|
|
1.IntroductionHuman malaria, a mosquito-borne infectious disease linked to two major species of protozoan parasites, Plasmodium falciparum and Plasmodium vivax, resulted in approximately 660,000 deaths in 2010 from among over 219 million documented episodes, particularly affecting young children in Africa.1 Parasites, transmitted by mosquitoes, invade and multiply inside the host red blood cells (RBC). During the 48-h intraerythrocytic cycle, the host RBCs undergo significant structural and biochemical modification regulated by complex molecular activities of the parasites. Our understanding of how malaria parasites orchestrate these mechanisms still remains incomplete. Direct imaging of the three-dimensional (3-D) structures and dynamics of the P. falciparum (Pf)-RBCs may hold the key to develop a better understanding of the process of malaria infection.2,3 Soft x-ray tomography provides structural information with extremely high spatial resolution,4,5 but it requires large-scale synchrotron x-ray facilities, and it does not access the dynamics of live cells. Recent advances in optical microscopy techniques have been employed for the study of structural and physical properties of RBCs altered by malaria parasitization.6 Fluorescence confocal microscopy, requiring the use of exogenous labeling agents rendering high-contrast molecular information, has also been used to visualize the 3-D shape of Pf-RBCs.7 However, its drawbacks include possible photo-bleaching, photo-toxicity, and interference with normal molecular activities. Recently, refractive index (RI), an intrinsic optical property of materials including biological cells and subcellular components, has been studied to link the structure of RBCs with their physical properties modulated by malarial infection.8,9 Measuring the RI offers a means to probe, noninvasively and quantitatively, the structural and chemical information of biological cells10 including morphology,11–14 dynamics of the cell membrane,15–17 dry mass of the cell,18,19 and concentration of specific molecules in the living cells.20,21 A series of optical tomographic techniques has been developed to measure the 3-D distributions of RI in biological cells. They include: (1) digital holographic microscopy or quantitative phase microscopy methods to measure multiple transmitted electric field (E-field) with various angles of illumination22–25 or with sample rotation26,27 and (2) optical sectioning methods based on low-coherent interference such as optical coherence microscopy28,29 and optical gating based on interferometric imaging with broadband fields such as spatial light interference tomography.30,31 Recent studies reported that RI invoked multiple wavelength digital holograms obtained by tomographic holography, which circumvents the need for mechanical scanning,32,33 as well as a noninterferometric imaging technique which quantified cellular RI and dry mass density using a standard trans-illumination optical microscope.34 Among these various techniques and applications, RBCs have been studied with 3-D RI tomography with unique advantages.8,9 RBCs have a relatively simple two-dimensional (2-D) cytoskeletal structure without a nucleus. Therefore, 3-D RI measurement can directly provide structural information about the cell membrane. RBC cytoplasm mainly consists of hemoglobin (Hb) protein, and the RI of RBC cytoplasm can be directly translated into chemical information about Hb inside the cell.8 Furthermore, the structural and chemical information probed by measuring the RI could be strongly correlated with RBC pathophysiology, including malaria infection6,8,9 and sickle cell disease,35–37 and with different biochemical conditions introduced by different levels of osmotic pressure17 and ATP metabolism.16,38,39 Recently, 3-D RI tomography has been employed for noninvasively measuring the structural and biochemical information of Pf-RBCs,8 and studying the mechanistic processes associated with parasite egress following the intraerythrocytic cycle.9 Tomographic phase microscopy has been utilized to measure the multiple E-fields measurements with various illumination angles from which the 3-D RI tomograms of the Pf-RBCs are reconstructed using a projection algorithm.8,40 Reconstruction using the projection algorithm, assuming no diffraction of light in the sample, is computationally simple but does not generally provide a precise 3-D RI distribution, especially for a sample with inhomogeneous distribution of RI values. The parasitization of RBCs results in the formation of several optically inhomogeneous structures including digestive food vacuoles (a major digestive organelle of the malaria parasite), hemozoin (a disposal product formed from the digestion of Hb by parasites), and knobs (nanometer-scale protuberances in the Pf-RBC membrane conferring adherence to vascular endothelia).41 All these structures exhibit highly inhomogeneous RI distribution, and thus can significantly cause refraction, reflection, and diffraction of light. Consequently, the 3-D RI tomograms of the Pf-RBCs extracted solely from optical projection have limitations in imaging quality and resolution. Here, we report high-resolution 3-D optical tomograms of Pf-RBCs constructed using a 3-D optical diffraction tomography algorithm. We show that compared with earlier reports based on the projection algorithm, the present 3-D RI tomograms extracted with a diffraction algorithm provide highly detailed structural information as well as more precise RI values. We compare the 3-D RI tomograms of the Pf-RBC with the entire 48-h intraerythrocytic cycle using both diffraction- and projection-based reconstruction algorithms. We report quantitative estimates of host Pf-RBC volume, parasitophorous vacuole volume, and cytoplasmic volume over the entire intraerythrocytic maturation stages of the parasite. We also investigate the structural and chemical characteristics of in situ hemozoin development inside the Pf-RBCs. 2.TheoryReconstruction of a 3-D tomogram from the multiple 2-D E-fields with various illumination angles is performed by two methods: the projection and the diffraction algorithms. Since the diffraction algorithm generally considers the effect of diffraction—the interaction between light and matter—it addresses the challenges present in the projection algorithm such as limited spatial resolution and low image quality.23,24,42,43 Figure 1 shows a schematic diagram explaining the reconstruction processes of the two algorithms. Fig. 1Comparison of various three-dimensional (3-D) reconstruction algorithms. (a) Quantitative phase images of a Plasmodium falciparum-red blood cells (Pf-RBC) in trophozoite stage measured at various illumination angles (left) and corresponding spectrum in Fourier space (right). (b–c) Object functions in Fourier space reconstructed by (b) the projection algorithm and (c) the diffraction algorithm. (b1) Amplitude distributions of the object function reconstructed by the projection algorithm in the plane with five representative illuminations angles and (b2) with the full illumination angles. The corresponding amplitude distributions in the plane are shown in (b3) and (b4), respectively. (c1) Amplitude distributions of the object function reconstructed by the diffraction algorithm in the plane with five representative illuminations angles and (c2) with the full illumination angles. The corresponding amplitude distributions in the plane are shown in (c3) and (c4), respectively. 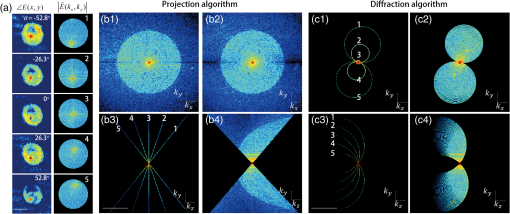 In the projection algorithm, diffraction of light in a sample is assumed to be negligible; light is assumed to propagate along straight lines with unchanged spatial frequency vectors, . The projection algorithm can be implemented by the filtered backpropagation method.40,44 A single 2-D E-field image from a specific illumination angle corresponds to an inclined plane, corresponding to the incident angle in Fourier space (Fourier slice theorem), and the center of the inclined plane is located at the origin of the Fourier space [Fig. 1(b)]. Consequently, in the projection algorithm, all the measured 2-D E-fields with various illumination angles will map a bow-tie-shaped object function distribution in Fourier space. Inverse 3-D Fourier transformation of the objective function then provides a 3-D optical tomogram of the sample. The mapping process in the projection algorithm can be easily performed by applying inverse Radon transformation.45 A set of 2-D E-field images with various illumination angles, a so-called sinogram, can be converted into a 3-D tomogram. Full details of the projection algorithm can be found elsewhere.40,44 In contrast, the diffraction tomography algorithm, originally established by Wolf,42 takes diffraction of light into account. Recently, the diffraction algorithm has been successfully employed for retrieving high-resolution 3-D structures of weakly scattering samples.23,42,43 The diffraction algorithm can be implemented with either the Born46 or Rytov47 approximations. The Born and the Rytov approximations both take the incident field as the driving field at each point of the scatterer. While both the Born and Rytov approximations consider the scattered E-field due to the presence of a sample, the Born approximation is valid for a weak scattering field from a relatively small sample. The Born approximation can be applied for a large object, provided the RI contrast is low. However, the Rytov approximation is valid as long as the phase gradient in the sample is small and is independent of the sample size. In this study, we used the Rytov approximation. Rytov approximation works independently of the size of an object, whereas Born approximation depends on the size of an object.42 In the diffraction algorithm with the Rytov approximation, the spatial frequencies of the scattered E-fields from one illumination angle cover a 2-D hemispheric surface, the so-called Ewald sphere [Fig. 1(c)]. The peak intensity of the surface, implying the lowest spatial frequency from an unscattered field, matches the origin at the Fourier space, and the range of the Ewald surface is limited by the numerical aperture (NA) of the optical imaging system.23,48 Thus, the scattered E-fields from all the possible illumination angles form a butterfly-shaped object transfer function. Inverse 3-D Fourier transformation of the object function is mapped via the diffraction algorithm, and then provides a 3-D tomogram of the sample. The details on the diffraction algorithm can be found elsewhere.43,48–50 3.Materials and Methods3.1.Experimental SetupThe detailed experimental setup for measuring E-fields with various illumination angles has been described in previous works.40,43 Briefly, complex E-fields images are recorded by Mach–Zehnder interferometry. A laser beam from a He-Ne laser (, 10 mW, unpolarized light, Throlabs, Newton, New Jersey) is split into two arms. One arm can be tilted by a galvano mirror (GVS012, Throlabs) and illuminates samples with various angles of illuminations. The diffracted beam from the sample is collected by a high NA objective lens (UPLSAPO, , , Olympus Inc., Center Valley, Pennsylvania) and a tube lens (). Then, the beam is further magnified by an additional lens system so that the total lateral magnification is . The diffracted beam from the sample interferes with a reference beam and generates spatially modulated interferograms, which are recorded by a high-speed CMOS camera (1024 PCI, Photron USA Inc., San Diego, California). Typically, 600 2-D holograms of the sample, illuminated by plane waves with illumination angles ( to 70 deg at the sample plane), are recorded for the reconstruction of one tomogram. The complex amplitude (amplitude and phase) of the E-fields images is then extracted from the measured interferograms using the field retrieval algorithm (see Ref. 51 for field retrieval algorithms). Diffraction tomograms are mapped in 3-D Fourier space using multiple 2-D E-field images, and then reconstructed by using a 3-D inverse Fourier transformation (refer to Supporting Materials for the MatLab code used for the reconstruction). The projection tomograms are reconstructed from the sequential E-fields maps with various illumination angles via inverse Radon transformation. Due to the limited angle of acceptance of the imaging system, or NA, there is missing information in the 3-D Fourier spectrum for both the projection and diffraction algorithms [Fig. 1(b) and 1(c)]. Even with the high NA objective lens, only a fraction of the scattered light from the sample can be utilized for optical imaging. To fill this information due to the limited NA, the iterative constraint algorithm43,52 was executed to the tomograms reconstructed from both the projection and diffraction algorithms. The following constraints were invoked in the interactive algorithm in the present study:
Applying the iterative algorithm fills the information corresponding to the missing angles with appropriate values. All data analysis was performed using custom-made scripts in MatLab (MathWorks Inc., Natick, MA) on a desktop computer (Inter Core i5-2600 CPU, 3.4 GHz, 8 GB RAM). The MatLab codes used for optical tomographic reconstruction based on diffraction algorithm are included in Supporting Information S1. The computation times for reconstructing a 3-D RI tomogram (size of matrix) are typically 100 and 200 s for the projection and diffraction algorithms, respectively. The iterative algorithm takes approximately 100 iterations for the diffraction algorithm, corresponding to -s computation time, whereas the projection algorithm takes approximately 15 iterations (3000-s computation time.) 3.2.Sample PreparationP. falciparum (3D7) parasites were maintained in leukocyte-free human erythrocytes (Research Blood Components, Boston, Massachusetts), stored at 4°C for no longer than two weeks under an atmosphere of 3% , 5% , and 92% in RPMI 1640 medium (Gibco Life Technologies, Carlsbad, California) supplemented with 25 mM HEPES (Sigma-Aldrich, St. Louis, Missouri), 200 mM hypoxanthine (Sigma-Aldrich), 0.209% (Sigma-Aldrich) and 0.25% albumax I (Gibco Life Technologies). Cultures were synchronized successively by a concentration of mature schizonts using plasmagel flotation53 and sorbitol lysis 2 h after merozoite invasion to remove residual schizonts.54 The sample preparation procedures and the methods were approved by the Institutional Review Board. Upon measurement, Pf-RBC samples were first diluted in phosphate-buffered saline (PBS) solution to approximately . Ten microliters of the diluted samples were sandwiched between two cover glasses. Measurements were performed at 14 to 20 h (ring stage), 20 to 36 h (trophozoite stage), and 36 to 48 h (schizont stage) following merozoite invasion. 4.Results4.1.3-D RI Tomograms of Pf-RBCs Based on Diffraction AlgorithmThe 3-D RI tomograms of 26 Pf-RBCs were measured throughout the full intraerythrocytic states (8, 13, and 5 cells for ring, trophozoite, and schizont stages, respectively) For each individual Pf-RBC, 600 E-field holograms with various illumination angles ( to ) were measured from which both projection and diffraction tomograms were reconstructed by respective algorithms. The projection and diffraction tomograms from the representative Pf-RBCs at each intraerythrocytic state are shown in Figs. 2Fig. 3–4. Fig. 2Refractive index (RI) maps of a Pf-RBC at the ring stage. (a–c) cross-sectional slices of RI distribution mapped with the projection algorithm at (a) 1 μm above the focal plane, (b) the focal plane, and (c) the plane at the center. (d–f) Slices of RI distribution mapped with the diffraction algorithm. Green arrow indicates malaria parasite. Each dashed line stands for corresponding slices. Scale bar indicates 5 μm. 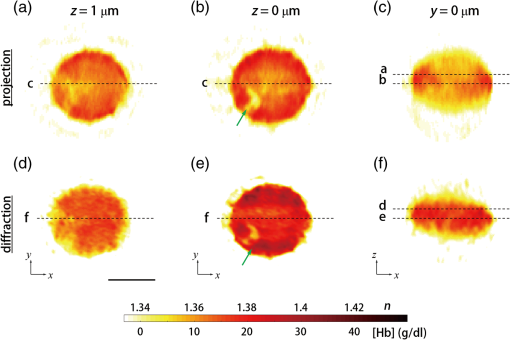 Fig. 3RI maps of a Pf-RBC at the trophozoite stage. (a–c) cross-sectional slices of RI distribution mapped with the projection algorithm at (a) 1 μm above the focal plane, (b) the focal plane, and (c) the plane at the center. (d–f) Slices of RI distribution mapped with the diffraction algorithm. Black arrow indicates hemozoin. Scale bar indicates 5 μm. 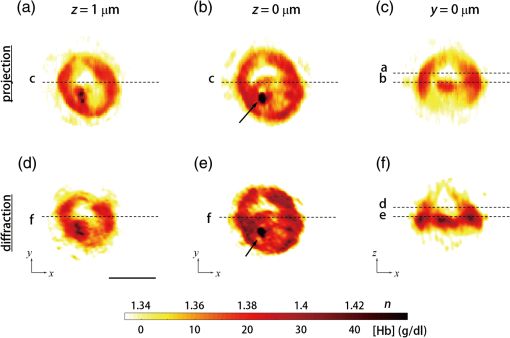 Fig. 4RI maps of a Pf-RBC at the schizont stage. (a–c) cross-sectional slices of RI distribution mapped with the projection algorithm at (a) 1 μm above the focal plane, (b) the focal plane, and (c) the plane at the center. (d–f) Slices of RI distribution mapped with the diffraction algorithm. Black arrow: hemozoin and blue arrow: plasmodium vacuoles. Scale bar indicates 5 μm. 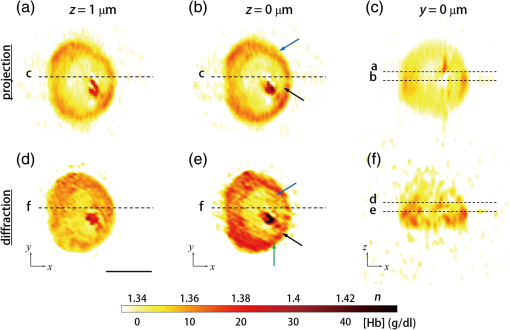 Each figure shows cross-sectional maps of the 3-D RI tomograms reconstructed by either the projection (upper row) or the diffraction algorithm (lower row) at various planes: 1 μm above the focal plane (left column), the focal plane (center column) along the axial direction, and plane at the center of the sample (right column). Since the cytoplasm of RBCs consists mainly of Hb, cytoplasmic Hb concentration for individual RBCs can be calculated from the measured RI.8 The RI of Hb solution is directly related to the concentration of Hb.55,56 The real part of the RI is proportional to the cytoplasmic Hb concentration as where is the RI of the surrounding medium ( at ) and is the specific refractive increment ( for both oxygenated and deoxygenated hemoglobin at ).8,20,55,56The 3-D RI tomograms of Pf-RBCs at the ring stage clearly show a ring-shaped internal structure inside the host Pf-RBC, which corresponds to an invaded parasite having an RI smaller than that of the cytoplasmic area (green arrows, Fig. 2). At the focal plane, both diffraction and projection algorithms provide comparable structures. However, above and below the focal plane, the tomogram reconstructed with the projection algorithm shows blurry cell boundaries that are extended in the axial direction, whereas the diffraction tomogram shows more distinct cell boundaries. More importantly, cross-section images reconstructed by the projection algorithm provide incorrectly elongated shapes in the -direction [Fig. 2(c)]. This is due to the omission of light diffraction in the projection algorithm; light diffraction at the RBC boundaries can be only taken into account in the diffraction reconstruction algorithm. Considering the complex shape of the RBC membrane and the RI difference between the surrounding medium and the cell cytoplasm ( to 0.06, depending on the Hb concentration), the plane wave impinging into the RBC may exhibit both refraction and reflection, thus resulting in significant diffraction of the transmitted beam. The RI tomogram of Pf-RBCs not only shows morphological information regarding the cell and its subcellular structures, but also enables us to obtain the biochemical information of the Pf-RBCs; the cytoplasmic RI can be directly translated into quantitative information about Hb concentration and total Hb contents.8 The effect of light diffraction becomes more significant in the later stages. The 3-D RI tomograms of the Pf-RBCs at the trophozoite stage show more difference between the reconstructions with the projection and diffraction algorithms (Fig. 3). Distinct differences can be observed, especially for the extremely high RI values in the cytoplasm, indicating the hemozoin structure (black arrows, Fig. 3). As they grow inside the host RBCs, the parasites metabolize Hb and produce hemozoin—an insoluble crystallized form of free heme, the nonprotein component of Hb, having a high RI (). At the trophozoite stage, the overall shape of the Pf-RBCs membrane becomes more complex, and malaria byproducts are newly generated as substructures with significantly high RI values and with a high spatial gradient. The 3-D RI tomograms of the Pf-RBCs in the schizont stage show significantly decreased RI in the cytoplasm (Fig. 4), which is consistent with a previous report.8 The 3-D RI tomograms reconstructed with the diffraction algorithm clearly show plasmodium vacuoles generated by the parasites (blue arrows). By contrast, the tomogram reconstructed with the projection algorithm does not provide high-resolution details on these subcellular structures. The structural and biochemical alterations resulting from the parasitization of the host Pf-RBC inevitably cause significant light diffraction from the sample. Severe structural alterations in the schizont stage Pf-RBCs can result in more light diffracted in the Pf-RBCs. In addition, significant reorganization of Hb proteins can also contribute more light diffraction; most of the cytoplasmic Hb is digested by the parasites, and then converted into hemozoin with high RI values. The use of the diffraction reconstruction algorithm could provide more precise 3-D RI tomograms. On the other hand, the projection algorithm does not support the appearance of light diffraction from the sample. As shown in Figs. 2–4, the reconstruction with the diffraction algorithm provides more precise 3-D RI tomograms of the Pf-RBCs with finer details. For comparison purposes, the projection and diffraction tomograms from healthy RBCs are also shown in Fig. 5. Similar to the Pf-RBCs, the tomograms of the healthy RBCs reconstructed by the diffraction algorithm show less distortion in their shapes than the ones obtained with the projection algorithm. Fig. 5RI maps of a healthy RBC. (a–c) cross-sectional slices of RI distribution mapped with the projection algorithm at (a) 1 μm above the focal plane, (b) the focal plane, and (c) the plane at the center. (d–f) Slices of RI distribution mapped with the diffraction algorithm. Scale bar indicates 5 μm. 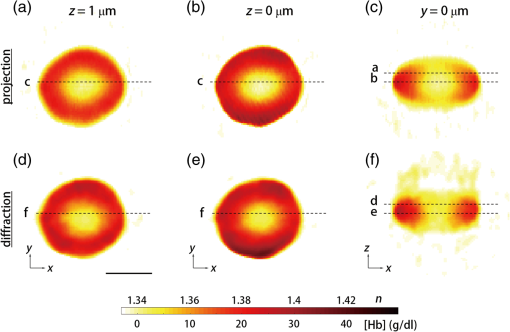 Based on the tomograms of the Pf-RBCs reconstructed by the diffraction algorithm, 3-D isosurfaces are rendered, as shown in Fig. 6 (see also Multimedia movies). The cell membranes of the Pf-RBCs are represented for the outer surface having the cytoplasmic RI; subcellular structures such as hemozoin, plasmodium vacuoles, and malaria parasites are also visualized as surfaces by connecting the specific refractive indices. The 3-D RI tomogram reconstructed with the diffraction algorithm shows the shapes and locations of the parasites as well as other associated structures in high detail. Rendering was performed by commercial software (Amira 5, Visage Imaging Inc., San Diego, California). The structures of Pf-RBCs depicted in 3-D RI tomograms are comparable with those obtained with high-resolution scanning electron microscopy.4,5 Fig. 6Rendering of the RI isosurfaces of Pf-RBCs at (a) the ring (Video 1, QuickTime, 4 MB) [URL: http://dx.doi.org/10.1117/1.JBO.19.1.011005.1], (b) the trophozoite (Video 2, QuickTime, 6 MB) [URL: http://dx.doi.org/10.1117/1.JBO.19.1.011005.2], and (c) the schizont stage (Video 3, QuickTime, 6 MB) [URL: http://dx.doi.org/10.1117/1.JBO.19.1.011005.3]. The length of arrows corresponds to 1 μm. 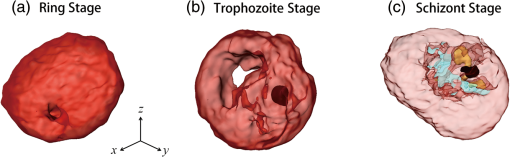 4.2.Cytoplasmic Volumes Calculated by Different Reconstruction AlgorithmsCompared with the projection tomogram, the diffraction tomograms of 3-D RI of the Pf-RBCs not only provide high-resolution structural details, but also enable the determination of biochemical information about the Hb content with higher precision. To quantitatively compare the effects of using the different reconstruction algorithms with respect to chemical information, we calculated the cytoplasmic volume and mean cellular Hb concentration (MCHC) values of the Pf-RBCs with both the projection and diffraction tomograms. The cytoplasmic volume and MCHC values are important parameters for hemodiagnosis. The MCHC values of each Pf-RBC calculated from the reconstructed RI tomograms do not show statistical differences between the projection and diffraction algorithms. This can be readily understood by the fact that, in principle, both algorithms should give comparable RI values of an object at the focal plane since the effect of diffraction of the propagated beam is minimized at the focal plane. The cytoplasmic volumes, however, show significant differences between the projection and diffraction algorithms (Fig. 7). Overall, the mean cytoplasmic volumes calculated from the projection algorithm are 33% higher than those calculated from the diffraction algorithm. In addition, the values retrieved with the diffraction algorithm showed narrower distribution among the Pf-RBCs. The difference in the values of cytoplasmic volume calculated by different reconstruction algorithm can be explained by the fact that, in the projection algorithm, the diffraction of light is not considered except at the focal plane, and thus, the reconstructed shapes of the RBC membrane can be severely distorted. Fig. 7Cytoplasmic volumes calculated by different reconstruction algorithms. Circular symbols represent individual Pf-RBCs. Dashed lines connect the values of the same cell calculated with the different algorithms. The cross symbols represent the median values, and the vertical lines indicate standard deviations. 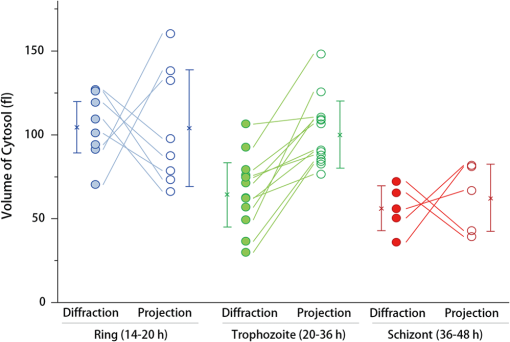 4.3.Volumes of Pf-RBC, Cytoplasm, and Parasitophorous Vacuole at Individual Cell LevelTo address the structural alterations of Pf-RBC during intraerythrocytic cycle, we calculate the volumes of host Pf-RBC, RBC cytoplasm, and parasite at individual cell levels, from the measured 3-D RI tomograms reconstructed by diffraction algorithm based on the Rytov approximation (Fig. 8). Since the 3-D RI tomograms provide quantitative information about morphologies of individual Pf-RBCs, the volumes of Pf-RBC, cytoplasm, and parasite are measured at the individual cell level. Fig. 8Measured values of (a) Pf-RBC volume, (b) cytoplasmic volume, and (c) parasite volume over intraerythrocytic cycle. Measured values from previous reports were also shown for comparison purpose. 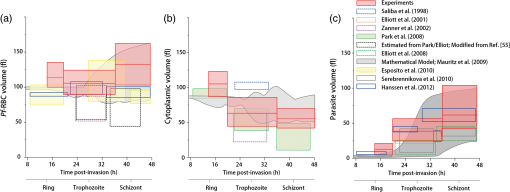 The whole host Pf-RBC volumes were calculated by integrating volumes inside individual Pf-RBCs. The cytoplasmic volumes were obtained by integrating volumes in which RI values are within a specific range () in order to select cytoplasm-containing Hb solution, and the space corresponding to parasitophorous vacuoles (with hemozoin inside) was excluded. Then, the volumes for parasitophorous vacuole were calculated by subtracting the whole host Pf-RBC volumes from cytoplasmic volumes. Despite the large cell-to-cell variations in the measurements, it is clear from the results that the total volumes of Pf-RBCs increase as the intraerythrocytic stage progresses. The mean values of the total Pf-RBCs volumes are fl (ring), fl (trophozoite), and fl (schizont) [Fig. 8(a)]. This increase of total Pf-RBC volumes is caused by the enlargement of parasite volumes even though cytoplasmic volume, corresponding to host cell water volume, decreases. Cytoplasmic volumes (host cell water volume) are measured to be fl (ring), fl (trophozoite), and fl (schizont) [Fig. 7(b)]. The mean values of the parasitophorous vacuole volumes are fl (ring), fl (trophozoite), and fl (schizont) [Fig. 7(c)]. Our results are in good agreement with the previous measurements obtained with independent techniques (Fig. 8). Zanner et al. reported cytoplasmic volumes and Pf-RBC volumes measuring diffusive water permeability using stopped-flow spectroscopy.57 Elliott et al. measured the transport of lactate and pyruvate in Pf-RBC,58 and employed serial thin-section electron microscopy and 3-D reconstruction59 in order to measure whole Pf-RBC volumes as well as parasite values. Recently, Esposito et al. measured total RBC volumes using confocal microscopy.60 More recently, Hanssen et al. determined parasitophorous volumes of Pf-RBCs using cryo x-ray tomography.4 Although the previous techniques have measured the values at different time scales with different sensitivities, their values of the total Pf-RBCs volumes are 75 to 100 fl (ring), 130 to 150 fl (trophozoite), and 50 to 160 fl (schizont). Previously measured cytoplasmic volumes are 75 to 100 fl (ring), 25 to 100 fl (trophozoite), and 10 to 80 fl (schizont) [Fig. 8(b)]. The previously measured values of the parasitophorous vacuole volumes are 5 to 20 fl (ring), 5 to 50 fl (trophozoite), and 30 to 100 fl (schizont). In addition, our measured values are also consistent with mathematical model simulations.61 4.4.3-D RI Tomograms of In Situ Formation of Hemozoin CrystalsTo further show the capability of 3-D optical tomograms with the diffraction reconstruction algorithm, we perform in situ 3-D imaging and structural analysis of the residual bodies containing subcellular hemozoin in Pf-RBCs. Hemozoin is a disposal product formed by the malaria parasites as a result of the metabolization of Hb. It accumulates as cytoplasmic crystals in the food vacuole of developing parasites. As Pf-RBCs mature from ring stage to trophozoite stage, hemozoins become detectable as brown birefringent crystals via light microscopy62. No hemozoin is observed at the ring stage. The shape and size of hemozoin provide metabolic information of individual Pf-RBCs.63 Different structural characteristics of hemozoin crystals have been observed for Plasmodium species.64 From the measured 3-D optical diffraction tomograms of the Pf-RBCs, we analyzed hemozoin structures and their optical properties in 16 Pf-RBCs (12 trophozoites and 4 schizonts). By selecting positions having RI values higher than a threshold value (), we retrieved the 3-D volumes of RI corresponding to the residual bodies containing hemozoin in the Pf-RBCs [Fig. 9(a)]. In order to investigate the structural characteristics of the hemozoin crystals quantitatively, we calculated the volume and surface area of hemozoin from the Pf-RBCs. The measured values for the volume and surface area are fl and , respectively, for trophozoite stage, and fl and , respectively, for schizont stage. These values are consistent with previous studies using independent experimental methods. Gligorijevic et al. used spinning disk confocal microscopy and measured the volumes of hemozoin crystals in the range 0.2 to 0.7 fl.63 Hanssen et al. employed x-ray microscopy and measured the volumes of hemozoin crystals during gametogenesis in the range of 0.6 to 0.8 fl.4 Although the magnitudes of the measured values for the volume and surface area are in agreement with the previous reports, we note that the decrease in the measured values from trophozoite to schizont stage is not consistent with previous works. This can be partly attributed to the limited number of samples ( and 4 for the number of cells in trophozoite and schizont stage, respectively). Moreover, the sphericity of hemozoin was found to be (trophozoite) and (schizont), which was calculated from the volume and surface area as . Fig. 9Characterizations of in situ hemozoins. (a) 3-D renderings of representative hemozoins inside Pf-RBCs. The length of arrows corresponds to 1 μm. Three isosurfaces with representative RI values are rendered according to the color bar. No hemozoin was observed during the ring stage. (b) Hb concentration in hemozoin. (c) Hemozoin volume. (d) Hemozoin contents. (e) Sphericity of the hemozoin. The cross symbols represent the median values, and the vertical lines indicate standard deviations. 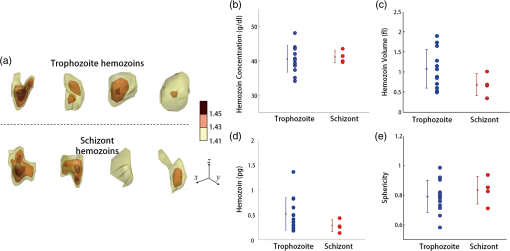 In addition, 3-D RI tomogram reconstructed with the diffraction algorithm provides complex RI values ; the real part of RI is a conventional value for RI that relates the speed of light in the material, whereas the imaginary part of RI corresponds to the absorption coefficient. Absorption coefficient , which describes how much a material absorbs light so that illuminated light intensity is attenuated to after passing a material with a thickness , . and are related by Beer–Lambert law as . The absorption coefficients of the hemozoins are calculated from the imaginary part of RI: of in situ hemozoin was found to be (trophozoite) and (schizont), while the rest of Pf-RBCs are nearly transparent at the wavelength used (). The measured is comparable with the previous report (; estimated from Ref. 65 with the size of hemozoin is ) where an UV-VIS absorption spectrometer was used. The concentration of hemozoin is calculated from the measured based on Beer–Lambert law: where is the molar extinction coefficient (molar absorptivity) of hemozoin which is estimated as at 633 nm from Refs. 65 and 66. The hemozoin concentrations were calculated as and for trophozoite and schizont stages, respectively. Then, by multiplying the molecular weight of hemozoin (), the hemozoin contents of Pf-RBCs are retrieved as and pg for trophozoite and schizont stages, respectively. Considering the limited number of samples, especially for the schizont stage, these assured values are also in agreement with previous studies that used UV-VIS absorption spectroscopy (0.72 to 0.76 pg for trophozoite and 0.76 to 0.84 pg for schizont stages).67 We would like to emphasize that the chemical and structural information of hemozoin were directly measured at the individual intact Pf-RBCs for the first time.5.ConclusionIn this work, we present high-resolution 3-D optical RI tomograms of Pf-RBCs employing the diffraction algorithm for tomogram reconstruction. The structural information about individual Pf-RBCs as well as parasitophorous vacuole and cytoplasm of the host Pf-RBCs was quantitatively measured. Furthermore, the in situ structural and chemical information of hemozoin inside Pf-RBCs was also investigated using the optical diffraction tomography. The multiple 2-D E-field images of individual RBCs were measured with various angles of illumination using quantitative phase microscopy from which 3-D tomograms were reconstructed via the diffraction algorithm with Rytov approximation. The RI tomograms of Pf-RBCs during the full intraerythrocytic cycle were presented with the diffraction algorithm. The 3-D RI tomograms of the same samples reconstructed by the projection algorithm were also presented and systematically compared. Our results show that the light diffraction effect is significant in Pf-RBCs; thus, the RI tomograms obtained with the diffraction algorithm were more precise than those obtained with the projection algorithm. Three-dimensional high-resolution imaging holds a key to revealing the mechanisms behind malaria pathophysiology, and is now accessible to direct experimental study without using exogenous labeling agents. Dynamic measurements of the RI tomograms of Pf-RBCs provide further opportunities for the determination of the dynamic properties of Pf-RBCs68 in order to better understand in vivo circulation through microcapillaries and spleen sinusoids. Although the particular focus of this paper is to demonstrate the reconstruction of 3-D RI tomograms of Pf-RBCs based on diffraction algorithm, the present approach is sufficiently broad and general and will directly find use in multiple studies. For example, the processes of invasion and egress of this particular parasite and the RBCs invaded with other species of protozoan parasites, such as P. vivax or Plasmodium ovale, can be investigated with the current approach. Furthermore, the other RBC-related diseases associated with significant morphological alteration including sickle cell diseases are now accessible to direct experimental study using optical tomography. AppendicesAppendix:MatLab Code for Optical Diffraction Tomography Algorithm% Optical diffraction tomography reconstruction algorithm % Programmed by Kyoohyun Kim (Biomedical Optics Lab, KAIST) 2012-8-9 % for further information, please refer to http://bmokaist.wordpress.com. % A raw-data file must contain following variables: % 1. EField(frame, x, x) is a 3-D complex amplitude (amplitude and phase) % matrix for multiple 2-D electric-field images % 2. u0_x(frame): x spatial frequency for illumination angle: [pixel] % 3. u0_y(frame): y spatial frequency for illumination angle: [pixel] %% initialization clear all n_m=1.334; % refractive index of medium, PBS=1.334 @ 633 nm lambda=0.633; % wavelength of the laser [μm] ZeroPadding_Real=1024; % # of pixels at image plane (2-D) after zero-padding ZeroPadding_Fourier3D=600; % # of pixels at Fourier plane (3-D) after zero-padding PixSize_CCD=16; % pixel size at the CCD plane [μm] Mag_total=210; % total magnification PixSize_Sample=PixSize_CCD/210; % pixel size at the sample plane [μm] %% File Import load(‘Field_Trophozoite_04.mat’); [thetaSize xSize ySize]=size(EField); %ex) xSize=380, ySize=380, thetaSize=600; %% fitting Incident illumination angles Frame_range=1:thetaSize; Frame_normal=300; % frame # for normal incident angle p1=polyfit (Frame_range, u0_x (Frame_range), 1); p2=polyfit (Frame_range, u0_y (Frame_range), 1); u0_x=p1(2)+p1(1)*Frame_range; u0_y=p2(2)+p2(1)*Frame_range; u0_x=(u0_x - u0_x(Frame_normal))*ZeroPadding_Real/xSize; %considering zero-padding u0_y=(u0_y - u0_y(Frame_normal))*ZeroPadding_Real/ySize; %% Set parameters k0=2*pi/lambda; % k vector (angular spatial frequency) of incident beam k0_x=2*pi/(PixSize_Sample*ZeroPadding_Real)*u0_x; k0_y=2*pi/(PixSize_Sample*ZeroPadding_Real)*u0_y; k0_z=sqrt ( (n_m*k0) ^2 - k0_x.^2 - k0_y.^2); kPixSize_Sample=2*pi/(PixSize_Sample*ZeroPadding_Real); %% k-space Mapping Frame_range=[65 512]; % select frame range for reconstruction Rytov_Tomo=zeros(ZeroPadding_Fourier3D,ZeroPadding_Fourier3D,ZeroPadding_Fourier3D,’single’); Count=zeros(ZeroPadding_Fourier3D,ZeroPadding_Fourier3D,ZeroPadding_Fourier3D,’single’); % to count the number of overlapped information. for kk=Frame_range(1):Frame_range(2) Rytov_2Dimage=log (squeeze (EField(kk,:,:)) ); %% set Rytov field padVal=(ySize*mean(Rytov_2Dimage(1,:)) + ySize*mean(Rytov_2Dimage(end,:)) + xSize*mean(Rytov_2Dimage(:,1)) + xSize*mean(Rytov_2Dimage(:,end)))/(2*(xSize+ySize)−4); Rytov_2Dimage=padarray (Rytov_2Dimage, round([(ZeroPadding_Real-xSize) (ZeroPadding_Real-ySize)]./2), padVal); %zero-padding at the sample plane Rytov_2DFourier=fftshift (fft2 (Rytov_2Dimage))/ZeroPadding_Real; Rytov_2DFourier=circshift (Rytov_2DFourier, round([u0_x(kk) u0_y(kk)])); %% mapping to Ewald’s sphere with a radius of n_m*k0 [ky kx]=meshgrid ( (kPixSize_Sample* (1:ZeroPadding_Real) -ZeroPadding_Real/2). *[1,1]); kz=real (sqrt ((n_m*k0)^2 - kx.^2 - ky.^2)); Ewald=1i.*kz.*Rytov_2DFourier/pi; % mapping Rytov field to Ewald sphere %% Select Rytov field for real k vectors [xtemp, ytemp]=find (kz>0); Xrange=min(xtemp): max(xtemp); Yrange=min(ytemp): max(ytemp); Ewald=Ewald(Xrange,Yrange); Kx=kx - k0_x(kk); Kx=Kx(Xrange,Yrange); Ky=ky - k0_y(kk); Ky=Ky(Xrange,Yrange); Kz=kz - k0_z(kk); Kz=Kz(Xrange,Yrange); %% Applying to 3D Rytov space Kx=round (Kx/kPixSize_Sample+ZeroPadding_Fourier3D/2); Ky=round (Ky/kPixSize_Sample+ZeroPadding_Fourier3D/2); Kz=round (Kz/kPixSize_Sample+ZeroPadding_Fourier3D/2); ZeroPadding_Real3=length(Kx); KZeroPadding_Real=(Kz-1)*ZeroPadding_Fourier3D^2+(Ky-1)*ZeroPadding_Fourier3D+Kx; KZeroPadding_Real=PixSize_Samplehape (KZeroPadding_Real, 1, ZeroPadding_Real3^2); % make indexing column vector temp=PixSize_Samplehape (Rytov_Tomo (KZeroPadding_Real), ZeroPadding_Real3, ZeroPadding_Real3); Rytov_Tomo (KZeroPadding_Real)=temp+Ewald; Count (KZeroPadding_Real)=Count (KZeroPadding_Real)+1; % count mapped 3-D Fourier space coordinates end Rytov_Tomo (Count>0)=Rytov_Tomo (Count>0)./Count (Count>0); % average for overlapped points ReconImg=ifftn (ifftshift (Rytov_Tomo)). * (ZeroPadding_Fourier3D^(3/2)); % transform to real space n=n_m*sqrt (1-ReconImg.* (lambda/(2*pi*n_m)) ^2); % convert to actual refractive index % Iterative algorithm for optical diffraction tomography % Programmed by Kyoohyun Kim, (Biomedical Optics Lab, KAIST) 2012-8-9 % for further information, please refer to http://bmokaist.wordpress.com Rytov_Tomo_index=(abs(Rytov_Tomo)==0); % locate k position for missing cone Iter=100; %total number of iteration for mm=1:Iter constn=real(n)<n_m; % criterion: nonnegative refractive index difference n(constn)=n_m+1i*angle(n(constn)); F=-(2*pi*n_m/lambda)^2.*(n.^2/n_m^2-1); temp=fftshift (fftn (F) )/(ZeroPadding_Fourier3D^ (3/2) ); % new 3D Fourier field Rytov_Tomo_new=temp.*Rytov_Tomo_index+Rytov_Tomo; % replace previously measured field Rytov_Tomo) F=ifftn (ifftshift (Rytov_Tomo_new) ) * (ZeroPadding_Fourier3D^ (3/2)); n=n_m*sqrt (1 - F.* (lambda/(n_m*2*pi) ) ^2); end AcknowledgmentsThe authors wish to acknowledge Professor Subra Suresh (MIT and NSF) for helpful discussions. This work was funded by KAIST, the Korean Ministry of Education, Science and Technology (MEST) Grant No. 2009-0087691 (BRL), National Research Foundation (NRF-2012R1A1A1009082, NRF-2012K1A31A1A09055128, and NRF-M3C1A1-048860). YongKeun Park acknowledges support from TJ ChungAm Foundation. Kyoohyun Kim is supported by Global Ph.D. Fellowship from National Research Foundation of Korea. Monica Diez-Silva and Ming Dao acknowledge support from National Institutes of Health (R01HL094270) and the Singapore-MIT Alliance for Research and Technology (SMART) Center. Ramachandra R. Dasari acknowledges support from National Institutes of Health (9P41-EB015871-26A1), the National Science Foundation (DBI-0754339). References
“World Malaria Report 2012,”
53
–62 Geneva
(2012). Google Scholar
L. Bannisteret al.,
“A brief illustrated guide to the ultrastructure of Plasmodium falciparum asexual blood stages,”
Parasitol. Today, 16
(10), 427
–433
(2000). http://dx.doi.org/10.1016/S0169-4758(00)01755-5 PATOE2 0169-4758 Google Scholar
F. Frischknecht,
“Imaging parasites at different scales,”
Cell Host Microbe, 8
(1), 16
–19
(2010). http://dx.doi.org/10.1016/j.chom.2010.06.013 CHMECB 1931-3128 Google Scholar
E. Hanssenet al.,
“Soft X-ray microscopy analysis of cell volume and hemoglobin content in erythrocytes infected with asexual and sexual stages of Plasmodium falciparum,”
J. Struct. Biol., 177
(2), 224
–232
(2012). http://dx.doi.org/10.1016/j.jsb.2011.09.003 JSBIEM 1047-8477 Google Scholar
C. Magowanet al.,
“Intracellular structures of normal and aberrant Plasmodium falciparum malaria parasites imaged by soft x-ray microscopy,”
Proc. Natl. Acad. Sci. U. S. A., 94
(12), 6222
–6227
(1997). http://dx.doi.org/10.1073/pnas.94.12.6222 1091-6490 Google Scholar
S. Choet al.,
“Optical imaging techniques for the study of malaria,”
Trends Biotechnol., 30
(2), 71
–79
(2012). http://dx.doi.org/10.1016/j.tibtech.2011.08.004 TRBIDM 0167-7799 Google Scholar
B. Gligorijevicet al.,
“Stage independent chloroquine resistance and chloroquine toxicity revealed via spinning disk confocal microscopy,”
Mol. Biochem. Parasitol., 159
(1), 7
–23
(2008). http://dx.doi.org/10.1016/j.molbiopara.2007.12.014 MBIPDP 0166-6851 Google Scholar
Y.-K. Parket al.,
“Refractive index maps and membrane dynamics of human red blood cells parasitized by Plasmodium falciparum,”
Proc. Natl. Acad. Sci. U. S. A., 105
(37), 13730
–13735
(2008). http://dx.doi.org/10.1073/pnas.0806100105 1091-6490 Google Scholar
R. Chandramohanadaset al.,
“Biophysics of malarial parasite exit from infected erythrocytes,”
PLoS ONE, 6
(6), e20869
(2011). http://dx.doi.org/10.1371/journal.pone.0020869 1932-6203 Google Scholar
K.-R. Leeet al.,
“Quantitative phase imaging techniques for the study of cell pathophysiology: from principles to applications,”
Sensors, 13
(4), 4170
–4191
(2013). http://dx.doi.org/10.3390/s130404170 SNSRES 0746-9462 Google Scholar
G. Popescuet al.,
“Diffraction phase microscopy for quantifying cell structure and dynamics,”
Opt. Lett., 31
(6), 775
–777
(2006). http://dx.doi.org/10.1364/OL.31.000775 OPLEDP 0146-9592 Google Scholar
T. Ikedaet al.,
“Hilbert phase microscopy for investigating fast dynamics in transparent systems,”
Opt. Lett., 30
(10), 1165
–1168
(2005). http://dx.doi.org/10.1364/OL.30.001165 OPLEDP 0146-9592 Google Scholar
Y. K. Parket al.,
“Diffraction phase and fluorescence microscopy,”
Opt. Express, 14
(18), 8263
–8268
(2006). http://dx.doi.org/10.1364/OE.14.008263 OPEXFF 1094-4087 Google Scholar
Y.-K. Parket al.,
“Speckle-field digital holographic microscopy,”
Opt. Express, 17
(15), 12285
–12292
(2009). http://dx.doi.org/10.1364/OE.17.012285 OPEXFF 1094-4087 Google Scholar
Y. Parket al.,
“Measurement of red blood cell mechanics during morphological changes,”
Proc. Natl. Acad. Sci. U. S. A., 107
(15), 6731
–6736
(2010). http://dx.doi.org/10.1073/pnas.0909533107 1091-6490 Google Scholar
Y. K. Parket al.,
“Metabolic remodeling of the human red blood cell membrane,”
Proc. Natl. Acad. Sci., 107
(4), 1289
–1294
(2010). http://dx.doi.org/10.1073/pnas.0910785107 PMASAX 0096-9206 Google Scholar
Y. K. Parket al.,
“Measurement of the nonlinear elasticity of red blood cell membranes,”
Phys. Rev. E, 83
(5), 051925
(2011). http://dx.doi.org/10.1103/PhysRevE.83.051925 PLEEE8 1063-651X Google Scholar
G. Popescuet al.,
“Optical imaging of cell mass and growth dynamics,”
Am. J. Physiol. Cell Physiol., 295
(2), C538
–C544
(2008). http://dx.doi.org/10.1152/ajpcell.00121.2008 0363-6143 Google Scholar
M. Miret al.,
“Optical measurement of cycle-dependent cell growth,”
Proc. Natl. Acad. Sci. U. S. A., 108
(32), 13124
–13129
(2011). http://dx.doi.org/10.1073/pnas.1100506108 1091-6490 Google Scholar
Y. Parket al.,
“Spectroscopic phase microscopy for quantifying hemoglobin concentrations in intact red blood cells,”
Opt. Lett., 34
(23), 3668
–3670
(2009). http://dx.doi.org/10.1364/OL.34.003668 OPLEDP 0146-9592 Google Scholar
Y. JangJ. JangY. K. Park,
“Dynamic spectroscopic phase microscopy for quantifying hemoglobin concentration and dynamic membrane fluctuation in red blood cells,”
Opt. Express, 20
(9), 9673
–9681
(2012). http://dx.doi.org/10.1364/OE.20.009673 OPEXFF 1094-4087 Google Scholar
G. G. Levinet al.,
“Three-dimensional limited-angle microtomography of blood cells: experimental results,”
Proc. SPIE, 3261 159
–164
(1998). http://dx.doi.org/10.1117/12.310549 Google Scholar
V. Lauer,
“New approach to optical diffraction tomography yielding a vector equation of diffraction tomography and a novel tomographic microscope,”
J. Microsc., 205
(2), 165
–176
(2002). http://dx.doi.org/10.1046/j.0022-2720.2001.00980.x JMICAR 0022-2720 Google Scholar
B. Simonet al.,
“Tomographic diffractive microscopy of transparent samples,”
Eur. Phys. J. Appl. Phys., 44
(1), 29
–35
(2008). http://dx.doi.org/10.1051/epjap:2008049 EPAPFV 1286-0050 Google Scholar
S. O. Isikmanet al.,
“Lens-free optical tomographic microscope with a large imaging volume on a chip,”
Proc. Natl. Acad. Sci., 108
(18), 7296
–7301
(2011). http://dx.doi.org/10.1073/pnas.1015638108 PMASAX 0096-9206 Google Scholar
F. Charrièreet al.,
“Living specimen tomography by digital holographic microscopy: morphometry of testate amoeba,”
Opt. Express, 14
(16), 7005
–7013
(2006). http://dx.doi.org/10.1364/OE.14.007005 OPEXFF 1094-4087 Google Scholar
M. Fauveret al.,
“Three-dimensional imaging of single isolated cell nuclei using optical projection tomography,”
Opt. Express, 13
(11), 4210
–4223
(2005). http://dx.doi.org/10.1364/OPEX.13.004210 OPEXFF 1094-4087 Google Scholar
J. Izattet al.,
“Optical coherence microscopy in scattering media,”
Opt. Letters, 19
(8), 590
–592
(1994). http://dx.doi.org/10.1364/OL.19.000590 OPLEDP 0146-9592 Google Scholar
M. Chomaet al.,
“Spectral-domain phase microscopy,”
Opt. Lett., 30
(10), 1162
–1164
(2005). http://dx.doi.org/10.1364/OL.30.001162 OPLEDP 0146-9592 Google Scholar
Z. Wanget al.,
“Spatial light interference tomography (SLIT),”
Opt. Express, 19
(21), 19907
–19918
(2011). http://dx.doi.org/10.1364/OE.19.019907 OPEXFF 1094-4087 Google Scholar
G. Popescu, Quantitative Phase Imaging of Cells and Tissues, McGraw-Hill, New York
(2011). Google Scholar
M. PotcoavaM. Kim,
“Optical tomography for biomedical applications by digital interference holography,”
Meas. Sci. Technol., 19
(7), 074010
(2008). http://dx.doi.org/10.1088/0957-0233/19/7/074010 MSTCEP 0957-0233 Google Scholar
J. Kühnet al.,
“Submicrometer tomography of cells by multiple-wavelength digital holographic microscopy in reflection,”
Opt. Lett., 34
(5), 653
–655
(2009). http://dx.doi.org/10.1364/OL.34.000653 OPLEDP 0146-9592 Google Scholar
K. G. PhillipsS. L. JacquesO. J. T. McCarty,
“Measurement of single cell refractive index, dry mass, volume, and density using a transillumination microscope,”
Phys. Rev. Lett., 109
(11), 118105
(2012). http://dx.doi.org/10.1103/PhysRevLett.109.118105 PRLTAO 0031-9007 Google Scholar
N. T. Shakedet al.,
“Quantitative microscopy and nanoscopy of sickle red blood cells performed by wide field digital interferometry,”
J. Biomed. Opt., 16
(3), 030506
(2011). http://dx.doi.org/10.1117/1.3556717 JBOPFO 1083-3668 Google Scholar
Y. Kimet al.,
“Anisotropic light scattering of individual sickle red blood cells,”
J. Biomed. Opt., 17
(4), 040501
(2012). http://dx.doi.org/10.1117/1.JBO.17.4.040501 JBOPFO 1083-3668 Google Scholar
H. S. Byunet al.,
“Optical measurement of biomechanical properties of individual erythrocytes from a sickle cell patient,”
Acta Biomater., 8
(11), 4130
–4138
(2012). http://dx.doi.org/ 10.1016/j.actbio.2012.07.011 ABCICB 1742-7061 Google Scholar
E. Ben-Isaacet al.,
“Effective temperature of red-blood-cell membrane fluctuations,”
Phys. Rev. Lett., 106
(23), 238103
(2011). http://dx.doi.org/10.1103/PhysRevLett.106.238103 PRLTAO 0031-9007 Google Scholar
Y. Parket al.,
“Light scattering of human red blood cells during metabolic remodeling of the membrane,”
J. Biomed. Opt., 16
(1), 011013
(2011). http://dx.doi.org/10.1117/1.3524509 JBOPFO 1083-3668 Google Scholar
W. Choiet al.,
“Tomographic phase microscopy,”
Nat. Methods, 4
(9), 717
–719
(2007). http://dx.doi.org/10.1038/nmeth1078 1548-7091 Google Scholar
A. G. Maieret al.,
“Malaria parasite proteins that remodel the host erythrocyte,”
Nat. Rev. Microbiol., 7
(5), 341
–354
(2009). http://dx.doi.org/10.1038/nrmicro2110 1740-1526 Google Scholar
E. Wolf,
“Three-dimensional structure determination of semi-transparent objects from holographic data,”
Opt. Commun., 1
(4), 153
–156
(1969). http://dx.doi.org/10.1016/0030-4018(69)90052-2 OPCOB8 0030-4018 Google Scholar
Y. Sunget al.,
“Optical diffraction tomography for high resolution live cell imaging,”
Opt. Express, 17
(1), 266
–277
(2009). http://dx.doi.org/10.1364/OE.17.000266 OPEXFF 1094-4087 Google Scholar
A. J. Devaney,
“A filtered back-propagation algorithm for diffraction tomography,”
Ultrason. Imag., 4
(4), 336
–350
(1982). http://dx.doi.org/10.1016/0161-7346(82)90017-7 ULIMD4 0161-7346 Google Scholar
K. C. TamV. Perezmendez,
“Tomographical imaging with limited-angle input,”
J. Opt. Soc. Am., 71
(5), 582
–592
(1981). http://dx.doi.org/10.1364/JOSA.71.000582 JOSAAH 0030-3941 Google Scholar
P. Goldieet al.,
“Biochemical characterization of Plasmodium falciparum hemozoin,”
Am. J. Trop. Med. Hyg., 43
(6), 584
–596
(1990). AJTHAB 0002-9637 Google Scholar
A. Devaney,
“Inverse-scattering theory within the Rytov approximation,”
Opt. Lett., 6
(8), 374
–376
(1981). http://dx.doi.org/10.1364/OL.6.000374 OPLEDP 0146-9592 Google Scholar
O. Haeberleet al.,
“Tomographic diffractive microscopy: basics, techniques and perspectives,”
J. Mod. Opt., 57
(9), 686
–699
(2010). http://dx.doi.org/10.1080/09500340.2010.493622 JMOPEW 0950-0340 Google Scholar
M. Debailleulet al.,
“Holographic microscopy and diffractive microtomography of transparent samples,”
Meas. Sci. Technol., 19
(7), 074009
(2008). http://dx.doi.org/10.1088/0957-0233/19/7/074009 MSTCEP 0957-0233 Google Scholar
S. S. KouC. J. R. Sheppard,
“Image formation in holographic tomography,”
Opt. Lett., 33
(20), 2362
–2364
(2008). http://dx.doi.org/10.1364/OL.33.002362 OPLEDP 0146-9592 Google Scholar
S. K. DebnathY. Park,
“Real-time quantitative phase imaging by spatial phase shifting algorithm,”
Opt. Lett., 36
(23), 4677
–4679
(2011). http://dx.doi.org/10.1364/OL.36.004677 OPLEDP 0146-9592 Google Scholar
B. P. Medoffet al.,
“Iterative convolution backprojection algorithms for image-reconstruction from limited data,”
J. Opt. Soc. Am., 73
(11), 1493
–1500
(1983). http://dx.doi.org/10.1364/JOSA.73.001493 JOSAAH 0030-3941 Google Scholar
G. Pasvolet al.,
“Separation of viable schizont-infected red cells of Plasmodium falciparum from human blood,”
Ann. Trop. Med. Parasitol., 72
(1), 87
–88
(1978). ATMPA2 1364-8594 Google Scholar
C. LambrosJ. P. Vanderberg,
“Synchronization of Plasmodium falciparum erythrocytic stages in culture,”
J. Parasitol., 65
(3), 418
–420
(1979). http://dx.doi.org/10.2307/3280287 0020-7519 Google Scholar
R. Barer,
“Refractometry and interferometry of living cells,”
J. Opt. Soc. Am., 47
(6), 545
–556
(1957). http://dx.doi.org/10.1364/JOSA.47.000545 JOSAAH 0030-3941 Google Scholar
O. Zhernovayaet al.,
“The refractive index of human hemoglobin in the visible range,”
Phys. Med. Biol., 56
(13), 4013
–4021
(2011). http://dx.doi.org/10.1088/0031-9155/56/13/017 PHMBA7 0031-9155 Google Scholar
M. A. Zanneret al.,
“Water and urea transport in human erythrocytes infected with the malaria parasite Plasmodium falciparum,”
Mol. Biochem. Parasitol., 40
(2), 269
–278
(1990). http://dx.doi.org/10.1016/0166-6851(90)90048-Q MBIPDP 0166-6851 Google Scholar
J. L. ElliottK. J. SalibaK. Kirk,
“Transport of lactate and pyruvate in the intraerythrocytic malaria parasite, Plasmodium falciparum,”
Biochem. J., 355
(Pt 3), 733
–739
(2001). BIJOAK 0264-6021 Google Scholar
D. A. Elliottet al.,
“Four distinct pathways of hemoglobin uptake in the malaria parasite Plasmodium falciparum,”
Proc. Natl. Acad. Sci., 105
(7), 2463
–2468
(2008). http://dx.doi.org/10.1073/pnas.0711067105 PMASAX 0096-9206 Google Scholar
A. Espositoet al.,
“Quantitative imaging of human red blood cells infected with Plasmodium falciparum,”
Biophys. J., 99
(3), 953
–960
(2010). http://dx.doi.org/10.1016/j.bpj.2010.04.065 BIOJAU 0006-3495 Google Scholar
J. M. A. Mauritzet al.,
“The homeostasis of Plasmodium falciparum-infected red blood cells,”
PLoS Comput. Biol., 5
(4), e1000339
(2009). http://dx.doi.org/10.1371/journal.pcbi.1000339 1553-734X Google Scholar
L. J. Bruce-Chwatt, Essential Malariology, William Heinemann Medical Books Ltd., London
(1980). Google Scholar
B. Gligorijevicet al.,
“Spinning disk confocal microscopy of live, intraerythrocytic malarial parasites. 1. Quantification of hemozoin development for drug sensitive versus resistant malaria,”
Biochemistry, 45
(41), 12400
–12410
(2006). http://dx.doi.org/10.1021/bi061033f MIRBD9 0144-0578 Google Scholar
G. S. NolandN. BrionesD. J. Sullivan,
“The shape and size of hemozoin crystals distinguishes diverse Plasmodium species,”
Mol. Biochem. Parasitol., 130
(2), 91
–99
(2003). http://dx.doi.org/10.1016/S0166-6851(03)00163-4 MBIPDP 0166-6851 Google Scholar
A. U. OrjihC. D. Fitch,
“Hemozoin production by plasmodium-falciparum—variation with strain and exposure to chloroquine,”
Biochim. Biophys. Acta, 1157
(2), 270
–274
(1993). http://dx.doi.org/10.1016/0304-4165(93)90109-L BBACAQ 0006-3002 Google Scholar
R. K. SahaS. KarmakarM. Roy,
“Computational investigation on the photoacoustics of malaria infected red blood cells,”
PLoS One, 7
(12), e51774
(2012). http://dx.doi.org/10.1371/journal.pone.0051774 1932-6203 Google Scholar
Y. M. Serebrennikovaet al.,
“Quantitative analysis of morphological alterations in Plasmodium falciparum infected red blood cells through theoretical interpretation of spectral measurements,”
J. Theor. Biol., 265
(4), 493
–500
(2010). http://dx.doi.org/10.1016/j.jtbi.2010.05.037 JTBIAP 0022-5193 Google Scholar
M. Diez-Silvaet al.,
“Pf155/RESA protein influences the dynamic microcirculatory behavior of ring-stage Plasmodium falciparum infected red blood cells,”
Sci. Rep., 2 614
(2012). http://dx.doi.org/10.1038/srep00614 2045-2322 Google Scholar
|

