|
|
1.IntroductionConfocal microscopy is a widely used tool for imaging 3-D structures within light-scattering specimens due to its superior axial and improved lateral resolution compared to traditional wide-field microscopy.1 The axial improvement allows for the isolation of individual optical sections in an object. By combining several high-resolution 2-D image sections collected at different focal planes, the full 3-D structure of an object can be reconstructed.2 In recent years, confocal microscopy has broadened its utility from pure structural imaging to functional imaging, particularly for applications in the life sciences, where both spatial and temporal resolution must be maximized. The ability to create axial optical sections is due to the combination of point illumination and point detection, which practically eliminates scattered light from within the focal plane and strongly rejects light from out-of-focus planes. One common method for achieving point illumination is to focus a laser beam onto the specimen using a microscope objective lens. The focused beam is positioned from pixel to pixel using a beam steering device, such as a pair of galvanometer-driven mirrors, to sequentially build up a complete image. In such mirror-based systems, the scanning mirrors also direct the fluorescence emission collected by the same objective lens back to the central optical axis, so that a stationary pinhole located in an image plane can act as a spatial filter for point detection. Despite the improvement in spatial resolution compared to traditional wide-field microscopes, there are several limitations with currently available confocal systems that limit their utility for fast functional imaging, such as the study of the electrical activity of neuronal or cardiac cells. One limitation arises from the fact that confocal microscopes typically raster scan a specimen with a single-point illumination source using galvanometer-driven mirrors. Because a confocal image must be built up on a pixel-by-pixel basis, raster scanning often takes per frame. This frame rate is orders of magnitude too slow for functional optical recordings of cellular electrical events, such as neuronal action potentials, which occur on a time scale of milliseconds. Some systems attempt to increase the scanning speed of the point source by operating the fast-axis galvanometer-driven mirror at its resonance frequency,3 or by replacing it with a rotating polygon mirror.4 These systems are successful at increasing the overall frame rate, but they remain too slow to measure fast electrical signals in cells. Moreover, this technique increases the frame rate solely at the expense of dwell time per pixel. Because the optical indicators used for functional biological measurements, such as voltage-sensitive or ion-sensitive dyes, often exhibit small fractional fluorescence changes,5, 6, 7 the total number of collectable emission photons corresponding to the signal of interest is limited. Thus, a decrease in the dwell time directly leads to poorer photon collection and results in degradation to the achievable signal-to-noise ratio. Another way to increase the frame rate of confocal microscopes, without drastically reducing the dwell time at any particular pixel, is by simultaneously scanning several sufficiently spaced spots, as is done with Nipkow disk-based scanners.8, 9 By scanning fixed-size pinholes, systems based on such scanners have reported frame rates of .10, 11, 12 However, a limitation in placing multiple excitation spots on the preparation is the resulting need for an imaging detector (e.g., a camera) instead of a single photodetector (e.g., a photodiode or photomultiplier tube) to simultaneously record the multiple fluorescence emission spots. Most cameras are limited to frame rates of several hundred hertz (the system reported by Tanaami 12 uses an unspecified camera capable of recording at ; however, its dynamic range is limited to ). At least one camera is available that can record at with of dynamic range,13 but it is only , thus limiting either the total field of view or the achievable spatial resolution, depending on the magnification used. Although the Nipkow disk systems allow for a significant improvement in scanning speed over the single-spot raster scan systems and excel in applications where faster imaging of full frames is needed, they are too slow to capture the majority of intracellular electrical signaling events such as neuronal action potentials. In addition, there is currently no way to optimize the size of the pinhole to the specific application with the Nipkow disk-based systems. One way that conventional galvanometer-based systems are successfully utilized for functional recordings is with the line scan technique.14 To do this, the slow axis of the scanner is held at a fixed position while the fast axis is allowed to scan at its maximal rate. By sacrificing the spatial information from one scan dimension, the frame rate can be increased to , which is suitable for recording certain biological signals such as intracellular calcium transients.15, 16 However, this frame rate still prevents accurate measurement of fast signals such as membrane potential changes in neuronal or cardiac cells. Moreover, line scans severely limit the region of study to sites located on a single line—it is impossible to study more than two sites on a specimen that are not collinear. For intricate biological structures such as the arborization of single neurons, it is often imperative to study sites distributed throughout the specimen. Recently, there has been renewed interest in systems that sweep a slit of light over the entire field of view using a single galvanometer-based scanner to closely match the effective temporal characteristics of line scanning systems without the sacrifice in spatial information.17, 18 Unlike the line scan systems, which scan a single point of light, the slit scan systems do not comply with the point illumination condition necessary for true confocal imaging, so the resolution in these systems is not isotropic. Although these systems have been shown to be useful extensions to traditional line scanning systems, the scanning speed is still too slow to measure fast signals such as membrane potential changes in neurons. In all of these techniques, time is spent interrogating regions of the preparation that do not contain any structures of interest. If the time spent on these regions is redistributed exclusively to areas that contain structures of interest, then the dwell time per useful pixel can be dramatically increased. Furthermore, by limiting the number of sites that are visited, the effective frame rate can also be significantly increased at a given dwell time per pixel. We accomplished both of these goals by combining a technique for random-access addressable point illumination with random-access addressable point detection, so that frames for functional data are made up of multiple sites of interest (SOIs), which can be recorded at high frame rates suitable for measuring fast biological activity with true confocality in all dimensions. These sites are selected from a full frame structural image recorded with the same system in raster scan mode. This enables an increase of at least one order of magnitude in the effective recording frame rate over currently available systems, while allowing the user to study intricate biological specimens, due to the flexibility in site selection. Additionally, because of the lack of inertia-limited grossly moving parts, our system allows for independent and adaptable control of the dwell time and frame rate at the selected sites to optimize the signal-to-noise ratio and recording speed for a given specimen. 2.Methods2.1.Point IlluminationTo provide for fast random access laser beam positioning, we used acousto-optic deflectors (AODs) instead of scanning mirrors to position the excitation light.19 AODs rely on diffraction of light, unlike mirrors that utilize reflection. Their operation is based on the fact that sound waves travel through an acousto-optic medium as a series of compressions and rarefactions. By doing so, the sound pattern creates a virtual diffraction grating with a grating constant that is directly proportional to the frequency of the sound wave [Fig. 1a ]. Generally, a light beam traveling perpendicular to the sound column will be diffracted into several discrete orders; however, when AODs are operated in the Bragg regime,20 most of the laser power is diffracted into the first order. Therefore, light diffracted into higher orders can be obstructed with minimal overall power loss, and only a single scanning spot is allowed to pass to the preparation. Fig. 1AOD-based scanner and DMD-based addressable spatial filter. (a) The angle of beam deflection using AODs is proportional to the frequency of the sound wave. For operation in the Bragg regime, the incident beam must strike the acousto-optic medium at an angle of , where is the wavelength of the incident light and is the wavelength of the sound wave in the medium. (b) By turning on one mirror or one group of mirrors on the DMD, the central in-focus spot will be sent to the detector (on), while out-of-focus emission light will be rejected (off).  By placing one deflector in the direction and another in the direction, any site on a 2-D area can be probed. We have previously shown that AODs can be used to implement a nonconfocal scanning microscope capable of recording both membrane potential and intracellular ion concentration at high rates in optically thin, nonscattering preparations.21, 22 Furthermore, since there are no moving parts in these deflectors, their positioning speed is not inertia limited as with galvanometer-driven mirrors. The settling time for AODs is mainly dependent on the width of the beam in the direction of deflection and the type of acousto-optic material used. The particular deflectors that we used for the current system (LS110A-XY, Isomet Corporation, Springfield, VA) combine two tellurium dioxide AODs in one package with a -diam active aperture. Assuming a beam that fills the entire aperture, the acousto-optic medium and active aperture size result in a total settling time of , which is independent of the positioning distance between consecutively visited sites. 2.2.Point DetectionIn traditional mirror-based scanning systems, the same mirrors that position the excitation light can also direct the fluorescence emission back to the central optical axis. Such “descanning” is straightforward with mirrors, since reflection is wavelength independent. However, AODs, which use wavelength-dependent diffraction, cannot be easily used for descanning because fluorescence is emitted at a longer wavelength than the excitation light and is not monochromatic. In addition, AODs have a diffraction efficiency of in each dimension; a loss of this much light is not tolerable in the photon-limited emission pathway, because it limits the achievable signal-to-noise ratio. The lack of descanning prevents the use of a stationary pinhole as in traditional systems. Instead, to enable confocal imaging, an addressable spatial filter is needed that can synchronously track the input excitation point in a conjugate image plane.23, 24 To create an addressable spatial filter, we used a digital micromirror device (DMD, Texas Instruments, Dallas, Texas) composed of an array of square mirrors that can be individually tilted to along an axis running diagonally across each mirror.25 The settling time of the DMD is regardless of the number of mirrors that are flipped in one update cycle. Individual mirrors can be used to independently direct emission light either toward (on) or away from (off) a single photodetector. This allows one or more “on” mirrors to define the pinhole, while all of the neighboring “off ” mirrors discard out-of-focus fluorescence [Fig. 1b]. With this scheme, the number of adjacent mirrors used to create each pinhole will affect the degree of confocality and the amount of measurable fluorescence in the same manner in which pinhole size effects traditional confocal microscopes.26 Additionally, unlike traditional systems, both the pinhole location and size can be rapidly updated to track the AOD scan pattern by changing the address and number of mirrors in the “on” position. 2.3.Registration AlgorithmTo ensure optimal confocality, it is crucial to keep the point illumination and point detection in exact register with one another in a 2-D space, since the AODs and DMD are addressed independently. To accomplish this task, we developed a registration algorithm based on a Nelder-Mead simplex search27, 28 that finds the AOD-based beam position that best matches every potential DMD-defined pinhole. This utilizes the fact that AODs have a continuous positioning resolution while a DMD is intrinsically discrete. For each potential pinhole position, the AOD-based beam address that results in collection of the maximal light intensity from a uniform reflective preparation is saved in a lookup table. Furthermore, the registration algorithm was designed to work on a dataset collected with several evenly spaced pinholes simultaneously in the “on” position to minimize the time needed to complete the process. This feature exploits the fact that a Nelder-Mead simplex can only search for local maxima rather than a single global maximum. By calculating initial guesses for each pinhole location, we could find the optimal AOD-based beam address for each site of interest without interference from other pinholes. The ability to run the registration in this parallel fashion tremendously decreased the time that it took for the algorithm to complete; with 64 simultaneous pinholes, the total time needed to register the entire field of view was . 2.4.Optical LayoutA schematic of the setup can be seen in Fig. 2 . For all experiments, we used either a tunable air-cooled argon laser (532-AP, Melles Griot, Carlsbad, CA) set to the line, or a multiline air-cooled argon laser (5425A, Ion Laser Technology, Salt Lake City, UT) with an appropriate laser line filter to select the line. Both lasers produce of continuous wave power at this wavelength, which is attenuated to minimize photodamage using the modulation input of the AODs. An inverted microscope (Axiovert 35, Zeiss, Thornwood, NY) was used for all studies. A water-immersion objective lens with a correction collar (c-apochromat, Zeiss) was used for the axial resolution plot, the surface reconstruction of the spiny pollen grain, and the neuronal recordings. All other experiments were done using a oil-immersion objective lens (plan neofluar, Zeiss). The optical signal was detected using a photomultiplier tube (H7112, Hamamatsu, Bridgewater, NJ) and digitized using either a , or a , analog-to-digital converter (PCI-6035E/PCI-6251, National Instruments, Austin, TX). Axial scanning was accomplished with an objective lens nanopositioner that allowed reproducible submicron steps over a range (P721.LLQ, Physik Instrumente, Irvine, CA). Fig. 2Schematic of optical layout. Excitation light (dashed line) from the laser is positioned with the AODs and is focused on the specimen with the objective lens. A dichroic mirror placed after the AODs allows for separation of the excitation light from the emission light (solid line) and prevents the emission light from passing back through the scanner. An addressable confocal pinhole is created with the DMD to track the fluorescence in a conjugate image plane and direct the in-focus signal toward a PMT. 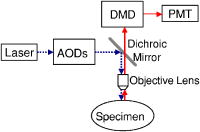 To fill the field of view with the scan pattern, a demagnification telescope was used after the deflectors to increase the scan angle by sacrificing some of the total beam diameter. However, despite the demagnification, we were able to fill the back focal aperture of the objective lens and place diffraction-limited spots on the specimen by utilizing the full aperture of the AODs. 2.5.Electrophysiology SetupThe preparations used to demonstrate a biological application were -thick acute brain slices from green fluorescent protein (GFP)-expressing transfected mice and cultured neurons from the hippocampi of wild-type mice using the procedure of Pyott and Rosenmund.29 Brain slices and cultures were prepared in accordance with the guidelines of the National Institute of Health, as approved by the animal care and use committee of Baylor College of Medicine. The cells were allowed to grow for at before recordings were made from them. Electrical stimulation and recordings were made using an Axopatch 200A patch clamp amplifier (Axon Instruments, Union City, CA). Borosilicate glass pipettes were pulled using a multistep protocol (P-87, Sutter Instruments, Novatp, CA). This resulted in pipettes with a tip diameter of and a tip resistance of . Cells were held in whole-cell current clamp for all recordings. 3.ResultsThe performance of the system was evaluated to assess its spatiotemporal resolution and signal-to-noise ratio as well as its ability to carry out confocal structural and functional imaging. 3.1.Temporal ResolutionThe maximal acquisition speed is limited by the total pixel dwell time, i.e., the combination of settling time and sampling time. As previously stated, the settling times of the AODs and the DMD are 15 and , respectively. Via custom software and control electronics, we can issue new addresses to both devices simultaneously from the lookup table created by the registration algorithm. Because both devices settle concurrently, we are able to update the site of interest at the slower DMD settling time of . After an address is updated, we allow another for photodetector settling and sampling. Therefore, the total time per recording site is and the maximum effective frame rate of the system is given by , where number of sites to be studied. The control electronics and software also allow for oversampling to reduce system noise. Each additional sample takes another , depending on the sampling rate of the analog-to-digital converter. 3.2.Axial ResolutionTo measure the axial resolution of the AOD/DMD system, we used a flat mirror in the specimen plane to simulate an infinitesimally thin object and then recorded the change in intensity of the reflected light along the axial direction for two pinhole sizes30 at a single preregistered site. A reflective preparation was used instead of a fluorescent preparation to eliminate any contribution from photobleaching on the measured axial intensity. Comparison of the axial sectioning capability of our system with an equivalent nonconfocal system was accomplished by using the AODs for point illumination with all of the mirrors on the DMD in the “on” position, so that there was no spatial filtering of the collected fluorescence. This procedure makes use of the fact that point illumination coupled with wide-field detection gives a similar response to a pure wide-field system.31 In all cases, images are normalized to account for brightness differences between confocal and nonconfocal imaging after ensuring that there was no saturation of the photodetector. By filling the back focal aperture of the objective lens, a diffraction-limited spot was placed on the preparation. For these tests, the two pinhole sizes that were used mapped to and at the specimen plane. With the pinhole, the full width at half maximum (FWHM) axial section size was , and for the pinhole it was . With nonconfocal imaging, the FWHM is greater than the range of the nanopositioner. Therefore, the confocal imaging case has an axial resolution that is the nonconfocal case. The full axial response for all three tested cases is shown in Fig. 3 . Fig. 3Axial resolution. Using mirrors to define each pinhole resulted in a specimen plane pinhole size of and a full width at half maximum (FWHM) axial resolution of (solid line). Using mirrors, the pinhole size is and the FWHM axial resolution was (dotted line). With all mirrors turned on, the system acts as a nonconfocal microscope. In this case, the FWHM is larger than (dashed line). 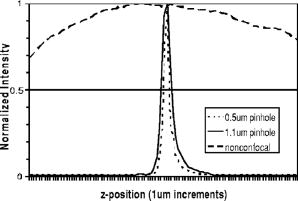 3.3.Signal-to-Noise RatioWe assessed the improvement in signal-to-noise ratio (SNR) based on the adaptive dwell time and frame rate of our system by comparing it to a commercial confocal laser scanning microscope (C1, Nikon, Melville, NY) operated in the line scan mode. To quantify the improvement, we compared the fluorescence signals from two sites on a sparse fluorescent polystyrene microsphere preparation (F24634, Molecular Probes, Eugene, OR)—one site on a microsphere and one site on the nonfluorescent background. With the Nikon C1 system, the line scan was set so that it crossed from the background through the microsphere; two sites could then be selected from the resulting dataset. With our system, the two sites were directly selected using the custom software interface. In its fastest mode of operation, the Nikon C1 is able to achieve a line frequency of . At this speed, the dwell time per pixel is . The mean fluorescence intensity at the site on the microsphere was with a standard deviation of 1.8, while the site on the background had a mean intensity of with a standard deviation of 1.1 [Fig. 4a ]. Therefore, the SNR for the fluorescent site is . Fig. 4Signal-to-noise ratio comparison. Measurement of fluorescent intensity at one site on a fluorescent microsphere (black) and one site on the nonfluorescent background (gray). (a) With the Nikon C1 operated at a line scan frequency of , the mean intensity at the fluorescent site was with a standard deviation of 1.8 (SNR ). (b) Using the AOD/DMD system at , the mean intensity at the fluorescent site was with a standard deviation of 0.1 (SNR ). (c) Using the AOD/DMD system at , the mean intensity at the fluorescent site was with a standard deviation of 0.4 (SNR ). 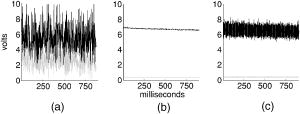 To achieve a frame rate comparable to that of the Nikon C1, we set the frame rate of our system to . At this frame rate, two sites can be studied using a dwell time of per pixel, which allows for an oversampling factor of 825. In this case, the mean fluorescence intensity of the site on the microsphere was with a standard deviation of , while the site on the background had a mean intensity of with a standard deviation of [Fig. 4b]. The SNR for the fluorescent site is , which is -fold greater than the commercial system. For comparison, we also repeated the same test using our system at a frame rate of with a dwell time of per pixel. At this rate, the oversampling factor is 5. The mean fluorescence intensity of the site on the microsphere was with a standard deviation of , while the site on the background had a mean intensity of with a standard deviation of [Fig. 4c]. The SNR for the fluorescent site is , which is -fold greater than the commercial system. 3.4.Structural Confocal ImagingFor additional demonstration of the confocality of the system, we compared confocal and nonconfocal images from fluorescent polystyrene microspheres (F8836, Molecular Probes) embedded in a light-scattering medium (agarose gel). Image stacks were collected as 100 optical sections spaced at intervals. The data are presented as a side-view maximum projection to allow evaluation of the optical sectioning capabilities. The images show a significant improvement in the optical sectioning between confocal and nonconfocal imaging modes (Fig. 5 ). In the confocal case, the polystyrene microsphere appears round and has a uniform fluorescence intensity. The nonconfocal case shows a dual cone of fluorescence from out-of-focus planes. The out-of-focus cones are the same intensity as the central microsphere despite normalization and avoidance of saturation. Fig. 5Confocal versus nonconfocal comparison. Side-view maximum projection of 100 optical sections taken at steps of a fluorescent polystyrene microsphere embedded in a light-scattering medium collected in confocal and nonconfocal mode. Both images are normalized to remove the influence of brightness and detector saturation on the comparison. The nonconfocal case is not able to resolve the shape of the microsphere. 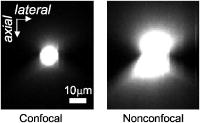 Further demonstration of the system performance was accomplished by collecting image stacks of fluorescent pollen grains (30-4264, Carolina Biological Supply, Burlington, NC) and creating reconstructions of the 3-D structure. The first test specimen was a lobular pollen grain that is roughly across. Figure 6a shows individual optical sections from three different focal planes that were recorded with a oil-immersion objective using a pinhole and a step size of between adjacent slices. The dwell time at each voxel was , which allowed for oversampling. We used the Amira software package (Mercury Computer Systems, Richmond, TX) to create a cut-away volume reconstruction that demonstrates the ability to image fluorescent structures on the interior of the specimen and hence optical sectioning. Fig. 6Structural imaging. (a) Individual optical sections of a lobular pollen grain collected at three different focal depths, along with a cut-away volume reconstruction that allows visualization of internal fluorescent structures formed from the lobe boundaries. (b) Individual optical sections of a spiny pollen grain along with a surface reconstruction allow for visualization of the spikes covering the pollen surface. 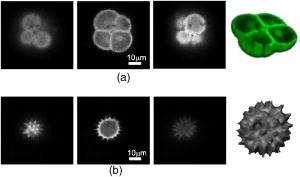 A second test specimen was a spiny pollen grain that is also across. This form of pollen is studded with spikes that are each long and have a subresolution diameter at the tip. Figure 6b shows optical sections of the spiny pollen grain along with a surface reconstruction. The data were collected and reconstructed in the same way as the lobular pollen grain, except a water-immersion lens was used with a pinhole. The surface reconstruction is used instead of the volume reconstruction to highlight the ability to resolve the spikes that cover the specimen. With confocal detection, the spikes on the pollen grain are clearly visible, even in individual optical sections. Finally, to demonstrate the ability of our system to record from living tissue, we collected image stacks of GFP-labeled neurons from -deep within -thick acute mouse brain slices in both confocal mode and nonconfocal mode. The stacks were collected over a range with a step size of between each image. Figure 7 shows the 3-D maximum projection of the images collected in confocal mode. Neuronal cell bodies and apical dendrites are clearly visible in these views. The nonconfocal images (data not shown) had almost uniform fluorescence across the entire field of view at every depth step with no visibly discernable neuronal structures. Fig. 7Imaging in a brain slice. Maximum projection images of GFP-labeled hippocampal neurons in -thick brain slices were collected in confocal and nonconfocal modes. In the confocal mode, neuronal structures are clearly seen deep within the slice preparation. The displayed volume measures . In nonconfocal mode (data not shown), there is almost uniform fluorescence across the field of view due to scattered light, and no neuronal structures can be visualized within the brain slice.  3.5.Functional Neuronal ImagingTo demonstrate the functional imaging properties of the system, we recorded calcium transients from several noncollinear sites on cultured hippocampal neurons filled with the fluorescent calcium indicator Oregon Green BAPTA-1. To elicit calcium changes throughout the dendritic arborization, we depolarized a neuron held in whole-cell current clamp with trains of five current injections at . Each current injection caused the cell to fire an action potential that back propagated throughout the dendrites and resulted in the opening of voltage-dependent calcium channels.32 Calcium ions entering the cell through these channels bind to the indicator molecules, and differential fluorescence signals between distinct neuronal compartments were measured. All sites were monitored for , with the first stimulation in the train occurring after recordings began. Cultured neurons were used instead of neurons in brain slices because it is not technically possible to patch cells in brain slices on an inverted microscope under visual control. The signals recorded from the neuronal preparations show that consecutive depolarizations each elicited a detectable calcium influx (Fig. 8 ). Furthermore, responses were not uniform throughout the cell and the differential signaling was visualized. For each depolarization, a stepwise increase in intracellular calcium concentration was seen in the optical signal . To allow for oversampling, the dwell time was set to at each site, thus the optical signal was measured at a pixel rate of . Fig. 8Functional imaging. Trains of five back-propagating action potentials (APs) were elicited with current injections in a cultured hippocampal neuron and were measured electrically through the patch pipette. The site of the patch pipette attachment is marked (P). Several sites distributed throughout the neuron (1 to 6) were simultaneously monitored to optically measure intracellular calcium transients. 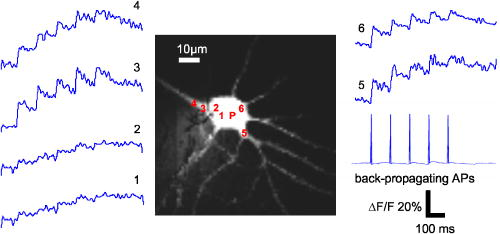 4.DiscussionWe have developed a novel addressable confocal microscope that allows targeted studies at user-selected sites of interest to dramatically increase the frame rate over that which can be achieved with raster scanning. The spatiotemporal resolution of this system is sufficient to measure several electrophysiologic parameters at multiple sites distributed throughout a living biological specimen. 4.1.Spatiotemporal ResolutionThe acquisition speed that we achieved is an order of magnitude faster than that of known commercially available devices. Also, our system is capable of true random-access scanning; therefore, the distance between any two consecutive SOIs is of no consequence to the positioning time. This creates a highly adaptive system that can be used to scan a few SOIs at a high frame rate, e.g., for membrane potential measurements, or many SOIs at lower frame rates, e.g., for intracellular calcium measurements. Additionally, the adaptability allows us to easily and rapidly switch between different modes of imaging, such as a fixed dwell time with a variable frame rate or vice versa. This is especially important when trying to maximize the recorded signal-to-noise ratio from weakly fluorescent optical indicators. A DMD can be used for point illumination in addition to point detection. However, this requires the entire face of the DMD to be flooded with excitation light so that sites anywhere on the specimen can be illuminated. This results in a loss of most of the excitation light, since only a few mirrors define each SOI. Although we found this mode to work in principle, the low excitation power at each site resulted in less fluorescence. This inefficiency forced a tradeoff in bandwidth for gain and prevented the measurement of the fast cellular parameters that we are interested in. Furthermore, by using the AODs as an independent means for positioning the point illumination, we are able to place diffraction-limited spots on the preparation, regardless of the pinhole size being used. In an earlier system, we showed that the confocal imaging case could achieve a improvement in axial resolution over the nonconfocal case.33 That system utilized AODs with a rectangular active aperture (LS55, Isomet Corporation). In the preliminary tests, we used a circular -diam beam that did not allow for placement of diffraction-limited spots on the specimen due to underfilling of the back focal aperture of the objective lens. To improve the resolution, we inserted anamorphic optics in front of the AODs to elongate the beam to utilize the available active aperture. Although there was an improvement in resolution, the aberrations introduced by the anamorphic optics ultimately limited the utility of that system. Because the current system uses an AOD package with a large circular active aperture, no beam shaping other than expansion is necessary. This simplifies the optical layout and greatly reduces the number of extraneous aberrations that are coupled into the system. The only disadvantage of using large aperture AODs is the resulting increase in settling time. The current AODs take to settle as opposed to for the previous rectangular aperture deflectors. However, this increase in positioning time is irrelevant to the final recording speed, since we are limited by the settling time of the DMD. Because both the AOD and DMD can be issued addresses simultaneously, the settling times are not additive and there is no change in the temporal dynamics of the complete AOD/DMD system. In the current system, there is no overlap between adjacent pinholes. To allow for accurate measurements of the lateral resolution, it is necessary to update the pinhole location on the DMD by a distance smaller than the diameter of a single pinhole. By sweeping over a subresolution object, a smooth function can then be measured as was done to measure axial resolution. Because multiple mirrors are used for each pinhole, it is possible to do this in the combined AOD/DMD system; however, limitations of the current software prevented testing of this feature. Currently, we use the size of the pinhole in the specimen plane to approximate the lateral resolution. Based on the physical dimension of an individual mirror on the DMD, we can probe a region on the specimen that is as small as using a objective lens; however, for all imaging tests, we used pinholes made up of mirrors, so that the region probed was . The mapping of a single mirror in the specimen plane also represents the minimum distance that a larger pinhole can be laterally translated. By filling the back focal aperture of the objective lens, we have shown that the axial resolution improvement between confocal imaging and nonconfocal imaging is greater than using a pinhole. Furthermore, the ratio of the measured axial resolution to the estimated lateral resolution was for both tested pinhole sizes, which closely matches the ratio predicted by theory. Using the equations given by Jonkman and Stelzer,34 the smallest lateral resolvable distance between two points in confocal microscopy is generally given by where is the wavelength of the excitation light and NA is the numerical aperture of the objective lens. Although many factors affect the axial resolution, the smallest axial resolvable distance between two points can be roughly approximated bywhere is the refractive index of the specimen. For biological specimens, the refractive index of water is a good approximation for . Therefore, the theoretical axial to lateral resolution ratio is to 3.8 if using objective lenses with NAs of 1.2 to 1.3 and is independent of .The use of a larger magnification objective results in the ability to map smaller pinholes in the specimen plane. We verified this by collecting similar axial resolution plots with the objective lens (data not shown). In that case, the best axial resolution of was achieved at a specimen plane pinhole size of . Although the resolution with the objective is superior, the objective lens is better suited to functional biological imaging due to the use of water as the immersion medium and the ability to view a larger field of the specimen plane. 4.2.Signal-to-Noise RatioWith our system, we are able to increase the effective frame rate, without any sacrifice to the dwell time, by using addressable elements that allow all of the allotted scanning/recording time to be spent at sites of interest. Other systems that seek to improve readout time by simply increasing the speed of scanning do so by sacrificing the dwell time at each pixel. These systems are often not suited to measure the small fluorescence changes found with optical indicators in neurons, since the measured signal-to-noise ratio is proportional to the square root of the total number of collected photons for Poisson-limited optical signals. The total number of collected photons can be increased by raising the fluorescence excitation intensity or by increasing the photon collection time; however, above a certain threshold, increased excitation intensity will result in rapid photodamage to living tissue. The dramatically longer dwell time of our system at comparable frame rates allows for collection of more signal photons through oversampling, thus reducing the variations in the measured signal. It is impractical to study the same living cell from a biological specimen on two independent systems. By doing these tests with a control preparation (e.g., fluorescent microspheres) rather than a biological specimen, we are able to accurately and quantitatively compare the responses of the different systems. Furthermore, for purpose of comparison, all of the presented data were recorded in single sweeps. With both systems, the SNR can be improved by averaging successive trials of the same experiment. Using the AOD/DMD system, there is a 22-fold improvement in the SNR compared to the Nikon C1 at a comparable frame rate of . At this frame rate, the C1 has a dwell time of , and our system has a total dwell time of , of which is used for oversampling. Therefore, the collection time of our system is greater than the Nikon system. This should correspond to an increase in the SNR of , which is close to the measured value. 4.3.Structural ImagingThe polystyrene microsphere imaging clearly shows that our system is rejecting out-of-focus light and thus conferring the main advantage of confocal microscopy. Both pollen grain image stacks highlight the ability of the system to collect optical sections of a specimen that can be used to create accurate 3-D reconstructions. Additionally, the single optical sections from the spiny pollen grain demonstrates that our system enables visualization of small features that are not visible with wide-field microscopy. The images collected from the GFP-labeled neurons deep within an acute brain slice show that the system can record in highly light-scattering preparations. They also show that the achievable resolution is sufficient to allow precise selection of sites for functional imaging from areas in the cell body as well as in smaller features such as neuronal dendrites. This ensures that functional traces accurately represent the small compartment from where they are being recorded rather than an aggregate response in larger cellular areas. The relative intensity of particular voxels during structural imaging gives an indication of the baseline fluorescence signal that can be expected from the same sites during functional imaging, since the same dwell time is used in both modes. 4.4.Functional ImagingThe data presented in the current study were all obtained with single sweeps. Typically, electrophysiology data are collected by giving the same stimulation protocol repeatedly to a cell after allowing rest periods between each acquisition. By doing this, several traces can be collected at each site and averaged to reduce noise. We chose to avoid this procedure to demonstrate a worst-case scenario for the system. Even with the single sweeps, calcium transients that resulted in changes in the optical signal could be detected in neurons. All functional data are filtered using forward and reverse digital filtering with a constrained least-squares linear phase finite impulse response (FIR) filter. The double-pass filtering scheme assures that there will be zero phase shift in the filtered signal relative to the raw signal. To record functional images from neurons in brain slices, it will be necessary to use an upright microscope. Neurons in slice preparations are preferable to neurons in culture for most studies, because the distribution of receptors and channels is more physiologic and because the cellular structure is better maintained. However, signals from neurons in brain slices occur on the same time scale as signals from neurons in culture, so there are no technical limitations to adapting our system based on recording speed. Furthermore, we have shown that our system is capable of imaging neurons in light-scattering preparations such as brain slices. 5.ConclusionsWe show that acousto-optic deflectors can be successfully combined with a digital micromirror device to form a high-speed addressable confocal microscope that allows for functional imaging from user-selected sites of interest. The effective frame rate of this system is an order of magnitude faster than commercially available systems while still maintaining a longer dwell time. The increased dwell time allows for collection of more emission photons, which leads to an improved signal-to-noise ratio when compared to the commercial systems. Furthermore, there are no constraints on site selection as in line scan systems. Another significant advantage is that the frame rate, dwell time, and number of sites can all be adaptively optimized for the particular parameter under study. For slower changing parameters, more sites can be selected, while for faster parameters, fewer sites can be targeted. This adaptability ensures that the addressable confocal imaging system has the spatiotemporal bandwidth to study many different parameters, including transients of cellular ion concentration and neuronal membrane potential. AcknowledgmentsWe are deeply indebted to Robert Nowak for supplying us with the DMD and Everett Taylor of Isomet Corporation for the long evaluation period of the large aperture AODs. We would like to thank Christian Rosenmund and his laboratory, especially Jayeeta Basu and Mingshan Xue, for supplying the cultured neurons. Finally, we thank the members of the Saggau laboratory-specifically Zanwen Chen, Brad Losavio, and James Mancuso for their assistance with the neuronal electrophysiology, Rob Gaddi and Rudy Fink for help updating the custom hardware and software, and Duemani Reddy for comments on the manuscript. ReferencesJ. B. Pawley, Handbook of Biological Confocal Microscopy, Plenum Press, New York
(1995). Google Scholar
M. Roy and
C. J. R. Sheppard,
“Effects of image processing on the noise properties of confocal images,”
Micron, 24 623
–635
(1993). 0968-4328 Google Scholar
G. Y. Fan,
H. Fujisaki,
A. Miyawaki,
R. K. Tsay,
R. Y. Tsien, and
M. H. Ellisman,
“Video-rate scanning two-photon excitation fluorescence microscopy and ratio imaging with cameleons,”
Biophys. J., 76 2412
–2420
(1999). 0006-3495 Google Scholar
M. Rajadhyaksha,
R. Rox-Anderson, and
R. H. Webb,
“Video-rate confocal scanning laser microscope for imaging human tissues in vivo,”
Appl. Opt., 38 2105
–2115
(1999). 0003-6935 Google Scholar
A. Grinvald,
R. D. Frostig,
E. Lieke, and
R. Hildesheim,
“Optical imaging of neuronal activity,”
Physiol. Rev., 68 1285
–1366
(1988). 0031-9333 Google Scholar
A. Grinvald,
B. M. Salzberg,
V. LevRam, and
R. Hildesheim,
“Optical recording of synaptic potentials from processes of single neurons using intracellular potentiometric dyes,”
Biophys. J., 51 643
–651
(1987). 0006-3495 Google Scholar
L. M. Loew,
“Potentiometric membrane dyes and imaging membrane potential in single cells,”
Fluorescent and Luminescent Probes for Biological Activity, 210
–221 2nd ed.Academic Press, San Diego
(1999). Google Scholar
M. Petran,
M. Hadravsky,
M. D. Egger, and
R. Galambos,
“Tandem scanning reflected light microscope,”
J. Opt. Soc. Am., 58 661
–664
(1968). 0030-3941 Google Scholar
M. Petran,
M. Hadravsky, and
A. Boyde,
“The tandem scanning reflected light microscope,”
Scanning, 7 97
–108
(1985). 0161-0457 Google Scholar
G. Q. Xiao,
T. R. Corle, and
G. S. Kino,
“Real-time confocal scanning optical microscope,”
Appl. Phys. Lett., 53 716
–718
(1988). https://doi.org/10.1063/1.99814 0003-6951 Google Scholar
G. S. Kino,
“Intermediate optics in Nipkow disk microscopes,”
Handbook of Biological Confocal Microscopy, 155
–165 2nd ed.Plenum, New York
(1995). Google Scholar
T. Tanaami,
S. Otsuki,
N. Tomosada,
Y. Kosugi,
M. Shimizu, and
H. Ishida,
“High-speed 1-frame/ms scanning confocal microscope with a microlens and Nipkow disks,”
Appl. Opt., 41 4704
–4708
(2002). 0003-6935 Google Scholar
S. D. Antic,
“Action potentials in basal and oblique dendrites of rat neocortical pyramidal neurons,”
J. Physiol. (London), 550 35
–50
(2003). 0022-3751 Google Scholar
A. Hernandezcruz,
F. Sala, and
P. R. Adams,
“Subcellular calcium transients visualized by confocal microscopy in a voltage-clamped vertebrate neuron,”
Science, 247 858
–862
(1990). 0036-8075 Google Scholar
M. Segal and
D. Manor,
“Confocal microscopic imaging of ⟨Ca2+⟩i in cultured rat hippocampal neurons following exposure to N-methyl-D-aspartate,”
J. Physiol. (London), 448 655
–676
(1992). 0022-3751 Google Scholar
T. M. Hoogland and
P. Saggau,
“Facilitation of L-type Ca2+ channels in dendritic spines by activation of beta2 adrenergic receptors,”
J. Neurosci., 24 8416
–8427
(2004). 0270-6474 Google Scholar
C. J. R. Sheppard and
X. Q. Mao,
“Confocal microscopes with slit apertures,”
J. Mod. Opt., 35 1169
–1185
(1988). 0950-0340 Google Scholar
C. J. Koester,
“Comparison of various optical sectioning methods—the scanning slit confocal microscope,”
Handbook of Biological Confocal Microscopy, 525
–534 2nd ed.Plenum, New York
(1995). Google Scholar
M. Gottlieb,
C. L. M. Ireland, and
J. M. Ley, Electro-Optic and Acousto-Optic Scanning and Deflection, Marcel Dekker, New York
(1983). Google Scholar
B. E. A. Saleh and
M. C. Teich, Fundamentals of Photonics, WileyNew York,1991). Google Scholar
A. Bullen and
P. Saggau,
“High-speed, random-access fluorescence microscopy: II. Fast quantitative measurements with voltage-sensitive dyes,”
Biophys. J., 76 2272
–2287
(1999). 0006-3495 Google Scholar
A. Bullen,
S. S. Patel, and
P. Saggau,
“High-speed, random-access fluorescence microscopy: I. High-resolution optical recording with voltage-sensitive dyes and ion indicators,”
Biophys. J., 73 477
–491
(1997). 0006-3495 Google Scholar
S. R. Goldstein,
T. Hubin,
S. Rosenthal, and
C. Washburn,
“A confocal video-rate laser-beam scanning reflected-light microscope with no moving parts,”
J. Microsc., 157
(1), 29
–38
(1990). 0022-2720 Google Scholar
S. R. Goldstein,
T. Hubin, and
T. G. Smith,
“An improved no-moving-parts video-rate confocal microscope,”
Micron Microsc. Acta, 23 437
–446
(1992). 0739-6260 Google Scholar
L. J. Hornbeck,
“Digital Light Processing™: A new MEMs-based display technology,”
(1996) Google Scholar
T. Wilson,
“The role of the pinhole in confocal imaging system,”
Handbook of Biological Confocal Microscopy, 167
–182 2nd ed.Plenum, New York
(1995). Google Scholar
J. A. Nelder and
R. Mead,
“A simplex method for function minimization,”
Comput. J., 7 308
–313
(1965). 0010-4620 Google Scholar
W. H. Press,
S. A. Teukolsky,
W. T. Vetterling, and
B. P. Flannery,
“Minimization or maximization of functions,”
Numerical Recipes in C, 394
–455 2nd ed.Press Syndicate Univ. Cambridge, Cambridge, MA
(1992). Google Scholar
S. J. Pyott and
C. Rosenmund,
“The effects of temperature on vesicular supply and release in autaptic cultures of rat and mouse hippocampal neurons,”
J. Physiol. (London), 539 523
–535
(2002). 0022-3751 Google Scholar
T. Wilson and
A. R. Carlini,
“Three-dimensional imaging in confocal imaging systems with finite sized detectors,”
J. Microsc., 149 51
–66
(1988). 0022-2720 Google Scholar
T. Wilson and
C. J. R. Sheppard, Theory and Practice of Scanning Optical Microscopy, Academic Press, London
(1984). Google Scholar
G. Stuart,
N. Spruston,
B. Sakmann, and
M. Hausser,
“Action potential initiation and backpropagation in neurons of the mammalian CNS,”
Trends Neurosci., 20 125
–131
(1997). 0166-2236 Google Scholar
V. Bansal,
S. S. Patel, and
P. Saggau,
“High speed confocal laser scanning microscopy using acousto-optic deflectors and a digital micromirror device,”
Proc. SPIE, 5324 47
–54
(2004). 0277-786X Google Scholar
J. E. N. Jonkman and
E. H. K. Stelzer,
“Resolution and contrast in confocal and two-photon microscopy,”
Confocal and Two-Photon Microscopy: Foundations, Applications, and Advances, 101
–125 Wiley-Liss, Inc., New York
(2002). Google Scholar
|

