼500 cells/μl when expressed in HEK293 cells. Our results demonstrate, however, that the use of bioluminescence rather than fluorescence as an imaging modality completely circumvents this problem. However, there is a large difference in the bioluminescent output levels and imaging strategies between the holux and Luc systems. The holux system has the advantage of not requiring addition of a substrate to elicit bioluminescent production, therefore allowing for completely autonomous bioluminescent readings that should routinely correlate with cell number, regardless of time. The disadvantage of the holux system is that it is significantly less efficient than the Luc system. While the average radiance of ∼1 × 106 holux cells had a peak value of 6400 p/s/cm2/sr, this is comparable to the peak average radiance of only ∼100 HEK-Luc cells/well (although this number of cells/well cannot be reliably detected following the initial bioluminescent burst following substrate amendment as shown in Table 2). Therefore, detection of small numbers of cells in culture is best suited to a Luc-based reporter system, especially if the production of light is only to be monitored over short time periods. However, if working with larger cell populations, the use of a holux-based reporter system gives the benefit of continuous bioluminescent output, and is not dependent on the addition of luciferin to the cell culture medium. Regardless of which reporter system is employed, the use of a bioluminescent system (either holux or Luc) has the advantage of low background detection when compared with the use of a fluorescent system such as GFP in a medium-based cell culture setting. When applied to small animal imaging, the same general benefits for each reporter system are reiterated. The major disadvantage of working with GFP or alternate fluorescent reporter systems in an animal model is the relatively high level of background fluorescence resulting from excitation of endogenous chromophoric material within the subject tissue. The use of a bioluminescent reporter helps to overcome this disadvantage due to the low levels of background autoluminescence in mammalian tissues.14 While Luc-based systems have most often been utilized for small animal imaging, the holux system provides a distinct advantage for near-surface target visualization. Although not as bright as the Luc system [total flux averaged 1.5 (± 0.2) × 105 p/s for a subcutaneous injection of ∼5 × 106 HEK293 holux cells versus an overall average total flux of 1.4 (± 0.2) × 108 p/s for a subcutaneous injection of ∼5 × 105 Luc cells], the bioluminescent profile of the holux-containing cells was relatively flat over the full course of the assay, while the bioluminescent profile of the Luc-containing cells varied greatly following substrate injection due to the nonstandard rate of substrate uptake by Luc-containing cells, the depletion of the administered luciferin substrate, and the lack of coordinated bioluminescent expression from reporter cells exposed to luciferin. In addition, the act of administering luciferin encompasses its own set of concerns. It has been well documented that the bioluminescent profile can be altered depending on the route of substrate administration for Luc-based systems,15 with each route having different uptake rates throughout the body.16 Also of concern, the process of substrate injection allows for the introduction of error due to differences in the efficiency of each injection and/or the possibility of potential injection failure (i.e., injection into the bowel during intraperitoneal administration).15 Any changes in the quality of the luciferin over time during multiple injections17 as well as the possible introduction of tissue damage that can prohibit further injections create additional concern.18 For large-scale experiments, the cost of luciferin must also be taken into consideration, as it is an expensive substrate. Therefore, the use of a holux-based reporter is more simplistic and economical and may provide more reliable results if relatively large numbers of cells are being imaged close to the surface of the subject. While previous reports have suggested that detection of a single cell expressing the luc reporter gene is possible in the 4T1 mouse mammary tumor line,19 we determined that a minimum of 2500 cells were required when the Luc system was expressed in HEK293 cells under the control of a CMV promoter [Fig. 5e]. Despite the increase in cells required for detection under our expression conditions, this number was still well below that required for detection of the holux-expressing cells [Fig. 5b]. The diminished performance of the holux cells compared to Luc-containing cells during both minimal detection level testing and intraperitoneal injection demonstrates that the associated benefits of the holux system are of little value if they cannot be easily detected under experimentally relevant imaging conditions. In cases where deep tissue imaging is required, the use of a Luc-based system can be advantageous despite the concerns associated with substrate addition, especially since its use in these types of experiments is widespread and well documented. Whether subcutaneous or intraperitoneal injection is chosen as the route of administration, it is important to realize that the decreased efficiency of the holux system as compared to the Luc system necessitates an increase in integration time to obtain similar detection levels (Fig. 6). The amount of time required for signal detection must be considered in the context of a given experiment to determine if detection of the holux signal at a level similar to what a researcher may be accustomed to using a Luc-based system is acceptable. The greatest advantages of the new holux system, however, are the ability for researchers to integrate its use alongside other established fluorescent and bioluminescent systems and the ability to exploit the unique autonomous nature of lux bioluminescent expression with novel detection methods. Because the presence of fluorescently labeled cells would not be detected under bioluminescent imaging conditions (i.e., in the absence of an excitation signal), the location and size of bioluminescent signals could be determined and then differentiated from any fluorescent signals detected following administration of the excitation signal. In addition, the holux signal could be determined prior to substrate injection in conjunction with alternative bioluminescent reporter systems to sequentially determine the location and size of differentially labeled cell populations within a living host. Alternatively, the autonomous nature of lux bioluminescent expression could allow it to be paired with miniaturized integrated circuit microluminometers20 that could one day be implanted under the skin of an animal subject, allowing for real-time detection of signal without the need for external imaging equipment. This possibility opens the door for development of integrated biofeedback circuits that can autonomously monitor and subsequently react to numerous in vivo disease conditions. So while the introduction of a holux imaging target certainly does not displace the use of currently available fluorescent and bioluminescent imaging targets, it can overcome some of the shortcomings of these systems and integrates well with them as an additional tool for noninvasive imaging. AcknowledgmentsThis work was supported by the National Institutes of Health, National Cancer Institute, Cancer Imaging Program, award number CA127745–01, the National Science Foundation Division of Chemical, Bioengineering, Environmental, and Transport Systems (CBET) under award number CBET-0853780, and the Army Defense University Research Instrumentation Program. ReferencesD. Close, S. Patterson, S. Ripp, S. Baek, J. Sanseverino, and
G. Sayler,
“Autonomous bioluminescent expression of the bacterial luciferase gene cassette (lux) in a mammalian cell line,”
PLoS ONE, 5
(8), e12441
(2010). https://doi.org/10.1371/journal.pone.0012441 Google Scholar
G. Choy, S. O’Connor, F. Diehn, N. Costouros, H. Alexander, P. Choyke, and
S. Libutti,
“Comparison of noninvasive fluorescent and bioluminescent small animal optical imaging,”
BioTechniques, 35
(5), 1022
–1031
(2003). https://doi.org/111200315 Google Scholar
E. Meighen,
“Molecular biology of bacterial bioluminescence,”
Microbiol. Rev., 55
(1), 123
–142
(1991). https://doi.org/10.1111/j.1751-1097.1995.tb08708.x Google Scholar
H. Seliger and
W. McElroy,
“Spectral emission and quantum yield of firefly bioluminescence,”
Arch. Biochem. Biophys., 88
(1), 136
–141
(1960). https://doi.org/10.1016/0003-9861(60)90208-3 Google Scholar
Y. Ando, K. Niwa, N. Yamada, T. Enomoto, T. Irie, H. Kubota, Y. Ohmiya, and
H. Akiyama,
“Firefly bioluminescence quantum yield and colour change by pH-sensitive green emission,”
Nature Photon., 2
(1), 44
–47
(2007). https://doi.org/10.1038/nphoton.2007.251 Google Scholar
, “Technical Manual: pGL4 Luciferase,”
(2009) Google Scholar
Y. Wang, Y. John, and
S. Chien,
“Fluorescence proteins, live-cell imaging, and mechanobiology: seeing is believing,”
Annu. Rev. Biomed. Eng., 10
(1), 1
–38
(2008). https://doi.org/10.1146/annurev.bioeng.010308.161731 Google Scholar
A. Crameri, E. Whitehorn, E. Tate, and
W. Stemmer,
“Improved green fluorescent protein by molecular evolution using DNA shuffling,”
Nat. Biotechnol., 14
(3), 315
–319
(1996). https://doi.org/10.1038/nbt0396-315 Google Scholar
T. Troy, D. Jekic-McMullen, L. Sambucetti, and
B. Rice,
“Quantitative comparison of the sensitivity of detection of fluorescent and bioluminescent reporters in animal models,”
Imaging, 3
(1), 9
–23
(2004). https://doi.org/10.1162/153535004773861688 Google Scholar
S. Hilderbrand and
R. Weissleder,
“Near-infrared fluorescence: application to in vivo molecular imaging,”
Curr. Opin. Chem. Biol., 14
(1), 71
–79
(2009). https://doi.org/10.1016/j.cbpa.2009.09.029 Google Scholar
J. Burdette,
“In vivo imaging of molecular targets and their function in endocrinology,”
J. Mol. Endocrinol., 40
(6), 253
–261
(2008). Google Scholar
J. Qin, L. Zhang, K. Clift, I. Hulur, A. Xiang, B. Ren, and
B. Lahn,
“Systematic Comparison of Constitutive Promoters and the Doxycycline-Inducible Promoter,”
PLoS ONE, 5
(5), e10611
(2010). https://doi.org/10.1371/journal.pone.0010611 Google Scholar
G. Caceres, X. Zhu, J. Jiao, R. Zankina, A. Aller, and
P. Andreotti,
“Imaging of luciferase and GFP transfected human tumours in nude mice,”
Luminescence, 18
(4), 218
–223
(2003). https://doi.org/10.1002/bio.729 Google Scholar
D. Welsh and
S. Kay,
“Bioluminescence imaging in living organisms,”
Curr. Opin. Biotechnol., 16
(1), 73
–78
(2005). https://doi.org/10.1016/j.copbio.2004.12.006 Google Scholar
Y. Inoue, S. Kiryu, K. Izawa, M. Watanabe, A. Tojo, and
K. Ohtomo,
“Comparison of subcutaneous and intraperitoneal injection of D-luciferin for in vivo bioluminescence imaging,”
Eur. J. Nucl. Med. Mol. Imaging, 36
(5), 771
–779
(2009). https://doi.org/10.1007/s00259-008-1022-8 Google Scholar
K. Lee, S. Byun, J. Paik, S. Lee, S. Song, Y. Choe, and
B. Kim,
“Cell uptake and tissue distribution of radioiodine labelled D-luciferin: implications for luciferase based gene imaging,”
Nucl. Med. Commun., 24
(9), 1003
–1009
(2003). https://doi.org/10.1097/00006231-200309000-00009 Google Scholar
S. Mohler,
“Tips on Buying and Working with Luciferin,”
Genetic Engineering and Biotechnology News, 30
(4), 20
–21
(2010). Google Scholar
K. O’Neill, S. Lyons, W. Gallagher, K. Curran, and
A. Byrne,
“Bioluminescent imaging: a critical tool in pre-clinical oncology research,”
J. Pathol., 220
(3), 317
–327
(2010). https://doi.org/10.1002/path.2656 Google Scholar
J. Kim, K. Urban, E. Cochran, S. Lee, A. Ang, B. Rice, A. Bata, K. Campbell, R. Coffee, and
A. Gorodinsky,
“Non-invasive detection of a small number of bioluminescent cancer cells in vivo,”
PLoS ONE, 5
(2), e9364
(2010). https://doi.org/10.1371/journal.pone.0009364 Google Scholar
S. Islam, R. Vijayaraghavan, M. Zhang, S. Ripp, S. Caylor, B. Weathers, S. Moser, S. Terry, B. Blalock, and
G. Sayler,
“Integrated circuit biosensors using living whole-cell bioreporters,”
IEEE Transactions on Circuits And Systems, 54
(1), 89
–98
(2007). https://doi.org/10.1109/TCSI.2006.887982 Google Scholar
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||